Abstract
Oligogalacturonide fragments that activate defensive genes in plant leaves heretofore have been thought to be generated only by pathogen-derived pectin-degrading enzymes, because polygalacturonase (PG) activity has not been reported in leaves. Here, we report that mRNAs encoding a PG catalytic subunit protein and its regulatory (β-subunit) protein are expressed in tomato leaves in response to wounding, systemin, and oligosaccharide elicitors. Synthesis of the two subunits in response to wounding is systemic and is accompanied by an increase in PG activity in extracts from both wounded and unwounded leaves. The finding that PG subunit mRNAs and PG enzyme activity are induced by wounding indicates that herbivore attacks can produce endogenous oligogalacturonide elicitors that may be involved in the local and systemic activation of defense responses against both herbivores and pathogens.
The pectin-degrading family members of plant polygalacturonase (PG; EC 3.2.1.15) have been studied extensively (1–3) for their roles in cell-wall degradation in fruit, abscission, pollen maturation and tube growth, pod and anther dehiscence, and growing and expanding tissues. In tomato fruit, the relationship of PG to ripening has been studied at both the transcriptional and translational levels, and the enzyme has been shown to exist in three isoforms called PG1, PG2A, and PG2B (4). PG2A and PG2B are products of the same gene but differ in their degrees of glycosylation. PG1 is a complex between either PG2A or PG2B (the catalytic subunits collectively called PGcat), and two catalytically inactive proteins called the β-subunits (5). The β-subunits are glycosylated and are the products of two genes called AroGP1 and AroGP2 (4). A third homologous gene, called AroGP3, that is not transcribed during the ripening process was identified; this gene was predicted to be either a pseudogene or a gene whose product has a physiological role in something other than fruit ripening (6).
All three isoenzymes of PG are active in vitro. A model for the interactions of the β-subunits with the PG2A and PG2B catalytic subunits and with plant pectin suggests that the β-subunits may alter the catalytic properties of the enzyme (4) and may play an important regulatory role in pectin degradation, although the data are still largely correlative.
During pathogenesis and infection, attacking fungi and bacteria secrete polygalacturonases that degrade the cell wall (7). In response, pectic fragments are released from plant cell walls and act as early signals to activate defensive genes near infection sites to protect the plants against pathogens (8, 9). The oligogalacturonide fragments produced in leaves are thought to be generated only by pathogen-derived pectin-degrading enzymes, because the presence of a PGcat subunit in plant leaf blades has not been reported previously, although low levels of the β-subunit mRNA have been detected in tomato leaves (6) and PG is present in leaf abscission zones.
In tomato leaves, an 18-aa polypeptide called systemin (10) signals the systemic synthesis of several wound-responsive proteins that seem to participate in plant defense and wound healing. Systemin is processed from the C terminus of a 200-aa precursor called prosystemin (11), and tomato plants transformed with the prosystemin cDNA under control of the cauliflower mosaic virus 35S promoter constitutively produce high levels of prosystemin mRNA throughout the plants (12). This constitutive expression is thought to result from the abnormal processing and release of systemin. This transgenic phenotype behaves as if it were in a permanently wounded state in the absence of wounding and results in permanently elevated levels of wound-inducible mRNAs and proteins (13). We report herein that among the elevated mRNAs in the unwounded transgenic plants are a PGcat subunit gene and a β-subunit gene; there is also PG activity in the leaves. mRNAs encoding both subunits were found to be wound-inducible in wild-type plants and were accompanied by an increase in PG activity in leaf extracts, both locally and systemically. The wound-inducible PGcat subunit gene is a distinct member of the family of PG genes. The β-subunit gene had been identified previously, although its expression and function had not been investigated. The wound-inducible PG activity in leaves may play a role in the degradation of leaf pectin to generate oligogalacturonic acid fragments as signals to activate defensive responses during herbivore and pathogen attacks.
MATERIALS AND METHODS
Isolation and Sequencing of Tomato Leaf PGcat cDNA.
Degenerate primers, based on the highly conserved PG active sites, were used in PCRs to amplify a 550-bp product from a cDNA library template prepared from transgenic plants overexpressing prosystemin (12) by using a λ ZAP II vector (Stratagene). The 550-bp product was confirmed by sequencing to be homologous with the PGcat previously isolated from tomato fruit (14) and abscission zones (15). The radiolabeled 550-bp fragment was used to screen the cDNA library, and several full-length cDNAs were isolated that were confirmed to encode an identical tomato leaf PGcat subunit protein. Both strands of a full-length PGcat cDNA clone were sequenced by using the fluorescent chain termination method and a Perkin–Elmer ABI model 373 sequencer; three randomly chosen positive clones were recovered and sequenced from their 5′ ends. The nucleotide sequence from each clone was identical. The sequence at the N terminus, which encoded the signal peptide and displayed the most divergence with the known tomato PGs, was confirmed by reverse transcription–PCR. The deduced amino acid sequence of the tomato PGcat subunit is shown in Fig. l.
RNA Analyses.
Leaf tissue was frozen in liquid nitrogen at intervals after treatment, and total RNA was extracted by using phenol/SDS. Leaf RNA was fractionated in 1.4% agarose gels with formaldehyde, blotted into nylon membranes, fixed by UV light with a Stratalinker (Stratagene), and incubated at 62°C for 1–2 h in 6× standard saline phosphate/EDTA (SSPE) hybridization buffer (20× SSPE stock = 175.3 g of NaCl/27.6 g of NaH2PO4⋅H2O/7.4 g of EDTA/1 liter of H2O, pH 7.4), 5× Denhardt’s solution (50× Denhardt’s stock = 5 g of Ficoll/5 g of BSA/5 g of polyvinylpyrrolidone/1 liter of H2O), 10% dextran sulfate, and 1.0% SDS. Probes were radiolabeled with [32P]dCTP by random priming, according to manufacturer’s instructions (Amersham Pharmacia), purified by spin chromatography (16), denatured by boiling, added to the hybridization buffer, and incubated with the blocked membranes overnight at 62°C. Membranes were washed with 4× SSPE and 1% SDS for 15 min at room temperature, 2× SSPE/SDS for 30 min at 62°C, and 1× SSPE/SDS for 20 min at 62°C, before exposing for 24–36 h on KODAK X-Omat film (Eastman Kodak).
Plant Propagation.
Tomato (Lycopersicon esculentum cv. Castlemart) plants were grown in peat pots and maintained under a day–night cycle of 17 h under light (30 μE⋅m−2⋅s−1) at 28°C and 7 h of darkness at 18°C. Tomato plants expressing a transgene consisting of a prosystemin cDNA under the control of the cauliflower mosaic virus 35S promoter (12) were grown under the same conditions. For all experiments, 15- to 18-day-old plants having two expanding leaves and a small apical leaf were used. To wound plants, the lowest leaf was crushed three to four times across the main vein with a hemostat. After treatments, plants were incubated under constant light for the duration of the experiments.
Enzyme and Protein Assays.
At times indicated in the figures, the wounded and unwounded leaves of treated plants were excised, and leaf extracts were prepared for PG assays. In a Sorvall Omnimixer (DuPont), 20 g of leaves were homogenized in 60 ml of ice-cold 0.1 M sodium citrate buffer, pH 6.0, containing 1 M NaCl, 4 mM ascorbic acid, 5 mM DTT, 2% polyvinylpyrrolidone, and 0.1% BSA. The extracts were filtered through four layers of cheesecloth, incubated at 4°C for 3 h, and then centrifuged at 10,000 × g for 20 min at 4°C. The supernatants were filtered through glass wool, and proteins were precipitated at 4°C by the slow addition of solid ammonium sulfate to a final saturation of 80% and stirred for 1 h at 4°C. The precipitates were recovered by centrifugation at 10,000 × g for 20 min at 4°C. The supernatant was discarded, and the pellet was dissolved in 20 ml of 1 M NaCl and dialyzed against 1 M NaCl at 4°C for 24 h. Aliquots equivalent to 0.1 g of fresh weight were assayed for PG (17).
RESULTS AND DISCUSSION
While searching for systemin-inducible mRNAs in a cDNA library derived from plants constitutively overexpressing the prosystemin transgene, a cDNA clone was identified and sequenced that exhibited 100% identity (data not shown) with the exon regions of the aromatic amino acid-rich AroGP3 gene from tomato plants. This gene, but not its function, had been reported previously (6). AroGP3 is homologous with two AroGP gene family members that are expressed in ripening tomato fruit. These two AroGP proteins, called β-subunits, have been hypothesized to interact with the fruit cell walls and with the PGcat subunit to regulate PG activity in ripening fruit (4).
Finding a wound-inducible β-subunit cDNA clone raised the question of whether there might also be a PGcat mRNA that is induced by wounding, because both may be involved with pectin degradation in tomato fruit. Conserved amino acid sequence regions of known PGcat enzymes were used to design degenerate primers for PCR amplification of potential PGcat subunit cDNAs in leaf tissue. By using the cDNA library prepared from leaves of transgenic tomato plants overexpressing prosystemin as a template, a 550-bp DNA fragment was generated and sequenced. A computer database comparison indicated that the amplified product shared moderate identity with the PGcat subunit cDNAs previously isolated from tomato fruit (14) and abscission zones (15). The partial sequence was radiolabeled and used as a probe to screen the transgenic cDNA library. Several clones were identified, all of which possessed identical full-length sequences encoding a tomato leaf PGcat subunit.
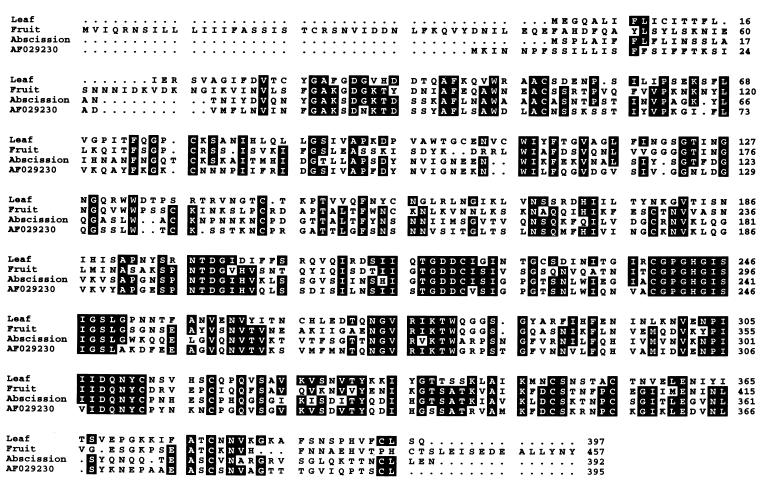
In Fig. 1, the deduced amino acid sequence of the tomato leaf PGcat polypeptide is compared with the PGcat polypeptides from tomato fruit PG, abscission zone PG, and a PG with unknown expression (PG AF029230). As with the other PGs, a consensus hydrophobic signal peptide was present at its N terminus, indicating that the enzyme is synthesized through the endoplasmic reticulum and Golgi apparatus. However, the N-terminal amino acid for any of the mature PG proteins is not known. In fruit, both the PGcat subunit and the β-subunit are glycosylated and secreted into the outer pericarp where pectin is degraded (17). In leaves, the tissue or cell-specific localization of neither PGcat nor the β-subunit is known. The amino acid sequence of the putative PGcat protein exhibits several potential N-linked glycosylation sites, but neither fruit nor abscission zone PGs have glycosylation sites at similar positions in their sequences in the alignments shown. All three PGs exhibit cysteines at identical sites at 10 positions, indicating that it is likely that some structural features are conserved.
Figure 1.
A comparison of the amino acid sequences of tomato leaf PGcat subunit with fruit PGcat subunit, tomato abscission zone PG, and a tomato PG with unknown expression.
Table 1 compares the percent identities of the deduced amino acid sequences between the PGcat subunits from tomato leaves, fruit, abscission zones, and PG AF029230. The leaf PGcat subunit was found to possess only 4l.5% amino acid identity to the fruit PGcat protein, 39.6% identity to the abscission zone PGcat protein, and 37.2% identity to the PG gene product from tomato plants whose expression characteristics are not known. Thus, the leaf gene is a member of a distinct subclass of wound-inducible leaf PGcat genes. The leaf β-subunit cDNA sequence bears 100% identity to the sequence of AroGP3 mRNA and is highly likely to be the same gene. The β-subunit mRNA had been reported previously to be weakly detectable in leaves (4); however, little significance was attributed to the finding, because PG activity had never been reported from leaves.
Table 1.
Comparisons of the percent identities of the amino acid sequence of tomato leaf PGcat subunit with tomato leaf PG, tomato fruit PG, tomato abscission zone PG, and a PG subunit of unknown expression
| Amino acid sequences | Percent identity |
|---|---|
| Leaf vs. leaf | 100 |
| Leaf vs. fruit | 41.5 |
| Leaf vs. abscission zone | 39.6 |
| Leaf vs. PG AF029230 | 37.2 |
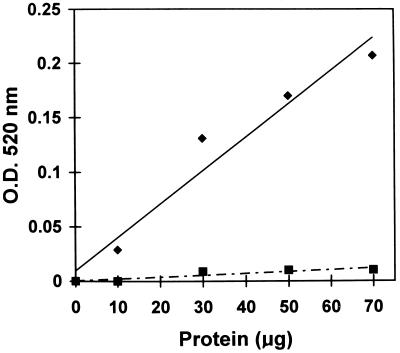
The presence of mRNAs coding for PGcat and the β-subunit in the transgenic plants was accompanied by the synthesis of an active PG enzyme. Leaf extract from these plants exhibited PG activity with a nearly linear correlation between the PG activity and the quantities of transgenic leaf protein used in the assays (Fig. 2).
Figure 2.
PG activity as a function of the quantities of soluble proteins from leaves of transgenic tomato plants overexpressing the prosystemin gene (diamonds) compared with extracts from leaves of wild-type plants (squares).
To determine whether PGcat and β-subunit mRNAs were systemically wound-inducible in wild-type plants, leaves of young wild-type tomato plants were assayed by Northern analysis of both the wounded leaves and upper, unwounded leaves before and after wounding. Fig. 3 shows that the levels of the PGcat mRNA increased in both the wounded and upper, unwounded leaves within 1 h after wounding, reaching a maximum at 6 h and declining slowly thereafter. The mRNA coding for the β-subunit increased only moderately during the first 6 h after wounding, and then increased for 12 h when the experiments were terminated. Although a low basal level of β-subunit mRNA was present in leaves of unwounded tomato plants, PGcat mRNA was not detected in leaves until after the plants were wounded.
Figure 3.
Northern analyses of the time course of the accumulation of leaf PGcat subunit and regulatory β-subunit in response to wounding. The lower leaves of 15- to 18-day-old tomato plants with two expanded leaves were wounded with a hemostat at time 0, and both lower, wounded leaves and the upper, unwounded leaves were assayed at the times indicated.
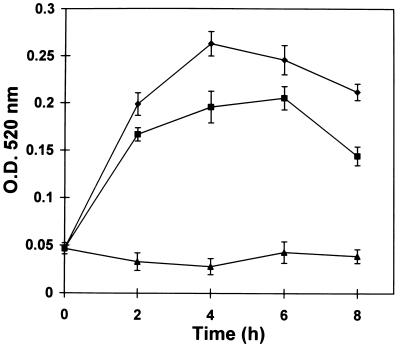
At intervals after wounding, the total soluble protein from both the wounded leaves and the unwounded leaves of the plant was extracted, partially purified, and assayed for PG activity. PG activity increased 4- to 5-fold within 2–4 h in wounded leaves (Fig. 4), and 3- to 4-fold in upper, unwounded leaves during the same period. After peaking at about 4 h, the activity in both wounded and unwounded leaves declined. This decline took place before the PGcat mRNA levels declined (compare with Fig. 3). Possible reasons for the decline in PG activity include increased protein degradation or modification and inhibition by an endogenous inhibitor. It is also possible that the β-subunit modulates PG activity, because the synthesis of the β-subunit mRNA was increasing at 9 and 12 h after wounding, at a time when the PG activity was decreasing. A somewhat analogous situation occurs during fruit ripening (4), where PG activity is highest when the β-subunit is at its lowest levels. The wound-inducible synthesis of both subunit mRNAs in response to wounding leads us to suspect that the β-subunit protein may play some role in regulating the activity, as the conditions during fruit ripening suggest (4).
Figure 4.
Time course of accumulation of PG activity in response to wounding. The lower leaves of 15- to 18-day-old tomato plants were wounded at time 0, and protein extracts from wounded leaves (diamonds) and upper unwounded leaves (squares) were assayed for PG activity (18) at the times indicated. Extracts from leaves of unwounded plants were assayed as controls (triangles).
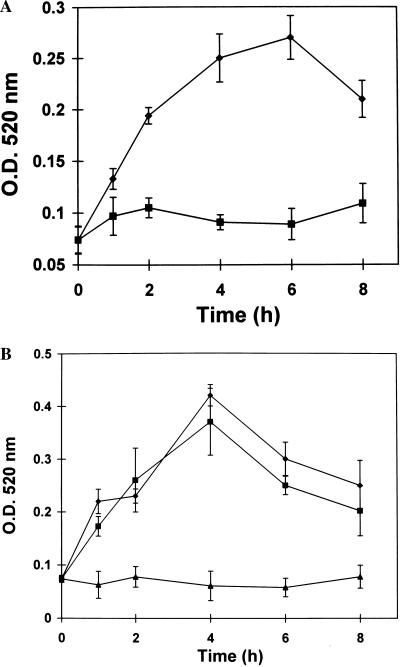
Supplying young excised tomato plants with systemin and the oligosaccharide elicitors oligogalacturonic acid and chitosan, all of which induce the synthesis of systemic wound-responsive proteins (18), induced PG activity, with induction kinetics similar to those observed after wounding (Fig. 5). By comparing the kinetics of PGcat mRNA and β-subunit mRNA with the wound-inducible and elicitor-inducible PG activities (Figs. 3 and 4), one can see that the enzyme activity in all cases declines after about 4 h.
Figure 5.
Time course of accumulation of PG activity in response to systemin, oligogalacturonic acid, and chitosan. Extracts were prepared from plants supplied with solutions of systemin (2.5 nM), oligogalacturonides (100 μM), or chitosan (250 μM) in 10−3 M potassium phosphate buffer, pH 6.0, for 40 min. After treatment with elicitors, the plants were incubated under light (15) for the times indicated, and the extracts were assayed for PG activity (18). Extracts from plants supplied with buffer alone were assayed as controls. (A) Systemin (diamonds); buffer control (squares). (B) Oligogalacturonides (squares); chitosan (diamonds); buffer controls (triangles).
The induction of PG activity by oligogalacturonides is somewhat perplexing. If PG produces pectic fragments that are elicitors of PG synthesis, then the process would be expected to be self-perpetuating, with the oligogalacturonide products eliciting more PG. However, this self-perpetuation does not seem to be the case, because the induction kinetics of PG activity in response to oligogalacturonide fragments are similar to those of systemin and chitosan. The leaf PGcat may be compartmentalized within the leaf cells and thus separated from pectin; if this is the case, the activity of PGcat would not be generated until the enzyme and its pectic substrates were mixed together at wound sites. This mixing would produce localized oligogalacturonides as signals that could activate a strong oxidative burst (19) to activate genes near the wound site for defense against pathogens as well as herbivores. A recent report has shown that systemin does not cause an oxidative burst in tomato cell suspension cultures, but systemin does potentiate the oxidative burst caused by oligogalacturonides (20). Within 9 h after exposure to systemin, tomato cells exhibited an oxidative burst 16-fold higher than cells not exposed to systemin. Thus, when tomato plants are wounded continually, the release of PG and the subsequent generation of oligogalacturonides near wounded tissues might activate a substantial oxidative burst that is antibiotic. On the other hand, the PG enzyme may be synthesized in specific cell types in response to wounding, producing oligogalacturonide signals that are involved in activating defense genes that do not include those of the PG enzyme.
Our finding that PG is wound-inducible in tomato leaves indicates that plants have the potential to produce pectic fragments during defense signaling in the absence of pathogens. Oligogalacturonide products of PG activity are well known elicitors of defense responses against attacks from both herbivores and pathogens. The data presented here, together with the recent report that systemin potentiates the oxidative burst in response to oligogalacturonide fragments, suggest that the signaling pathway that activates defense genes against herbivores has components in common with the pathways that activate defenses against pathogens. The significance of these observations and their relationships to defense responses found in other plant species must be investigated. Biochemical characterization of this PG enzyme would be necessary to gain a clear understanding of its functional role in tomato leaves.
Acknowledgments
We thank Thom Koehler and Sue Vogtman for growing our plants. This research was supported in part by Project 1791 of the College of Agriculture and Home Economics of Washington State University, National Science Foundation Grant IBN 9184542, U.S. Department of Agriculture Competitive Grants Program Grant 98-35301-6046, a National Institutes of Health grant to D.R.B., and a Graduate Fellowship from Conselho Nacional de Pesquisas (Brazil) to D.S.M.
ABBREVIATIONS
- PG
polygalacturonase
- PGcat
PG catalytic subunit
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF118567).
References
- 1.Fischer R L, Bennett A B. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:675–703. [Google Scholar]
- 2.Gray J, Picton S, Shabbeer J, Schuch W, Grierson D. Plant Mol Biol. 1992;19:69–87. doi: 10.1007/BF00015607. [DOI] [PubMed] [Google Scholar]
- 3.Hadfield K A, Bennett A B. Plant Physiol. 1998;117:337–343. doi: 10.1104/pp.117.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DellaPenna D, Watson C, Liu J P, Schuchman D. In: Pectins and Pectinesterases. Visser J, Boragen A G J, editors. Amsterdam: Elsevier Science; 1996. pp. 247–262. [Google Scholar]
- 5.Knegt E, Vermeer E, Bruinsma J. Physiol Plant. 1988;72:108–114. [Google Scholar]
- 6.Zheng L, Heupel R C, DellaPenna D. Plant Cell. 1992;4:1147–1156. doi: 10.1105/tpc.4.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collmer A, Keen N T. Annu Rev Phytopathol. 1986;24:383–409. [Google Scholar]
- 8.Ryan C A, Farmer E E. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:651–674. [Google Scholar]
- 9.John M, Rohrig G, Schmidt J, Walden R, Schell J. Trends Plant Sci. 1997;2:111–115. [Google Scholar]
- 10.Ryan C A, Pearce G. Annu Rev Cell Dev Biol. 1998;14:1–17. doi: 10.1146/annurev.cellbio.14.1.1. [DOI] [PubMed] [Google Scholar]
- 11.McGurl B, Pearce G, Orozco-Cardenas M, Ryan C A. Science. 1992;255:1570–1573. doi: 10.1126/science.1549783. [DOI] [PubMed] [Google Scholar]
- 12.McGurl B, Orozco-Cardenas M, Pearce G, Ryan C A. Proc Natl Acad Sci USA. 1994;91:9799–9802. doi: 10.1073/pnas.91.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergey D R, Howe G A, Ryan C A. Proc Natl Acad Sci USA. 1995;93:12053–12058. doi: 10.1073/pnas.93.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grierson D, Tucker G A, Keen J, Bird C R, Schuch W. Nucleic Acids Res. 1986;14:8595–8603. doi: 10.1093/nar/14.21.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalaitzis P, Koehler S M, Tucker M L. Plant Mol Biol. 1995;28:647–656. doi: 10.1007/BF00021190. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Pressey R. Hortic Sci. 1986;21:490–492. [Google Scholar]
- 18.Doares S, Syrovets T, Weiler E W, Ryan C A. Proc Natl Acad Sci USA. 1995;92:4095–4098. doi: 10.1073/pnas.92.10.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Low P S, Merida J R. Physiol Plant. 1996;96:533–542. [Google Scholar]
- 20.Stennis M J, Chandra S, Ryan C A, Low P S. Plant Physiol. 1998;117:1031–1036. doi: 10.1104/pp.117.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]