Abstract
Mycobacterium tuberculosis is an important pathogen of mammals that relies on 2-hydroxyphenyloxazoline-containing siderophore molecules called mycobactins for the acquisition of iron in the restrictive environment of the mammalian macrophage. These compounds have been proposed to be biosynthesized through the action of a cluster of genes that include both nonribosomal peptide synthase and polyketide synthase components. One of these genes encodes a protein, MbtB, that putatively couples activated salicylic acid with serine or threonine and then cyclizes this precursor to the phenyloxazoline ring system. We have used gene replacement through homologous recombination to delete the mbtB gene and replace this with a hygromycin-resistance cassette in the virulent strain of M. tuberculosis H37Rv. The resulting mutant is restricted for growth in iron-limited media but grows normally in iron-replete media. Analysis of siderophore production by this organism revealed that the biosynthesis of all salicylate-derived siderophores was interrupted. The mutant was found to be impaired for growth in macrophage-like THP-1 cells, suggesting that siderophore production is required for virulence of M. tuberculosis. These results provide conclusive evidence linking this genetic locus to siderophore production.
Mycobacterium tuberculosis is an obligate pathogen of mammals and is responsible for an incredible toll of human life every year. Even among pathogens, M. tuberculosis is noteworthy for having selected a particularly difficult lifestyle, inhabiting one of the most inhospitable cell types, the alveolar macrophage. Among the various defensive mechanisms expressed by the eukaryotic host are a potent burst of oxygen-derived radical species, acidification and digestion of phagocytosed organisms, and a dramatic restriction of available iron to support microbial growth (1, 2). Both pathogenic and saprophytic microbes have evolved sophisticated iron-acquisition systems to overcome iron deficiency imposed by a host defensive mechanism or their environment. At the core of such systems is the production of small molecules called siderophores, which are secreted into the extracellular space, tightly bind available iron, and then are reinternalized with their bound iron through specific cell surface receptors (3, 4).
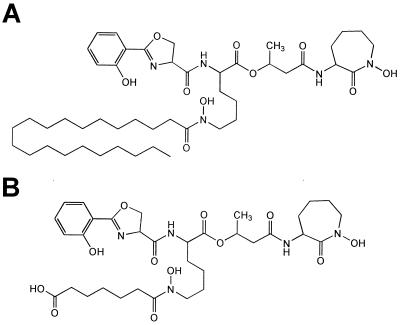
Mycobacteria produce a variety of siderophore-like substances that can be divided into two structural classes based on the presence or absence of a 2-hydroxyphenyloxazoline-ring system (5). M. tuberculosis produces only the mycobactin class of siderophore molecule, which contains this salicylic acid-derived moiety. Saprophytic mycobacteria such as M. smegmatis produce both this and a peptidic siderophore, called an exochelin, which coordinates iron through hydroxamic acids derived from the amides of the molecule. M. tuberculosis synthesizes two forms of mycobactin that differ in the nature of a single acyl group R1 (Fig. 1). These two forms have been proposed to have different physical properties, particularly regarding aqueous solubility, leading to the proposition that they have discrete biological roles. The hydrophilic mycobactin T (Fig. 1B) has been proposed to be secreted actively into the aqueous growth medium for direct competition with iron-binding molecules of the environment. The more lipophilic mycobactin T (Fig. 1A) has been proposed to operate as an ionophore to shuttle iron across the extremely hydrophobic cell wall of this organism after transfer of the iron from the extracellular siderophore (6, 7).
Figure 1.
Chemical structures of siderophores produced by M. tuberculosis. Both structures are of mycobactin T. In A, the structure of the more lipophilic mycobactin T is shown, whereas in B, the structure of the more hydrophilic mycobactin T is shown. The only chemical difference between the two is the nature of the N-acyl chain on the hydroxylated lysine in the middle of the molecule.
The core hydroxyphenyloxazoline structure of mycobactins is thought to be derived from salicylic acid after condensation with a serine residue, analogous to the biosynthetic pathway for both yersiniabactin and pyochelin, which contain hydroxyphenylthiazoline cores (8–10). Activation of the salicylate and condensation with an activated cysteine in these cases is followed by cyclization on the peptide synthase to give the final product (10). During annotation of the genome of M. tuberculosis H37Rv (11) we noted the presence of an enzyme, MbtB, with homology to proteins involved in hydroxyphenylthiazoline biosynthesis. MbtB was found to be 35% identical (46% similar) to Ybt (HMWP2) from Yersinia pestis, the enzyme involved in the formation of the yersiniabactin core structure (10). MbtB also was found to be 36% identical (48% similar) to PchE, an enzyme from Pseudomonas aeruginosa that also has been proposed to be involved in pyochelin biosynthesis (9). This enzyme occurred clustered with two other nonribosomal peptide synthase homologs as well as an unusual polyketide synthase. This genetic locus, therefore, appears to encode all of the relevant biosynthetic machinery for the production of the mycobactins, which contain, in addition to the hydroxyphenyloxazoline, two lysine residues and a short polyketide fragment (5). To test the role of these enzymes in mycobactin biosynthesis, as well as to test the biogenic relationship between the two classes of mycobactin produced in this organism, we constructed a targeted genetic deletion of mbtB.
Experimental Procedures
mbtB Gene Replacement.
M. tuberculosis strain H37Rv (ATCC 27294) was maintained in roller flasks in Middlebrook 7H9/ADC/Tween or on solid 7H11 medium as described previously (12). Where indicated, antibiotics were included at the following concentrations: kanamycin (Sigma), 25 μg/ml; and hygromycin (Calbiochem), 50 μg/ml. The cosmid Y22H8 was a kind gift of Stewart Cole of the Institute Pasteur (Paris). Routine molecular biological operations were carried out according to standard procedures (13).
Y22H8 was digested with SphI and the 3′ overhangs were removed by using the exonuclease activity of T4 DNA polymerase to create a blunt-ended, 5,281-nt fragment, which was gel-purified and then cloned into pCR-Blunt (Invitrogen). Removal of an internal, 1,273-nt BstEII fragment and the filling in of the resulting overhangs with the Klenow fragment of DNA polymerase allowed insertional replacement of mbtB coding sequence with a blunt-ended marker encoding hygromycin resistance from Streptomyces hygroscopicus (14, 15). Escherichia coli colonies were selected for hygromycin and kanamycin resistance. Restriction analysis verified the orientation was correct. The deletion-replacement fragment was removed from pCR-Blunt by digestion with NotI and SpeI and, after gel purification, was ligated to NotI- and SpeI-digested pMJ10 (a gift of Vladimir Pelicic, Institute Pasteur) (16). This plasmid was transformed into M. tuberculosis H37Rv by electroporation, and colonies were selected for hygromycin and kanamycin resistance at 32°C. Single colonies were obtained after 8 weeks of incubation, and these were analyzed for sensitivity to 2% sucrose on solid medium as described (16). A sucrose-sensitive single colony was grown in liquid medium to near saturation at 32°C. Serial dilutions of this culture were made and plated onto hygromycin and 2% sucrose at 39°C. After 6 weeks of incubation, hygromycin-resistant bacilli were obtained that were analyzed for kanamycin resistance. Ten different kanamycin-sensitive, hygromycin- and sucrose-resistant isolates were grown in 10 ml of liquid 7H9 at 37°C in the presence of hygromycin. Chromosomal DNA was prepared from these cultures as described previously (17) and analyzed by Southern blotting by using the following probes: (i) the internal, 1.3-kb BstEII fragment isolated by gel electrophoresis from cosmid Y22H8, (ii) the hygromycin-resistance cassette from p16R1 digested with SmaI and BspHI, and (iii) the cma1 gene encoded on a BamHI-to-PstI fragment described previously (18).
Siderophore Production Assays and Labeling Procedures.
Low-iron GAST was normal pH 6.6 glycerol–alanine–salts (GAS) medium [per liter: 0.3 g of Bacto Casitone (Difco), 0.05 g of ferric ammonium citrate, 4.0 g of dibasic potassium phosphate, 2.0 g of citric acid, 1.0 g of l-alanine, 1.2 g of magnesium chloride hexahydrate, 0.6 g of potassium sulfate, 2.0 g of ammonium chloride, 1.80 ml of 10 sodium hydroxide, and 10.0 ml of glycerol], except ferric ammonium citrate was omitted and Tween 80 was added to 0.05%. Low-iron GAST (50 ml) was inoculated with H37Rv or ΔmbtB∷hyg H37Rv to an OD650 of 0.05 and incubated at 37°C in a 250-ml roller bottle. After 3 days of growth [7-14C]salicylic acid (Moravek Biochemicals, Brea, CA) was added to the cultures to a final concentration of 3 μg/ml and final activity of 1.25 μCi/ml. After 10 days of incubation, the OD650 was 3.2 for H37Rv and 0.15 for ΔmbtB∷hyg H37Rv. At this point, the cultures were centrifuged for 10 min at 5,000 × g and the supernatants and cell pellets were collected. The supernatants were filtered through a 0.22-μm filter (Steriflip; Millipore). Ferric chloride (20 mg/ml FeCl3⋅6H2O in ethanol) was added to sterile filtrates to a final concentration of 555 μM. After incubation at room temperature for 1 hr, the solution was extracted twice with 75 ml of chloroform. The organic fraction was dried by rotary evaporation, resuspended in 1 ml of chloroform, and subjected to TLC on 10 cm × 10 cm, 250-μm-thick silica gel 60 (Merck) developed in petroleum ether/n-butanol/ethyl acetate, 2:3:3 (19). Plates were dried and analyzed by PhosphorImager. The cell pellets were resuspended in 10 ml of ethanol and incubated with agitation overnight at 37°C. After centrifugation for 10 min at 5,000 × g, the supernatant was filtered through a 0.22-μm filter and brought to 2.2 mM ferric ion with the addition of FeCl3 as above. After incubation at room temperature for 1 hr, the solutions were diluted with 10 ml of water and extracted twice with 10 ml of chloroform. The organic fraction was dried by rotary evaporation, resuspended in 1 ml of chloroform, and analyzed by TLC as above. In both cases, the amount analyzed by TLC was corrected for the OD650 of the cultures at the time of harvest.
Construction of Mycobacterial Luciferase Reporters.
The promoterless firefly luciferase reporter plasmid pMV306_luc was created by subcloning the 2,177-bp NotI-to-SalI fragment from pGL3-Basic (Promega) containing luc into the same sites of pMV306 (20). A 431-bp KpnI-to-HindIII fragment from pMV261 (14) containing the hsp60 promoter then was cloned into the same sites of pMV306_luc to create pMV306_hsp60_luc.
Macrophage Infection Assays.
THP-1 cells (ATCC 45503) were obtained from the American Type Culture Collection (Manassas, VA) and propagated in RPMI 1640 medium supplemented with 2 mM l-glutamine, 55 μM 2-mercaptoethanol, and 10% FBS as described previously (21). Two days before infections, 24-well plates were seeded with 1 × 106 cells in RPMI-10 containing 50 ng/ml phorbol 12-myristate 13-acetate. All media components were from Life Technologies (Gaithersburg, MD). THP-1 monolayers were washed once with 1 ml of RPMI-10 and then infected with 0.25 ml of diluted bacteria for 1 hr at 37°C, 5% CO2. Bacteria used for THP-1 infections were from fresh cultures. Bacteria were grown in 7H9 medium from a starting OD (at 650 nm) of between 0.05 and 0.15, such that on the day of infection both wild-type and mutant cultures had an OD of approximately 0.4. Cultures were adjusted to OD 0.4 with 7H9 and diluted 1:1,600 in RPMI-10 (see below). For inocula grown in various iron concentrations, bacteria were grown in GAST to midexponential phase (OD 0.5), washed once with GAST-Fe, and used to inoculate GAST-Fe medium supplemented with 0, 0.5, 5, or 500 μM additional FeCl3 to an optical density of 0.1. Two days later cultures were adjusted to OD 0.2 with GAST-Fe and diluted 1:400 in RPMI-10. Infected monolayers then were washed four times with 1 ml of RPMI and fed 1 ml of RPMI-10. Medium was replaced with fresh RPMI-10 2 and 5 days after infection. Three wells were used for each time point, and each well was assayed in triplicate for luciferase activity or colony-forming units (cfu) (see below). Before performing luciferase assays or cfu determinations, the medium was removed by aspiration and 0.25 ml of 1% Triton X-100 was added to lyse the THP-1 cells. After a 5-min incubation, 10-fold serial dilutions were made in 7H9 medium and 0.1-ml aliquots were plated on 7H11 agar. Alternatively, 50 μl of lysate was mixed with 50 μl of LucLite substrate (Packard) in an opaque, 96-well plate (Dynex Technologies, Chantilly, VA), incubated in the dark for 10 min, and assayed on a Top Count luminescence reader (Packard). Input relative light units were determined in quadruplicate directly from the diluted bacteria used for infection.
Results
Identification and Deletion of mbtB Encoding a Phenyloxazoline Synthase.
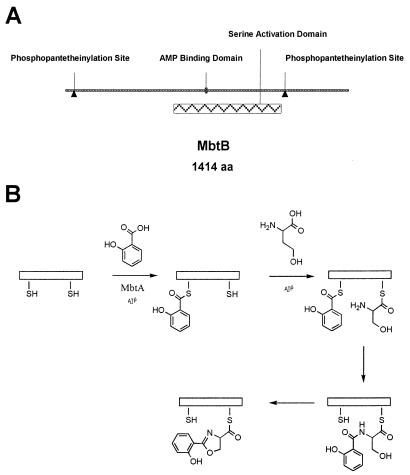
Mycobactin T can be considered to be a modified peptide constructed from serine, two lysine residues, and a β-hydroxyacid, wherein the N-terminal serine has been N-acylated with salicylic acid and cyclized to form the characteristic 2-hydroxyphenyloxazoline-ring system. By analogy to other siderophores and small peptides, the amide bond connecting serine and salicylate in this structure has been proposed to be formed nonribosomally through an activation and condensation sequence mediated by a peptide synthase (22). These enzymes are large, modular proteins that contain a phosphopantetheine moiety as a site of attachment for activated amino acid intermediates before condensation. These proteins also typically contain adenosine monophosphate-binding domains that are utilized in the initial activation of the amino acid by formation of an aminoacyl AMP ester to facilitate attachment of the amino acid to the thiol of the prosthetic group. An examination of the genome of M. tuberculosis H37Rv revealed relatively few nonribosomal peptide synthases. One gene cluster, designated the mbt locus, has been proposed on the basis of homology to encode the enzyme that initiates amide bond formation in mycobactin biosynthesis (5, 8, 11). Biochemical evidence for enzymatic phosphopantetheinylation of the predicted aryl carrier protein site in MbtB (Fig. 2A) recently has been reported, as has evidence for acylation of this domain with salicylate (8). Thus, mbtB appears to encode an enzyme capable of amide bond formation between the carboxylic acid of salicylate and the α-amino group of serine (Fig. 2B).
Figure 2.
Predicted protein structure and chemical reaction catalyzed by MbtB. In A, the predicted domains relevant to catalysis by MbtB are shown. The two predicted phosphopantetheinylation (ppant) sites are covalently modified by a ppant transferase; experimental evidence for this has been obtained in heterologous systems (8). In B, the biosynthetic reaction proposed for MbtB is shown in detail. Initial activation of salicylate to the AMP ester is catalyzed by MbtA. The activated salicylate then is transferred to the N-terminal ppant, where it is putatively condensed with serine that has been attached to the C-terminal ppant site by the activation domain of MbtB. The resulting amide is cyclized and then finally dehydrated to form the 2-hydroxyphenyloxazoline nucleus of the mycobactins.
To assess directly the role of MbtB in mycobactin T biosynthesis we obtained an H37Rv cosmid encoding the entire operon and subcloned a fragment containing only the coding sequence for MbtB (Fig. 3). A portion of the coding sequence for MbtB was removed and replaced with a hygromycin resistance cassette. This deletion construct was cloned into pMJ10, which contains a thermosensitive mycobacterial origin of replication, a sacB gene, and a kanamycin resistance determinant stable at 32°C (16). Subsequent selection at restrictive temperature (39°C) under conditions that select against the SacB protein allowed for efficient identification of double-homologous recombinants. At a frequency of approximately 1 × 10−6 to 1 × 10−7 we obtained colonies resistant to sucrose after 8 weeks of incubation at the restrictive temperature. Individual transformants were analyzed by Southern blot analysis of digested chromosomal DNA by using probes to both the hygromycin resistance cassette and the mbtB gene sequence, which was being deleted. Ten of 10 colonies were positive for hybridization with the hygromycin probe whereas 9 of these were also positive with the mbtB probe. The remaining colony lacked reactivity with mbtB and gave appropriate-sized signals in Southern blots probed with hyg. DNA from this organism also gave a positive Southern band for cma1, a gene that uniquely identifies the pathogenic mycobacteria (data not shown) (18).
Figure 3.
Construction of the targeted replacement construct for mbtB deletion. A 10-kb portion of the mbt locus that has been proposed to be involved in mycobactin synthesis. Below this is the 5,281-nt SphI fragment, from which an internal, 1.3-kb fragment was removed and replaced with the hygromycin resistance cassette as shown at the bottom.
The ΔmbtB∷hyg Strain Does Not Produce Siderophore.
To assess the effect of deletion of mbtB we examined various growth conditions for the production of siderophore in both mutant and wild-type strains. Although siderophore production has been reported for various mycobacterial species by using a wide variety of media, in general this has not been commonly reported for pathogenic mycobacteria (23). Surveying several media for production of detectable siderophore allowed us to achieve iron-dependent production of siderophore in GAS medium, from which we omitted ferric ammonium citrate. To establish that the mutant was incapable of production of mycobactin or any other siderophore-like molecule, we performed functional assays that measure dissociation of an iron-dye complex by iron-binding molecules [a chrome azurol S (CAS) shuttle assay (24)]. This assay relies on the formation of a colored iron complex of a commercial dye (CAS). Removal of iron from this complex by desferrisiderophore (or any other molecule with an affinity for iron higher than that of the dye) results in a decrease in absorbance. This assay revealed that wild-type H37Rv possessed iron-scavenging ability presumably associated with mycobactin T production whereas the mutant strain deleted for mbtB did not, confirming that this strain does not produce a siderophore-like molecule (Fig. 4B).
Figure 4.
Phenotypic characterization of the ΔmbtB∷hyg H37Rv strain. (A) TLC of siderophores extracted from wild-type (lanes 1 and 3) and mutant (lanes 2 and 4) cultures labeled with [7-14C]salicylic acid. Lanes 1 and 2 represent siderophore associated with the medium whereas lanes 3 and 4 are cell-associated siderophore. (B) CAS shuttle assay monitoring production of iron-binding components produced by the cells. The mutant strain produces negative values in this assay.
To specifically label any mycobactins in both the wild-type and mutant strains of M. tuberculosis we then incubated cultures in iron-deficient GAST medium in the presence of [7-14C]salicylic acid. To examine both the cell-associated and the secreted forms of mycobactin T we separately extracted the cell supernatant and the cell pellet after iron loading and analyzed these by TLC (Fig. 4A). Extraction of the cell pellet of wild-type bacilli showed that label was incorporated into organic extractable material with an Rf value identical to that reported previously for mycobactin T obtained by extraction of the whole cells (25). As expected for the gene deletion, label was not incorporated by the mutant into any extractable material from the whole cells. Extraction of the wild-type pellet produced two closely migrating spots that may correspond in structure to mycobactin T and the water-soluble form of mycobactin T in order of decreasing Rf, although definitive assignment awaits further structural work, which is in progress. Comparison of the salicylate-labeled material in the cell supernatant with that in the cell pellet from the wild-type strain revealed no major differences in the distribution of the two species produced. Although several different growth media with and without detergent present were surveyed, these two species did not appear to segregate with regard to cell or media association (data not shown). Interestingly, the mutant does appear to accumulate small quantities of a salicylate-labeled metabolite that can be extracted from the medium in the same manner as mycobactin T. Although this metabolite does appear to be close in retention time to the more polar siderophore in the wild-type strain, the results of the CAS shuttle assay suggest that it does not possess iron-binding capabilities.
The ΔmbtB∷hyg Strain Is Deficient for Growth in Low-Iron Media and Macrophages.
To assess the effects of lack of siderophore production on growth and metabolism of M. tuberculosis, we first measured growth in iron-limited GAS medium at concentrations of iron just above where we have noted growth inhibition of the wild-type strain. Under conditions of iron sufficiency for both strains (170 μM iron in normal GAST medium) there was little observable difference in growth rate between wild type and the mbtB knockout strain. There was a reproducible slight increase in lag time for entry into exponential growth for the mutant strain, but the ultimate growth rate achieved was similar to wild type in iron-rich medium. When iron was removed from the medium, the strain deficient in siderophore production displayed a pronounced defect in growth (Fig. 5A). This result represents the first definitive correlation between the production of iron-scavenging molecules and growth in the pathogenic mycobacteria.
Figure 5.
Growth in vitro and in vivo of wild-type and siderophore-deficient H37Rv. (A) Growth curves of wild-type H37Rv in low-iron GAST (●) and ΔmbtB∷hyg in low-iron GAST medium supplemented with 5 μM iron, 0.5 μM iron, and 0 μM iron (□, ▵, and ○, respectively). The growth curve for wild type in any of these iron concentrations is superimposable with the iron-deficient growth curve. (B) Growth of wild-type (●) and ΔmbtB∷hyg (○) H37Rv in the human macrophage-like cell line THP-1 over a 9-day infection. Inset demonstrates that the number of input organisms and the number of organisms internalized after 2 hr of infection were similar between the wild type (open bars) and mutant (shaded bars). (C) Growth of wild-type and ΔmbtB∷hyg H37Rv when the inocula were prepared from medium containing different concentrations of iron. The first four bars at each time point represent wild type, and the second four bars represent mutant. In each series the iron concentration increases from left to right as 0, 0.5, 5, and 500 μM iron. The organisms were passed for 2 days in medium of the indicated iron concentration before infection.
To determine whether this defect in iron acquisition also was related to the ability of such organisms to survive within the iron-restricted environment of the human macrophage, we then measured growth rates in the macrophage-like cell line THP-1. To facilitate accurate and rapid determination of the growth of the organism, we first integrated an hsp60 promoter-driven luciferase construct into the chromosome of both strains at the L5 phage-integration site (26, 27). Bioluminescence was found to be strongly correlated with cfu for both strains (data not shown). By monitoring bioluminescence during the infection, we observed that wild-type organisms grew profusely over the course of a 9-day infection. The siderophore-deficient mutant, however, was significantly retarded for ability to grow in this cell line (Fig. 5B). This difference was not due to efficiency of cell entry because the input and 2-hr time-point RLU values were nearly identical for the two strains (Fig. 5B Inset). These results were confirmed by direct determination of CFU from infected macrophage lysates with the same results (data not shown). To assess whether the siderophore deficiency could be overcome during in vivo growth by preloading the cells with iron we prepared inocula of wild-type and mutant organisms in medium with varying concentrations of iron. These then were used to infect THP-1 cells. Although there was a slight enhancement of growth of the wild-type strain prepared in iron-excess conditions (500 μM), there was no effect of iron starvation of the inoculum. There was also no rescue of the growth defect of the siderophore mutant by growth in iron-rich medium (Fig. 5C). Control experiments demonstrated that iron deficiency did not negatively effect luciferase activity from these strains.
Discussion
The role of siderophore molecules in microbial virulence is well established in many pathogenic bacterial systems and has been inferred for M. tuberculosis for many years by analogy to these more tractable systems. Recent advances in the ability to make targeted genetic insertions and deletions in the chromosome of slow-growing pathogenic mycobacteria have facilitated the direct examination of many putative virulence factors in M. tuberculosis (16, 28). The recent completion of the entire genomic DNA sequence of M. tuberculosis H37Rv has expanded our ability to interpolate biochemical function directly from sequence information (11). Using this information we and others predicted that the mbt gene locus would encode the appropriate enzymes for production of the salicylate containing siderophores of M. tuberculosis (8, 11). Some suggestive biochemical information supported this assignment, but definitive proof was hampered by extremely poor expression of the relevant enzymes in heterologous systems (8). We now have succeeded in linking the production of mycobactin T to the mbt gene locus by deletion of mbtB. The mbtB gene encodes an enzyme proposed to catalyze the initial steps in the mycobactin T biosynthetic pathway, the condensation of salicylic acid with serine, and the formation of the hydroxyphenyloxazoline nucleus of these siderophores (5).
Confusion in the literature has arisen surrounding the two “classes” of mycobacterial siderophore produced by many species in the genus. “Exochelin” was a phrase first used specifically to represent the peptidic siderophores originally isolated from M. smegmatis that coordinate iron through hydroxamic acids derived from the amide nitrogens of the peptide backbone (29, 30). The genes encoding the biosynthetic enzymes for these siderophores have been identified from M. smegmatis (31, 32). Exochelins, in this sense, are not produced by the pathogenic mycobacteria. “Mycobactin” has been applied to the more lipophilic, cell-associated siderophore molecules, which have as their core structure the hydroxyphenyloxazoline ring derived from salicylate. Water-soluble mycobactins also have been described [so-called “carboxymycobactins” (33, 34)]. Unfortunately, “exochelin” has been used recently to refer to the secreted, water-soluble mycobactins produced by mycobacteria (6, 35). Nomenclature aside, the biological roles of the two broad classes of siderophore molecules (cell-associated and -secreted) are thought to be complementary, with the secreted molecule competing for available iron in the growth milieu and the cell-associated molecule acting as an ionophore to facilitate transport of the iron across the virtually impermeable mycobacterial cell wall (6, 36). In M. tuberculosis, it has been suggested that these two roles are fulfilled by molecules that are intimately related in chemical structure, mycobactin T and water-soluble mycobactin T. These two molecules differ only by the nature of the fatty acyl side chain linked to the ɛ-amino group of N–hydroxy lysine, and the biosynthesis of both of these molecules is interrupted in the mbtB deletion strain. We have observed two forms of mycobactin T by radiosalicylate labeling. These may correspond to the two classes described above. Further studies are needed to determine their precise chemical structure.
Disruption of the biosynthesis of mycobactin T has allowed us to establish that, under conditions of iron limitation, siderophore production is essential for optimal growth of M. tuberculosis. In addition, siderophore production appears to enhance growth in macrophages, the preferred cell type for growth of the pathogenic mycobacteria in humans. The linkage between iron availability and growth or virulence of M. tuberculosis is supported further by ongoing clinical studies of dietary iron overload in Africa, where a strong association between elevated iron and tuberculosis has been observed (37, 38). Together these two observations suggest that the enzymes involved in siderophore biosynthesis, secretion, and uptake or transport after iron chelation represent outstanding candidates as targets for the development of new antibiotics for the treatment of tuberculosis and related mycobacterial diseases. Existing evidence suggests that p-aminosalicylic acid, a little-used second-line antitubercular, in fact may target mycobactin synthesis (39).
Acknowledgments
We thank Dr. Stewart Cole for providing the cosmid Y22H8. We also thank Dr. Vladimir Pelicic for providing the gene disruption system and Deborah Crane for technical assistance. This work was supported by National Health and Medical Research Council Grant 961249 to J.D.V. and by a Francine Kroesen Travelling Fellowship to K.R.
Abbreviations
- cfu
colony-forming unit
- CAS
chrome azurol S
- GAS
glycerol–alanine–salts
- low-iron GAST
GAS without added iron and containing Tween 80
- ppant
phosphopantetheine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kontoghiorghes G J, Weinberg E D. Blood Rev. 1995;9:33–45. doi: 10.1016/0268-960x(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg E D. Science. 1974;184:952–956. doi: 10.1126/science.184.4140.952. [DOI] [PubMed] [Google Scholar]
- 3.Braun V, Hantke K, Koster W. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- 4.Byers B R, Arceneaux J E. Met Ions Biol Syst. 1998;35:37–66. [PubMed] [Google Scholar]
- 5.De Voss J J, Rutter K, Schroeder B G, Barry C E., III J Bacteriol. 1999;181:4443–4451. doi: 10.1128/jb.181.15.4443-4451.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gobin J, Horwitz M A. J Exp Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golden C A, Kochan I, Spriggs D R. Infect Immunity. 1974;9:34–40. doi: 10.1128/iai.9.1.34-40.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quadri L E, Sello J, Keating T A, Weinreb P H, Walsh C T. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 9.Reimmann C, Serino L, Beyeler M, Haas D. Microbiology. 1998;144:3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 10.Gehring A M, Mori I, Perry R D, Walsh C T. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 11.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Y, Crane D D, Barry C E., III J Bacteriol. 1996;178:4484–4492. doi: 10.1128/jb.178.15.4484-4492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 14.George K M, Yuan Y, Sherman D R, Barry C E., III J Biol Chem. 1995;270:27292–27298. doi: 10.1074/jbc.270.45.27292. [DOI] [PubMed] [Google Scholar]
- 15.Garbe T R, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, Mukherjee R, Young D B. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 16.Pelicic V, Jackson M, Reyrat J M, Jacobs W R, Jr, Gicquel B, Guilhot C. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bose M, Chander A, Das R H. Nucleic Acids Res. 1993;21:2529–2530. doi: 10.1093/nar/21.10.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan Y, Lee R E, Besra G S, Belisle J T, Barry C E., III Proc Natl Acad Sci USA. 1995;92:6630–6634. doi: 10.1073/pnas.92.14.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macham L P, Ratledge C, Nocton J C. Infect Immun. 1975;12:1242–1251. doi: 10.1128/iai.12.6.1242-1251.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover C K, Bansal G P, Hanson M S, Brulein J E, Palaszynski S R, Young J F, Koenig S, Young D B, Sadziene A, Barbour A G. J Exp Med. 1993;178:197–209. doi: 10.1084/jem.178.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y, Crane D D, Simpson R M, Zhu Y Q, Hickey M J, Sherman D R, Barry C E., III Proc Natl Acad Sci USA. 1998;95:9578–9583. doi: 10.1073/pnas.95.16.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marahiel M A, Stachelhaus T, Mootz H D. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 23.Ratledge C. In: The Biology of the Mycobacteria. Ratledge C, Stanford J, editors. Vol. 1. San Diego: Academic; 1982. pp. 185–271. [Google Scholar]
- 24.Schwyn B, Neilands J B. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 25.Leite C Q F, Barreto A M W, Leite S R A. Rev Microbiol. 1995;26:192–199. [Google Scholar]
- 26.Mdluli K, Sherman D R, Hickey M J, Kreiswirth B N, Morris S, Stover C K, Barry C E., III J Infect Dis. 1996;174:1085–1090. doi: 10.1093/infdis/174.5.1085. [DOI] [PubMed] [Google Scholar]
- 27.Snewin V A, Gares M P, Gaora P O, Hasan Z, Brown I N, Young D B. Infect Immun. 1999;67:4586–4593. doi: 10.1128/iai.67.9.4586-4593.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam R A, Bloom B R, Hatfull G F, Jacobs W R., Jr Proc Natl Acad Sci USA. 1997;94:10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharman G J, Williams D H, Ewing D F, Ratledge C. Chem Biol. 1995;2:553–561. doi: 10.1016/1074-5521(95)90189-2. [DOI] [PubMed] [Google Scholar]
- 30.Sharman G J, Williams D H, Ewing D F, Ratledge C. Biochem J. 1995;305:187–196. doi: 10.1042/bj3050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu W, Arceneaux J E L, Beggs M L, Byers B R, Eisenach K D, Lundrigan M D. Mol Microbiol. 1998;29:629–639. doi: 10.1046/j.1365-2958.1998.00961.x. [DOI] [PubMed] [Google Scholar]
- 32.Yu S, Fiss E, Jacobs W R., Jr J Bacteriol. 1998;180:4676–4685. doi: 10.1128/jb.180.17.4676-4685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane S J, Marshall P S, Upton R J, Ratledge C, Ewing M. Tetrahedron Lett. 1995;36:4129–4132. [Google Scholar]
- 34.Ratledge C, Ewing M. Microbiology. 1996;142:2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- 35.Gobin J, Wong D K, Gibson B W, Horwitz M A. Infect Immun. 1999;67:2035–2039. doi: 10.1128/iai.67.4.2035-2039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ratledge C. Microbiol Ser. 1984;15:603–627. [Google Scholar]
- 37.Moyo V M, Gangaidzo I T, Gordeuk V R, Kiire C F, Macphail A P. Cent Afr J Med. 1997;43:334–339. [PubMed] [Google Scholar]
- 38.Murray M J, Murray A B, Murray M B, Murray C J. Br Med J. 1978;2:1113–1115. doi: 10.1136/bmj.2.6145.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown K A, Ratledge C. Biochim Biophys Acta. 1975;385:207–220. doi: 10.1016/0304-4165(75)90349-9. [DOI] [PubMed] [Google Scholar]