Abstract
Reversibly glycosylated polypeptides (RGPs) have been implicated in polysaccharide biosynthesis. In plants, these proteins may function, for example, in cell wall synthesis and/or in synthesis of starch. We have isolated wheat (Triticum aestivum) and rice (Oryza sativa) Rgp cDNA clones to study the function of RGPs. Sequence comparisons showed the existence of two classes of RGP proteins, designated RGP1 and RGP2. Glucosylation activity of RGP1 and RGP2 from wheat and rice was studied. After separate expression of Rgp1 and Rgp2 in Escherichia coli or yeast (Saccharomyces cerevisiae), only RGP1 showed self-glucosylation. In Superose 12 fractions from wheat endosperm extract, a polypeptide with a molecular mass of about 40 kD is glucosylated by UDP-glucose. Transgenic tobacco (Nicotiana tabacum) plants, overexpressing either wheat Rgp1 or Rgp2, were generated. Subsequent glucosylation assays revealed that in RGP1-containing tobacco extracts as well as in RGP2-containing tobacco extracts UDP-glucose is incorporated, indicating that an RGP2-containing complex is active. Gel filtration experiments with wheat endosperm extracts and extracts from transgenic tobacco plants, overexpressing either wheat Rgp1 or Rgp2, showed the presence of RGP1 and RGP2 in high-molecular mass complexes. Yeast two-hybrid studies indicated that RGP1 and RGP2 form homo- and heterodimers. Screening of a cDNA library using the yeast two-hybrid system and purification of the complex by an antibody affinity column did not reveal the presence of other proteins in the RGP complexes. Taken together, these results suggest the presence of active RGP1 and RGP2 homo- and heteromultimers in wheat endosperm.
Reversibly glycosylated polypeptides (RGPs) are thought to be involved in polysaccharide metabolism. Polysaccharides are the main components of the plant cell wall. In dicots, 20% of the primary cell wall consists of the polysaccharide xyloglucan, whereas in monocots, this hemicellulose makes up 2% of the primary cell wall (Darvill et al., 1980). Xyloglucan consists of a β-1,4-glucan backbone with xylosyl and xylosyl-galactosyl-fucosyl side chains (Hayashi, 1989). The transferases responsible for the addition of these side chains are localized to the Golgi apparatus (Brummell et al., 1990; Driouich et al., 1993; Staehelin and Moore, 1995). RGP1 has been implicated in xyloglucan biosynthesis in pea (Pisum sativum; Dhugga et al., 1997). This protein, a 40-kD doublet, which could be glycosylated with radiolabeled UDP-Glc in the presence of Mn2+ or Mg2+, is associated with Golgi membranes as shown by density gradient centrifugation (Dhugga et al., 1991). Immunolocalization experiments showed RGP1 to be present in trans-Golgi dictyosomal cisternae (Dhugga et al., 1997). An Arabidopsis homologue was found to be mostly soluble, but also to be associated with membranes (Delgado et al., 1998). Glycosylation of the pea protein with UDP-Glc, UDP-Xyl, and UDP-Gal in a ratio similar to that of the typical sugar composition of xyloglucan (UDP-Glc:UDP-Xyl:UDP-Gal = 10:7:3) suggested its involvement in xyloglucan synthesis.
Besides being components of the cell wall, polysaccharides are also major energy reserves. In plants, Glc is mainly stored as starch, an α-1,4-linked Glc polymer with α-1,6 branches. Although considerable progress has been made in the identification of the enzymes and the biosynthetic steps leading to the formation of the glucan polymers, the way in which starch synthesis and granule formation are initiated is still an enigma. In mammalian systems, a 38-kD protein has been identified as the primer for the biosynthesis of the soluble α-1,4-glucan polymer glycogen and was named glycogenin (Lomako et al., 1988). This self-glucosylating protein utilizes UDP-Glc as the Glc donor to elongate an α-1,4-glucan chain covalently linked to the polypeptide through a single Tyr residue in an Mn2+-dependent reaction (Alonso et al., 1995a). Likewise, starch biosynthesis has been suggested to be initiated on a protein primer. The enzyme catalyzing the Glc-protein linkage was previously termed UDP-Glc:protein transglucosylase (UPTG; Lavintman and Cardini, 1973). In vitro studies showed self-glucosylation of this protein in an Mn2+-dependent reaction, resulting in the hypothesis that glucosylated UPTG might represent the primer for enzymatic glucan chain elongation (Moreno et al., 1987). In a self-glucosylating protein from sweet corn, Glc was found to be bound to a single Arg residue via a novel Glc-protein bond (Singh et al., 1995). Sequence analysis of this protein revealed a high homology to RGP1. Molecular cloning of the UPTG from potato (Solanum tuberosum) also revealed a high homology (86%–93% identity at amino acid level) with RGP1 from several species (Bocca et al., 1999).
Until now, the relation between the different self-glucosylating proteins described is unclear. Because RGP1 may function in cell wall polysaccharide biosynthesis and a related RGP2 has been found in rice (Oryza sativa; Dhugga et al., 1997), we adopted the working hypothesis that RGP1-related (RGP2) proteins function in initiation of starch biosynthesis.
RGP-homologous cDNAs were isolated from wheat (Triticum aestivum) and rice endosperm. Our results show that two classes of RGP homologs can be distinguished, as judged from their amino acid sequence. In a first attempt to find indications about their role in polysaccharide synthesis, glucosylation assays were performed with extracts from Escherichia coli and tobacco (Nicotiana tabacum), each overexpressing Rgp1 or Rgp2. Furthermore, the presence of RGP complexes in wheat endosperm was studied by Superose 12 gel filtration experiments. The interaction of RGP proteins with other proteins was tested using a yeast (Saccharomyces cerevisiae) two-hybrid system and by affinity purification.
RESULTS
cDNA Clones Encoding RGP Homologs from Wheat and Rice
To study the function of RGPs, cDNAs homologous to published self-glucosylating proteins, termed RGPs, were cloned (Dhugga et al., 1997; Delgado et al., 1998). Rgp cDNAs and rice expressed sequence tags (ESTs) were compared, and primers designed on conserved DNA sequences were used to produce a PCR probe. A cDNA library made of 10-DPA wheat endosperm was screened with this PCR probe. Twenty positive clones were sequenced from the 5′ and 3′ ends and each was found to encode the same protein. By comparison of the rice EST sequences, two classes could be identified, one of which is homologous to the RGP1 clone described by Dhugga et al. (1997). Each of the isolated wheat cDNA clones is homologous to the other class, designated RGP2. To obtain cDNA clones corresponding to RGP1, a new specific antisense primer was designed. Using this primer, an Rgp1-homologous PCR probe was obtained and used to isolate Rgp1 clones. The complete sequence of the longest clone from each class was determined.
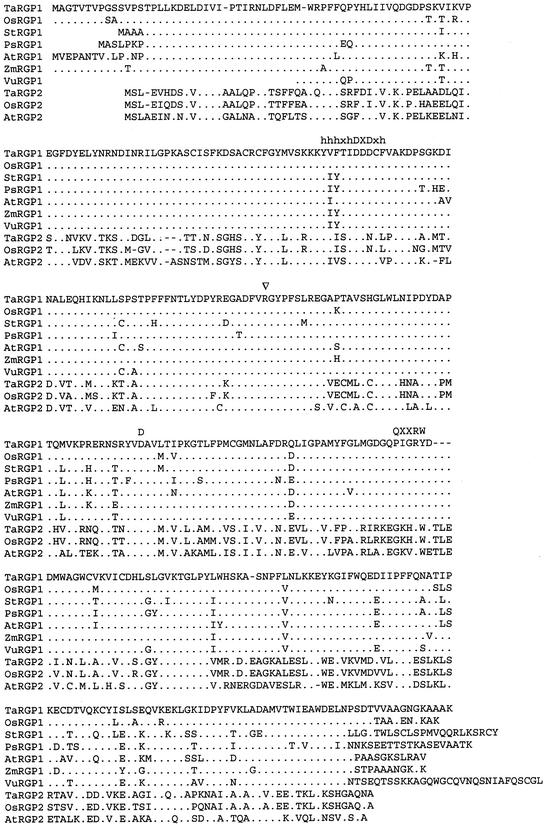
To obtain full-length rice cDNAs from both classes, rice cDNA libraries from etiolated shoot and 7-d-old somatic embryo were screened with two EST clones corresponding to both classes. From both classes, one cDNA clone containing the complete coding region was sequenced. The deduced amino acid sequences of both cDNA clones from wheat and rice are shown in Figure 1 in comparison with the amino acid sequences of homologous proteins from some other plant species. Both RGPs from the first class have a deduced molecular mass of 41 kD and a pI of 6.1. The proteins from the second class have a molecular mass of 39 kD and pI of 6.6 for the wheat protein and 6.4 for the rice protein. The region around the Arg residue that was determined to be the glucosylation site (Singh et al., 1995) is highly conserved in all proteins. The overall homology between the proteins is given in Table I. These data clearly show that the sequences fall into two classes. The class consisting of the RGP1 proteins is 87% to 93% identical (90%–96% similarity), whereas the RGP2 class is 63% to 88% identical (69%–91% similarity). Proteins from different classes are comparatively less homologous and 47% to 51% identical (57%–61% similarity).
Figure 1.
Sequence comparison of RGP protein sequences from wheat, rice, potato, pea, Arabidopsis, maize (Zea mays), and cowpea (Vigna unguiculata). Wheat RGP has been used as a reference. Dots indicate identical amino acids and dashes indicate gaps. The DXD motif flanked by hydrophobic residues (Wiggins and Munro, 1998) and the conserved D and QXXRW motif (Saxena et al., 1995) are indicated. The Arg that was shown to become glucosylated (Singh et al., 1995) is indicated with an arrowhead. Database accession numbers of the sequences used for the comparison are: Y18626 (TaRGP1), Y18624 (OsRGP1), AJ223252 (StRGP1), U31565 (PsRGP1), AF013627 (AtRGP1), U89897 (ZmRGP1), AF005279 (VuRGP1), Y18625 (TaRGP2), Y18623 (OsRGP2), and AB005242 (AtRGP2).
Table I.
Homology between RGP1 and RGP2 proteins of different species
| TaRGP1 | OsRGP1 | ZmRGP1 | StRGP1 | PsRGP1 | VuRGP1 | AtRGP1 | TaRGP2 | OsRGP2 | |
|---|---|---|---|---|---|---|---|---|---|
| OsRGP1 | 94 (97) | ||||||||
| ZmRGP1 | 93 (96) | 90 (93) | |||||||
| StRGP1 | 90 (95) | 91 (94) | 90 (95) | ||||||
| PsRGP1 | 89 (95) | 86 (94) | 89 (93) | 89 (95) | |||||
| VuRGP1 | 92 (96) | 88 (95) | 89 (93) | 92 (96) | 90 (94) | ||||
| AtRGP1 | 87 (92) | 87 (92) | 87 (90) | 90 (93) | 89 (94) | 90 (94) | |||
| TaRGP2 | 47 (68) | 47 (68) | 46 (67) | 46 (68) | 47 (69) | 47 (68) | 50 (70) | ||
| OsRGP2 | 47 (66) | 47 (66) | 46 (66) | 46 (66) | 48 (68) | 48 (67) | 49 (68) | 87 (93) | |
| AtRGP2 | 49 (69) | 49 (69) | 49 (69) | 49 (69) | 51 (71) | 51 (71) | 51 (71) | 61 (76) | 63 (76) |
For name abbreviations, see Figure 1. The percentage identity and similarity (between brackets) are given.
We performed Southern-blot analysis to estimate the number of genes in wheat that are homologous to Rgp1 (Fig. 2A) and Rgp2 (Fig. 2B). Multiple DNA fragments were hybridizing with Rgp1, indicating that it corresponds to a gene family. Probing the same blot with Rgp2 resulted in two to four bands in each lane. Considering the hexaploidy of wheat, we assume that Rgp2 is present as a single-copy gene per haploid genome.
Figure 2.
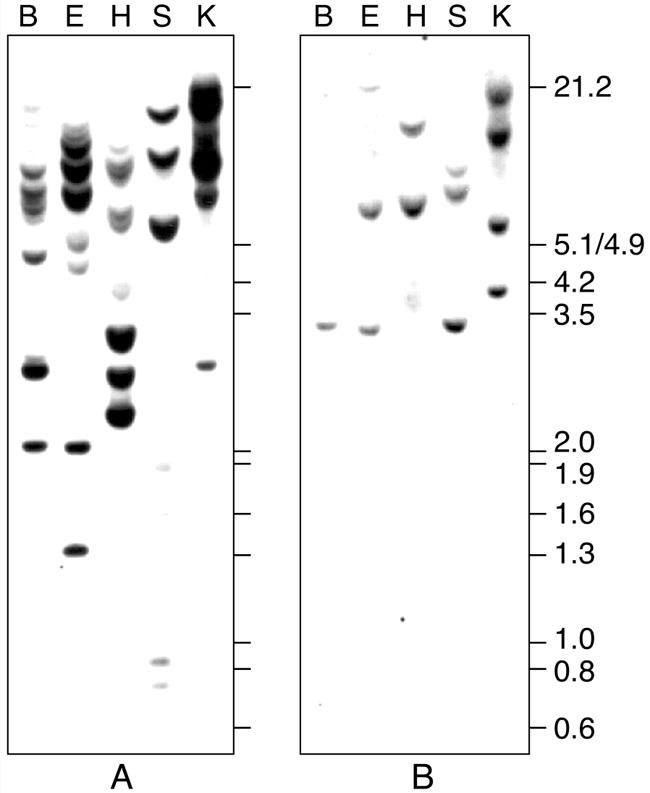
Southern-blot analysis of Rgp1 and Rgp2. Wheat genomic DNA was digested with BamHI (B), EcoRI (E), HindIII (H), SacI (S), and KpnI (K), electrophoresed, blotted, and hybridized with Rgp1 (A) or Rgp2 (B) cDNA inserts. Positions and sizes in kb of EcoRI- and HindIII-digested lambda DNA fragments are indicated.
Only RGP1 Shows Self-Glucosylating Activity When Produced in E. coli or in Yeast
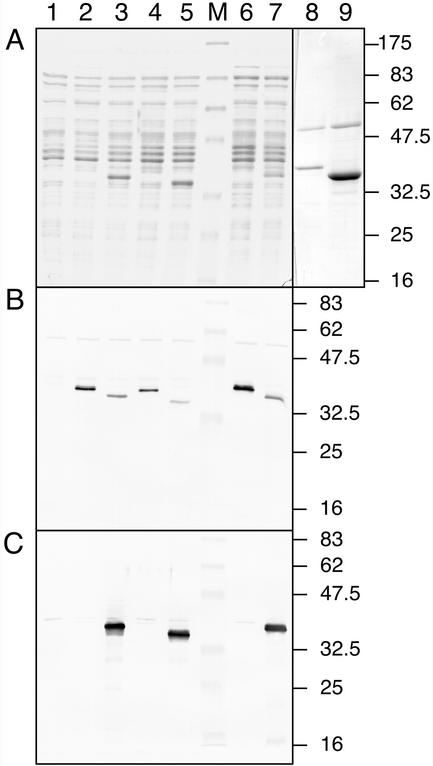
RGPs from several plant species have been shown to react with UDP sugars in an autocatalytic manner (Dhugga et al., 1997; Delgado et al., 1998). To test whether RGP1 and RGP2 from wheat and rice are autocatalytic self-glucosylating proteins, these proteins were separately produced in E. coli using the pET expression system. After induction, crude extracts were analyzed by SDS-PAGE (Fig. 3A) for the presence of the cloned gene products. The soluble extracts from E. coli expressing Rgp1 contain proteins of the correct size, but these were present in very small amounts (Fig. 3A, lanes 2 and 4). An appropriate band was visible in gels loaded with the soluble protein extracts containing RGP2 from wheat or rice (Fig. 3A, lanes 3 and 5, respectively). Large amounts of RGP2 were present in the insoluble fractions, whereas RGP1 was not detected in the insoluble fractions (results not shown).
Figure 3.
RGP1 and RGP2 produced in E. coli. Soluble proteins were isolated from E. coli expressing RGP1 or RGP2 from wheat or rice and separated by SDS-PAGE. Proteins were stained with Coomassie Brilliant Blue (A) or blotted onto nitrocellulose and analyzed for the presence of recombinant protein with anti-RGP1 antiserum (B) or anti-RGP2 antiserum (C). The following protein preparations were used: control extract containing plasmid pET29b (lane 1), extracts expressing wheat RGP1 (lane 2), wheat RGP2 (lane 3), rice RGP1 (lane 4), rice RGP2 (lane 5), wheat His-RGP1 (lane 6), wheat His-RGP2 (lane 7), purified wheat His-RGP1 (lanes 8), and purified wheat His-RGP2 (lane 9). Positions and sizes in kD of prestained molecular mass marker proteins (New England Biolabs, Beverly, MA; lane M) are indicated.
In addition, the extracts were analyzed by western blotting using anti-RGP1 antiserum (Fig. 3B) or anti-RGP2 antiserum (Fig. 3C). A few minor cross-reactive polypeptides were detected in all extracts, including the control extract not expressing RGP1 or RGP2, indicating that the antisera cross-reacted with E. coli proteins. However, the overexpressed RGP1 (Fig. 3B) and RGP2 (Fig. 3C) proteins were clearly detected at the correct mass value. The anti-RGP1 antiserum cross-reacted with the thick RGP2 bands (Fig. 3B, lanes 3, 5, and 7). Subsequently, the extracts were tested for glucosylation activity with UDP-[14C]Glc (Fig. 4). Control extract contained radiolabeled material, but this was running with the bromphenol blue tracking dye and is smaller than the 16.5-kD polypeptide of the molecular mass marker (Fig. 4, lane 1). The extracts containing wheat or rice RGP1 clearly showed a radiolabeled polypeptide of the correct size (Fig. 4, lanes 2 and 4, respectively). In contrast, the extracts containing wheat or rice RGP2 did not show a polypeptide that was labeled by UDP-[14C]Glc (Fig. 4, lanes 3 and 5), although RGP2 was present in considerably larger amounts than was RGP1.
Figure 4.
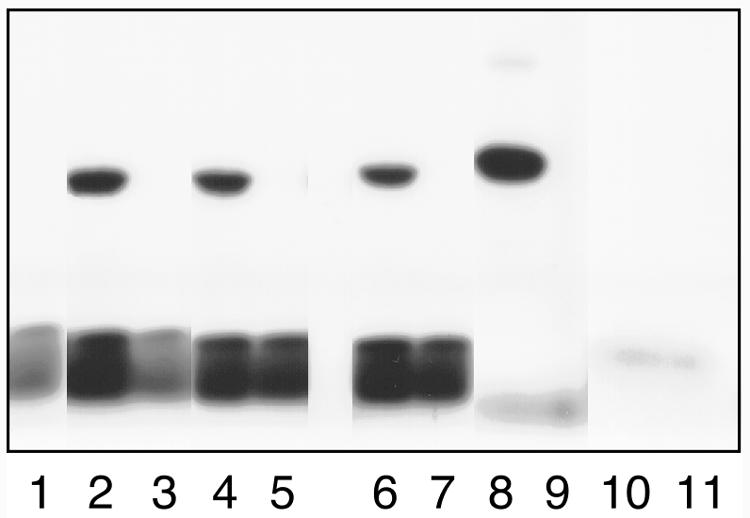
Glucosylation of RGP1 and RGP2 produced in E. coli. For testing of glucosylation activity, soluble proteins were incubated with UDP-[14C]Glc (lanes 1–9) or ADP-[14C]Glc (lanes 10–11) before SDS-PAGE and fluorography. The same protein preparations were used in lanes 1 through 9 as shown in Figure 3. For lanes 10 and 11, purified wheat His-RGP1 and purified wheat His-RGP2 were used, respectively.
To test whether RGP1 is an autocatalytic protein that is not using components from E. coli and that glucosylation of RGP2 was not inhibited by E. coli components, wheat Rgp1 and Rgp2 were expressed as His-tag fusions and purified on His-Bind resin (Novagen, Madison, WI). The extracts were analyzed by SDS-PAGE and western blotting (Fig. 3). Both His-RGP1 and His-RGP2 were present in the soluble extracts. Purification on His-Bind resin resulted in almost pure preparations (Fig. 3A). One additional polypeptide of about 55 kD was detected in both preparations. This polypeptide was not purified from control extract (results not shown). It is probable that it interacted with RGP1 and RGP2 and not with the His-Bind resin. These extracts and purified proteins were used for glucosylation assays. Both the crude extract containing His-RGP1 and the purified His-RGP1 preparation showed self-glucosylation (Fig. 4, lanes 6 and 8). In contrast, glucosylation was not observed with His-RGP2 (Fig. 4, lanes 7 and 9).
Because it is possible that RGP2 uses a substrate other than UDP-Glc, glucosylation assays were performed with ADP-[14C]Glc, the substrate for starch synthases. Crude E. coli control extract incorporated all ADP-[14C]Glc in high-molecular mass material that does not enter the running gel (results not shown). Therefore, purified His-RGP1 and His-RGP2 were used in this assay. Neither protein incorporated ADP-[14C]Glc (Fig. 4, lanes 10 and 11).
To test the possibilities that RGP2 produced in E. coli is not folded correctly, needs to be incorporated in membrane, or should be posttranscriptionally modified for its activity, yeast was used to generate both proteins. Both RGP1 and RGP2 were produced, but only preparations containing RGP1 incorporated UDP-[14C]Glc (results not shown). This suggests that RGP2 does not have glucosylating activity or needs to be expressed in plant cells for its activity. Therefore, we extended the study of RGP activity to plants.
Wheat Endosperm Superose 12 Fractions Show Reversibly Glycosylating Activity
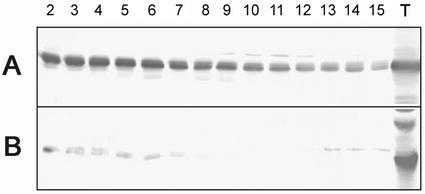
Total protein extract from 12-DPA wheat endosperm did not incorporate radiolabeled UDP-Glc. Because an inhibitor might interfere with RGP activity, total protein extract from wheat endosperm was partly purified on a Superose 12 gel filtration column and eluted fractions were analyzed by western blotting (Fig. 5). RGP1 appeared to be present in all fractions examined, but more abundantly in the high-molecular mass fractions 2 through 6 (Fig. 5, upper). RGP2 was present as a monomer (fractions 13–15) as well as part of a larger complex (Fig. 5, lower). By calibrating the Superose 12 gel filtration column with molecular mass markers, we estimate the RGP complexes to consist of about six subunits. This is consistent with the findings of Bocca et al. (1997), who predicted the RGP1 complex in potato to be a pentamer or a hexamer.
Figure 5.
Western-blot analysis of wheat endosperm fractions obtained by Superose 12 gel filtration. Fractions 2 through 15 and total protein (T) were used. A, Anti-RGP1 antibodies. B, Anti-RGP2 antibodies.
Several fractions of this wheat endosperm extract were tested for glucosylation activity with UDP-[14C]Glc, and they all show a radioactive polypeptide of the correct size (Fig. 6). Chase experiments were performed using pooled fractions 2 through 4 in which unlabeled UDP-Glc (Fig. 7, lane 2), UDP-Xyl (Fig. 7, lane 3), UDP-Gal (Fig. 7, lane 4), or ADP-Glc (Fig. 7, lane 5) was added to the radioactive labeled proteins. The label was chased by each UDP sugar tested, but not by ADP-Glc.
Figure 6.
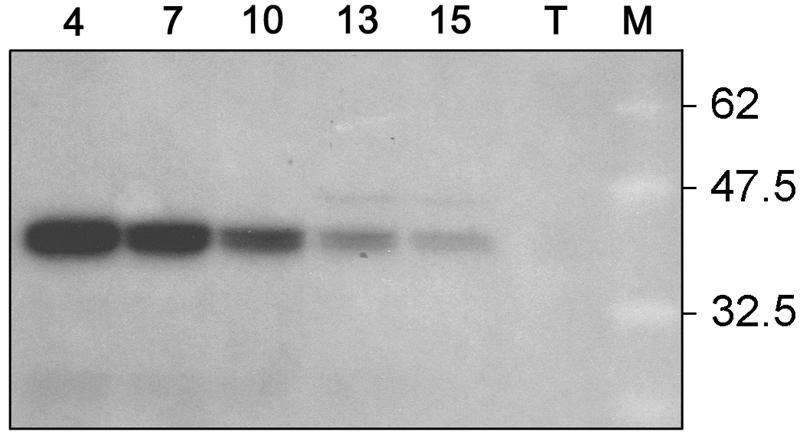
Glucosylation of wheat endosperm fractions 4, 7, 10, 13, and 15 obtained by Superose 12 gel filtration. T, Total protein extract. Positions and sizes in kD of prestained molecular mass marker proteins (New England Biolabs; lane M) are indicated.
Figure 7.
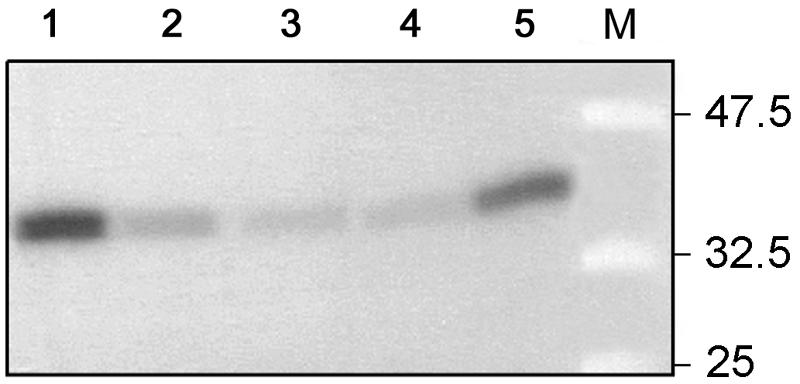
Chase of the glucosylated pooled fractions 2 through 4 of wheat endosperm (lane 1) with unlabeled UDP-Glc (lane 2), UDP-Xyl (lane 3), UDP-Gal (lane 4) or ADP-Glc (lane 5). Positions and sizes in kD of prestained molecular mass marker proteins (New England Biolabs; lane M) are indicated.
RGP2-Containing Complexes Are Active
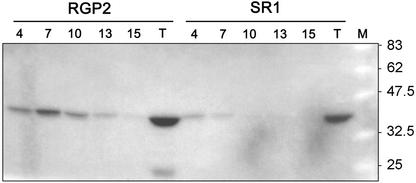
Because both RGP1 and RGP2 are present in wheat endosperm and could not be distinguished by their molecular masses, their separate activities could not be identified. Because in tobacco leaves endogenous RGP2 is not detected (not shown), tobacco was transformed with either wheat Rgp1 or Rgp2. Neither the 13 independent tobacco lines expressing Rgp1 nor the 16 independent tobacco lines expressing Rgp2 showed a phenotype concerning the following characteristics: plant development, growth rate (in Murashige and Skoog medium with different Suc concentrations), and size/shape of starch granules in chloroplasts. Leaf extracts from wild-type SR1-, RGP1-, and RGP2-transformed tobacco plants were partly purified on a Superose 12 gel filtration column and the eluted fractions were analyzed by western blotting (Fig. 8). In tobacco wild-type SR1 plants, RGP1 and RGP2 were not detected (Fig. 8, B and D). In tobacco plants overexpressing Rgp1 or Rgp2, the respective proteins were present in low as well as in high-molecular mass fractions (Fig. 8, A and C). Several fractions containing the RGP2 proteins were subjected to glucosylation tests using UDP-[14C]Glc and compared with similar fractions from SR1 plants (Fig. 9). The low-molecular mass fractions 13 and 15 of RGP2 plants hardly show glucosylation, suggesting that RGP2 monomers are inactive. In contrast, high-molecular mass fractions of RGP2 plants did show glucosylation, whereas only light-radioactive bands were detected in fractions 4 and 7 of the SR1 plants. These results show that RGP2-containing complexes are active in glucosylation.
Figure 8.
Western-blot analysis of tobacco leaf fractions 2 through 15 obtained by Superose 12 gel filtration. Extracts of tobacco plants overexpressing Rgp1 (A; anti-RGP1 antibodies) or Rgp2 (C; anti-RGP2 antibodies) were compared with extract from wild-type SR1 plants, which were challenged with anti-RGP1 antibodies (B) and anti-RGP2 antibodies (D).
Figure 9.
Glucosylation of Superose 12 fractions 4, 7, 10, 13, and 15 and total protein (T) of leaf extracts from tobacco plants overexpressing Rgp2 and from wild-type SR1 tobacco plants.
RGP1 and RGP2 Are Able to Form Homo- and Heterodimers
Because gel filtration experiments with wheat endosperm as well as tobacco leaf extracts suggested the presence of RGP1 and RGP2 in high-molecular mass complexes, our research was extended to complex formation of RGPs and the analysis of these complexes in vivo. The ability of RGP1 and RGP2 to form dimers was analyzed using a yeast two-hybrid system. Specific complex formation of RGPs was studied by expression of RGP1 and RGP2 fusion proteins with the GAL4 DNA binding domain or activation domain in yeast. Table II shows that RGP1 as well as RGP2 are able to form homodimers and heterodimers.
Table II.
Yeast two-hybrid analysis of RGP1 and RGP2 proteins
| Binding Domain | Activation Domain | Growth |
|---|---|---|
| RGP1 | – | – |
| – | RGP1 | – |
| RGP1 | RGP1 | + |
| RGP2 | – | – |
| – | RGP2 | – |
| RGP2 | RGP2 | + |
| RGP1 | RGP2 | + |
| RGP2 | RGP1 | + |
Binding domain of pAS2-1 and activation domain of pACTII were each fused to the coding regions of Rgp1 and Rgp2. Growth was tested in yeast strain CG1945 on plates lacking Trp, Leu, and His.
To reveal whether RGPs interact with other proteins that are possibly part of the high-molecular mass RGP complexes, a wheat endosperm cDNA library was screened using RGP2 as bait in the yeast two-hybrid system. Of the 100,000 colonies tested, 117 were found to be positive when tested for the three different reporter genes. Of the 25 clones, of which expression was dependent on RGP2, nine were identified as RGP1 and three as RGP2. Three clones were identified as prohibitin. Prohibitin has been described to be a part of a complex present in mitochondria (Nijtmans et al., 2000). Because RGP2 and prohibitin are localized in different cellular compartments (not shown), prohibitin was discarded as a possible candidate for the functional RGP2 complex. Ten clones containing 3′ non-coding regions or vector contamination were considered to be artifacts.
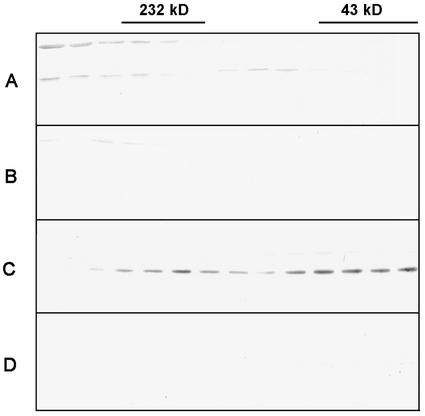
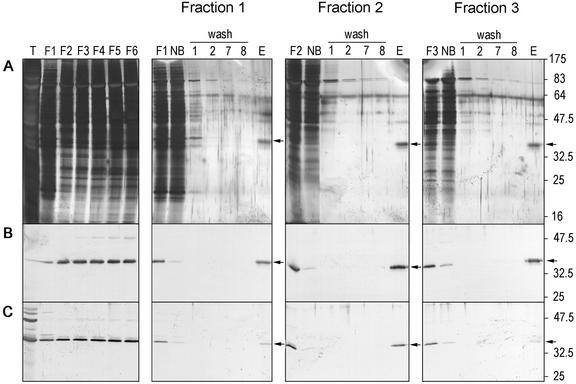
The native wheat complex was isolated using an antibody affinity column. Polyclonal RGP antibodies were covalently linked to immobilized protein A. The columns were tested with RGP1 and RGP2 produced in E. coli (results not shown). Native RGP1 binds to both columns, whereas native RGP2 hardly binds to the RGP1 antibody column. Therefore, the latter column was used to analyze endosperm complexes. The column material was incubated with different Superose 12 endosperm fractions. After several wash steps, the bound proteins were eluted and non-bound, wash, and elution fractions were analyzed by SDS-PAGE and western blotting (Fig. 10). The latter technique showed that RGP1 (Fig. 10B), but also RGP2 proteins (Fig. 10C), were detected in the eluted fractions. The silver-stained gels showed bands of 38 to 40, 47, and 64 kD in the eluted fractions. The 47-kD band binds to the RGP1 antibodies and therefore is not considered to be part of the complex. Because the 64-kD band is present in all lanes, it was considered to be an artifact. The 38- to 40-kD doublet corresponds to RGP1 and RGP2. Taken together, these results suggest that RGP complexes in wheat endosperm do not seem to contain other proteins besides RGP1 and/or RGP2. Because RGP1 and RGP2 are able to form heterodimers in yeast and RGP2 was isolated on an anti-RGP1 column, both proteins are likely to be present in the same complex.
Figure 10.
Affinity purification of the wheat RGP1 complex using anti-RGP1 antibodies covalently linked to protein A-coated beads. A, Silver-stained gel. B and C, Western-blot analysis of several purification steps of Superose 12 fractions 1, 2, and 3, challenged with anti-RGP1 antibodies (B) and anti-RGP2 antibodies (C). T, Total wheat endosperm protein; F1 through F6, Superose 12 fractions 1 through 6; NB, non-bound protein; wash 1, 2, 7, 8, proteins in first two and last two wash steps; E, eluted proteins. Positions and sizes in kD of prestained molecular mass marker proteins (New England Biolabs; lane M) are indicated.
DISCUSSION
To study the function of RGPs, we have isolated RGP-like cDNAs from wheat and rice. Comparison with related sequences from other species showed that two classes of RGP proteins can be distinguished, RGP1 and RGP2. The RGP described by Dhugga et al. (1997) belongs to the first class. Their results suggest that this class is involved in cell wall synthesis. We adopted the working hypothesis that RGP2 has a different function and is possibly involved in initiation of starch biosynthesis. Below we discuss possible functions for RGP1 and RGP2.
The results presented here indicate that both proteins have self-glycosylating activity. However, RGP2 activity could only be detected in transgenic tobacco plants overexpressing RGP2 and not in E. coli or yeast. RGP2 either needs additional plant factors for activity or is not correctly folded in E. coli and yeast. The possibility exists that because low RGP1 activity is present in the wild-type SR1 background, the presence of RGP2 may stimulate the endogenous RGP1 activity. Chase experiments with 12-DPA wheat endosperm showed that glucosylation of RGP is reversible. This is consistent with a role in cell wall synthesis, which confirms the results of Dhugga et al. (1991, 1997). However, this activity is mainly derived from RGP1 because much more RGP1 is present in these endosperm extracts compared with RGP2.
Both RGP1 and RGP2 are present in high-molecular mass fractions. The results of the yeast two-hybrid experiments and the antibody columns suggest that the high-molecular mass complexes contain both RGP1 and RGP2. RGP proteins only seem to be active when present in a complex. This is in accordance with the results from Alonso et al. (1995b), who suggested that glycogenin is active as a dimer in which one subunit may glucosylate the other. The mixed complexes could point at a similar function for both proteins. Alternatively, the composition of the RGP multimer complexes may influence their function: Relatively more RGP1 results in a certain activity and relatively more RGP2 in another one.
RGP proteins have partial homology with cellulose synthases and other β-glycosyltransferases, suggesting that RGPs may be non-processive β-glycosyltransferases, functioning as intermediates in the transfer of a single sugar residue from a nucleotide sugar to an acceptor molecule (Saxena and Brown, 1999). Using hydrophobic cluster analysis (Gaboriaud et al., 1987), processive β-glycosyltransferases were shown to contain two conserved domains, designated domains A and B, whereas non-processive glycosyltransferases carry only domain A (Saxena et al., 1995). Domain A usually consists of alternating α-helices with β-strands, the latter containing the DXD motif. This motif, consisting of two aspartic acids flanked by hydrophobic residues, seems to be conserved among RGPs (Fig. 1). A diverse range of glycosyltransferase families was shown to contain this motif, although in some families the second Asp is lacking (Wiggins and Munro, 1998). In addition to domain A, processive β-glycosyltransferases contain domain B, a single conserved Asp together with the motif QXXRW, although the Q is less conserved (Saxena et al., 1995). This motif may be required for activity of processive β-glycosyltransferases (Kamst and Spaink, 1999). RGP1 does not contain this motif, as was observed by Saxena and Brown (1999). However, RGP2 as well as glycogenin contain an RW motif. Because glycogenin is known to be a processive glucosyltransferase, this suggests that RGP2 also might be able to transfer more than one Glc moiety to an acceptor molecule. Furthermore, this indicates that RGP1 and RGP2 may have different functions in plants.
RGP proteins apparently do not contain membrane-spanning domains (Von Heijne, 1994) or N-terminal protein-sorting signals (Claros et al., 1997). This means that RGP proteins are not imbedded in membranes or translocated into plastids. Subcellular localization experiments confirm that RGPs are not transported into amyloplasts (results not shown). Therefore, it is unlikely that RGP2 has functional homology with glycogenin. This primer of glycogen synthesis remains attached to glycogen. Because of the observed similarities of RGP2 with NodC, which is a chitin oligosaccharide synthase (Kamst and Spaink, 1999), it is tempting to speculate about the priming activity of RGP2 in polysaccharide synthesis by generating oligosaccharides. However, a mechanism for transporting the oligosaccharides into the amyloplast is unknown.
A protein inhibiting glycosylating activity in maize endosperm was identified with high homology with Suc synthase (Rothschild et al., 1996; Wald et al., 1998). In our glycosylation experiments, total protein extracts of wheat endosperm also did not show activity, suggesting the presence of an inhibitor in our wheat endosperm extracts. Two isozymes of Suc synthase have been identified in maize endosperm, one involved in cell wall integrity and the other in starch biosynthesis (Choury et al., 1998). Either one of the isozymes possibly interact with one of the RGP protein classes. However, neither affinity purification of the complex nor yeast two-hybrid analysis did reveal the presence of the 80-kD Suc synthase-like inhibitor protein. If a physical interaction between Suc synthase and RGP exists, we consider this interaction to be weak. Alternatively, it is possible that not the Suc synthase itself but the produced unlabeled UDP-Glc is the cause of the “inhibitory effect” observed in maize endosperm. UDP-Glc levels of 0.425 μmol g fresh weight−1 have been measured in wheat endosperm (Beckles et al., 2001), suggesting that the “lack” of glucosylation may be caused by large amounts of unlabeled UDP-Glc. In contrast, in tobacco leaves a concentration of 7.4 μmol m−2 has been reported (Herbers et al., 1997) that, in our calculation, equals about 25 nmol g fresh weight−1. The small amount of endogenous UDP-Glc therefore may explain the glucosylation observed using total extract from tobacco.
In conclusion, it seems unlikely that RGP2 functions as a primer for starch synthesis in a way similar to the functioning of glycogenin as a primer for glycogen synthesis. The sequence analysis suggests that RGP1 and RGP2 have different functions. This is corroborated by the observation that their expression is differentially induced by several Glc analogs (results not shown). Our present results are more consistent with a role of both proteins in a common process, such as biosynthesis of components of the extracellular matrix, to which RGP1 and RGP2 contribute in a different way. Continuing research is needed to provide an adequate understanding of the functions of RGPs. In this regard, the manipulation of RGP expression levels in starch-storing crops may reveal the involvement of RGPs in polysaccharide synthesis in plants.
MATERIALS AND METHODS
cDNA Cloning
Rgp probes were obtained by reverse transcriptase-PCR using endosperm RNA isolated 10 DPA from wheat (Triticum aestivum cv Minaret). After denaturing 10 μg of total RNA for 10 min at 60°C, cDNA was made in a final volume of 10 μL containing 1 mm dNTPs, 2 mm T20, 200 units of Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) in PCR buffer for 1 h at 37°C. Rgp-homologous sequences were amplified after addition of 15 μL of 1.6 mm SP76 (GCGCCTCGAGCTGACTTTGTCCGTGGTTACC) and 1.6 mm SP90 (GCCACAGGCCGTGAG) for Rgp1, or 1.6 mm SP76 (GCGCCTCGAGCTGACTTTGTCCGTGGTTACC) and 1.6 mm SP79 (GGCCGAGCTCATCACAGCATCCACATAC) primers for Rgp2 and 2.5 units of Taq DNA polymerase in PCR buffer during 35 cycles of 1 min at 94°C, 2 min at 60°C, and 1 min at 72°C. The resulting fragments were labeled in a second PCR reaction containing 10 μm dCTP (0.5 mCi [32P]dCTP) in 200 μL, and used for screening a cDNA library in λ-uni-ZAP XR (Stratagene, La Jolla, CA) made on poly(A+) RNA from 10-DPA wheat endosperm. Lambda phages (50,000 plaque-forming units/150-mm plate) were grown on XL1-Blue MRF′ for 8 h at 37°C. Plaques were lifted on Hybond N+ filters (Amersham Pharmacia Biotech, Uppsala), DNA was denatured in 0.5 m NaOH and 1.5 m NaCl, and the filters were neutralized in 0.5 m Tris/HCl (pH 7.5), 1.5 m NaCl, and washed in 2× SSPE (20× SSPE: 3.6 m NaCl; 0.2 m NaH2PO4, pH 6.5; and 20 mm EDTA). Subsequently, the filters were exposed to UV light for 2 min, prehybridized, hybridized, and washed as described previously (Memelink et al., 1994). The resulting positive plaques were purified by a second and third screening.
To obtain full-length rice (Oryza sativa) Rgp1 and Rgp2 cDNA clones, cDNA libraries from etiolated shoot (Meijer et al., 1997) and 7-d-old somatic embryo (Postma-Haarsma et al., 1999) were screened with rice ESTs S5091 and C2546, respectively.
Sequence Analysis
After in vivo excision, the cDNA inserts were sequenced from the 5′ and 3′ ends with the T7 sequencing kit (Amersham Pharmacia Biotech). The complete sequences of the clones with the longest inserts were determined and analyzed with a VAX computer using the Genetics Computer Group Sequence Analysis Software Package (Genetics Computer Group, 1994). Sequence comparisons were performed using BLAST (Tatusova and Madden, 1999).
Plant Material
Wheat plants were grown as described by Langeveld et al. (2000).
Blotting and Hybridization
For genomic Southern-blot analysis, 10 μg of DNA was digested, electrophoresed on a 0.8% (w/v) agarose gel, blotted, and hybridized as described (Memelink et al., 1994). As probes, randomly labeled cDNA inserts were used. Blots were washed with 0.1× SSPE and 0.1% (w/v) SDS at 65°C.
Constructs and Expression in Escherichia coli
For cloning of wheat and rice Rgp coding regions in pET29b and pET16b (Novagen), the sequences 5′ of the ATG start codons were modified by PCR to obtain NdeI restriction sites. The following primers were used for PCR reactions: SP91 (sense; GATCGGTACCATATGGCAGGGACGGTGAC) and SP90 (antisense; GCCACAGGCCGTGAG) for wheat Rgp1, SP80 (sense; CCGGCCATGGAGTAACATATGTCTTTGGAGGTTCAC) and SP79 (antisense; GGCCGAGCTCATCACAGCATCCACATAC) for wheat Rgp2, SP91 and SP79 for rice Rg1p, and SP93 (sense; CCGGCATATGTCTTTGGAAATTCAGG) and SP79 for rice. The 5′ region of wheat Rgp1 was cloned in pET29b as NdeI-SacI PCR fragment. The 3′ region was cloned into the resulting plasmid as XhoI fragment. A His-tag fusion was constructed by cloning the NdeI-SacI fragment in pET16h that is a derivative of pET16b containing the BamHI-BglII polylinker fragment of pIC20H in the BamHI site. The coding region was completed by addition of the 3′ SacI-KpnI Rgp1 fragment. The 5′ region of wheat Rgp2 was cloned as NdeI-KpnI PCR fragment in pET29b and the 3′ region as KpnI-XhoI fragment. The NdeI-XhoI complete coding region was cloned in pET16b to construct a His-tag fusion. The 5′ region of rice Rgp1 was cloned in pET29b as NdeI-BamHI PCR fragment and the 3′ region as BamHI-XhoI fragment. The 5′ region of rice Rgp2 was cloned as NdeI PCR fragment in pET29b and the 3′ region as KpnI fragment. Sequence analysis confirmed that no mutations had been introduced during the PCR reactions. The His-tag of pET29b was not fused to the Rgp coding regions because the stop codons of the introduced coding regions were not removed.
The resulting plasmids were introduced into E. coli CGSC4954(DE3) (Alonso et al., 1994). For protein production, overnight cultures grown in Luria-Bertani medium containing 50 μg of kanamycin (pET29b derivatives) or 100 μg of ampicillin (pET16b derivatives) were diluted 20 times in the same growth media and incubated with shaking at 37°C until the optical density at 600 nm reached 0.6. Isopropylthio-β-galactoside was added to a final concentration of 1 mm and incubation was continued for 3 h. Cells were harvested by centrifugation, washed with 50 mm Tris/HCl (pH 7.5), and resuspended in one-tenth of the culture volume of cold 50 mm Tris/HCl (pH 7.5) containing Complete proteinase inhibitor cocktail (Boehringer Mannheim/Roche, Basel). Cells were sonicated two times for 4 s and centrifuged 10 min at 4°C. Protein concentrations in supernatants were determined by the method of Bradford (1976).
Protein Purification and Antibody Production
His-tagged RGP1 and RGP2 were expressed in E. coli GCSC4954(DE3) for activity tests and in E. coli Bl21(DE3) pLysS for antibody production. Fusion proteins were affinity purified on His-Bind resin according to the manufacturers instructions. For antibody production, His-tagged proteins were further purified by SDS-PAGE. The desired band was cut from the gel and electro-eluted as described (Hunkapiller et al., 1983). This preparation was then used to raise antibodies in a rabbit (performed by Eurogentec, Seraing, Belgium).
Protein Analysis
All experiments regarding protein analysis were performed at least in duplo with independent samples.
Crude protein extracts were analyzed by SDS-PAGE followed by staining with Coomassie Brilliant Blue or by western blotting as described (Memelink et al., 1994).
Glucosylation activity was tested with 40 to 60 μg of protein extract or 1 to 5 μg of purified protein and 2 μm UDP-[14C]Glc or ADP-[14C]Glc as a substrate in 50 mm Tris/HCl, pH 7.5, and 5 mm MnCl2 for 30 min at 30°C. The chase experiments included an additional 30-min incubation with 2 mm UDP-Glc, UDP-Xyl, UDP-Gal, or ADP-Glc. After separation of the mixtures by SDS-PAGE, gels were fixed in 25% (v/v) isopropanol and 10% (v/v) acetic acid, soaked in Amplify (Amersham Pharmacia Biotech), dried, and exposed for fluorography.
Superose 12 Gel Filtration
Total protein was extracted from 250 mg of young, in vitro-grown, tobacco (Nicotiana tabacum) leaves or 12-DPA wheat endosperm in 500 μL of buffer (50 mm Tris/HCl, pH 7.5; 2 mm EDTA; and 0.2 mm phenylmethylsulfonyl fluoride) containing Complete proteinase inhibitor cocktail (Boehringer Mannheim/Roche) by mechanical disruption of the tissue on ice using a potter device. Extracts were centrifuged two times for 5 min at 4°C, the second time through a 0.22-μm filter.
Total protein extract (200 μL) was fractionated on a Superose 12 HR 10/30 column at a flow rate of 0.5 mL min−1 using 50 mm Tris/HCl, pH 7.5, and 2 mm EDTA, generating 200-μL fractions. Fractions were taken after 20 min. Aliquots of each fraction (20 μL) were analyzed by western blotting and compared with 5 μL of the total protein extract. The column was calibrated with a molecular mass marker mixture containing ferritin (440 kD), catalase (232 kD), aldolase (167 kD), and bovine serum albumin (67 kD).
Tobacco Transformation
For tobacco transformation, a BamHI-EcoRV fragment containing the coding region of Rgp1 and a SalI-HindIII fragment encoding Rgp2 were separately cloned in vectors containing the double 35S promoter (van der Fits and Memelink, 1997) and the RbcS-3C terminator. The vectors were transferred to Agrobacterium tumefaciens strain MOG101 (Hood et al., 1993). Tobacco cv Petit Havana SR1 was transformed with the MOG101 derivatives by the leaf disc transformation method (Horsch et al., 1985).
Yeast Two Hybrid
Wheat Rgp1 and Rgp2 were separately cloned in pAS2-1 containing the GAL 4 DNA binding domain and in pACTII, containing the GAL 4 activation domain (CLONTECH Laboratories, Palo Alto, CA). pET16h-RGP1 was digested with NdeI. For cloning in pACTII, blunt ends were produced using the Klenow fragment of DNA polymerase I. The linear plasmids were digested with BamHI and the Rgp1 encoding cDNA fragments were cloned in pAS2-1 and pACTII digested with NdeI-BamHI or SmaI-BamHI, respectively. An NcoI site was introduced at the ATG of Rgp2 by PCR and the stop codon was replaced by an NcoI site using a linker. The NcoI fragment was cloned into the NcoI sites of pAS2-1 and pACTII. The orientations were checked by restriction analysis.
pAS2-1-RGP1 or pAS2-1-RGP2 and pACTII-RGP1 or pACTII-RGP2 were introduced in yeast (Saccharomyces cerevisiae) strain CG1945 according to Gietz et al. (1992). The transformed yeast was tested for growth on plates lacking Trp, Leu, and His. Positive colonies were transferred to plates lacking Leu and containing cycloheximide. Loss of the pAS2-1 vector was checked by growing the strains on plates lacking Leu and Trp, Leu, and His, and Leu only. Colonies only growing on the latter were retransformed with pAS2-1-RGP1 or pAS2-1-RGP2 to test whether growth was dependent on RGP1 or RGP2, respectively.
To isolate other proteins that bind to RGP2, a 10-DPA endosperm cDNA library was cloned into λACTII (Memelink, 1997) using the EcoRI and XhoI restriction sites. Screening of the cDNA library was performed in the yeast strain PJ69-4a (James et al., 1996).
Affinity Purification
An ImmunoPure IgG Orientation Kit (Pierce Chemical, Rockford, IL) was used to covalently bind anti-RGP1 or anti-RGP2 antibodies to protein A-coated beads. Superose 12 fractions (600 μL) were incubated overnight at 4°C with 100-μL beads in buffer (20 mm Tris/HCl, pH 7.5; 150 mm NaCl; and 0.5% [v/v] NP40) containing Complete proteinase inhibitor cocktail (Boehringer Mannheim/Roche) and 0.8 mm phenylmethylsulfonyl fluoride under constant inversion. Beads were consecutively washed six times with 1 mL of buffer and two times with 0.2 mL of buffer. Bound proteins were eluted with 200 μL of 0.1 m Gly, pH 2.8.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor. The amount of RGP1 and RGP2 antibodies is limited.
ACKNOWLEDGMENTS
We thank Dr. William J. Whelan (University of Miami School of Medicine, Miami) and his lab members for their hospitality and advice on the activity tests.
Footnotes
This work was partly financially supported by the European Union Eureka Program (grant no. EU–169311).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010720.
LITERATURE CITED
- Alonso MD, Lomako J, Lomako WM, Whelan WJ. A new look at the biogenesis of glycogen. FASEB J. 1995a;9:1126–1137. doi: 10.1096/fasebj.9.12.7672505. [DOI] [PubMed] [Google Scholar]
- Alonso MD, Lomako J, Lomako WM, Whelan WJ. Catalytic activities of glycogenin additional to autocatalytic self-glucosylation. J Biol Chem. 1995b;270:15315–15319. doi: 10.1074/jbc.270.25.15315. [DOI] [PubMed] [Google Scholar]
- Alonso MD, Lomako J, Lomako WM, Whelan WJ, Preiss J. Properties of carbohydrate-free recombinant glycogenin expressed in an Escherichia coli mutant lacking UDP-glucose pyrophosphorylase activity. FEBS Lett. 1994;352:222–226. doi: 10.1016/0014-5793(94)00962-7. [DOI] [PubMed] [Google Scholar]
- Beckles DM, Smith AM, ap Rees T. A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol. 2001;125:818–827. doi: 10.1104/pp.125.2.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocca SN, Kissen R, Rojas-Beltrán JA, Noël F, Gebhardt C, Moreno S, du Jardin P, Tandecarz JS. Molecular cloning and characterization of the enzyme UDP-glucose:protein transglucosylase from potato. Plant Physiol Biochem. 1999;37:809–819. doi: 10.1016/s0981-9428(99)00117-5. [DOI] [PubMed] [Google Scholar]
- Bocca SN, Rothschild A, Tandecarz JS. Initiation of starch biosynthesis: purification and characterization of UDP-glucose:protein transglucosylase from potato tubers. Plant Physiol Biochem. 1997;35:205–212. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Camirand A, Maclachlan GA. Differential distribution of xyloglucan glycosyl transferases in pea Golgi dictyosome and secretory vesicles. J Cell Sci. 1990;96:705–710. [Google Scholar]
- Choury PS, Taliercio EW, Carlson SJ, Ruan Y-L. Genetic evidence that two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet. 1998;259:88–96. doi: 10.1007/s004380050792. [DOI] [PubMed] [Google Scholar]
- Claros MG, Brunak S, von Heijne G. Prediction of N-terminal protein sorting signals. Curr Opin Struct Biol. 1997;7:394–398. doi: 10.1016/s0959-440x(97)80057-7. [DOI] [PubMed] [Google Scholar]
- Darvill AG, McNeil M, Albersheim P, Delmer DP. The primary cell walls of flowering plants. In: Tolbert N, editor. The Biochemistry of Plants. Vol. 1. New York: Academic Press; 1980. pp. 91–162. [Google Scholar]
- Delgado IJ, Wang Z, de Rocher A, Keegstra K, Raikhel NV. Cloning and characterization of AtRGP1. A reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol. 1998;116:1339–1350. doi: 10.1104/pp.116.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM. A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA. 1997;94:7679–7684. doi: 10.1073/pnas.94.14.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Ulvskov P, Gallagher SR, Ray PM. Plant polypeptides reversibly glycosylated by UDP-glucose. Possible components of Golgi beta-glucan synthase in pea cells. J Biol Chem. 1991;266:21977–21984. [PubMed] [Google Scholar]
- Driouich A, Faye L, Staehelin LA. The plant Golgi apparatus: a factory for complex polysaccharides and glycoproteins. Trends Biochem Sci. 1993;18:210–214. doi: 10.1016/0968-0004(93)90191-o. [DOI] [PubMed] [Google Scholar]
- Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyze amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Genetics Computer Group. Program Manual for Wisconsin Package, Version 8. Madison, WI: Genetics Computer Group; 1994. [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:139–168. [Google Scholar]
- Herbers K, Tacke E, Hazirezaei M, Krause K-P, Melzer M, Rohde W, Sonnewald U. Expression of a luteoviral movement protein in transgenic plants leads to carbohydrate accumulation and reduced photosynthetic capacity in source leaves. Plant J. 1997;12:1045–1056. doi: 10.1046/j.1365-313x.1997.12051045.x. [DOI] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgen Res. 1993;2:208–218. [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Hunkapiller MW, Lujan E, Ostrander F, Hood LE. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamst E, Spaink HP. Functional domains in the chitin oligosaccharide synthase NodC and related β-polysaccharide synthases. Trends Glycosci Glycotechnol. 1999;11:187–199. [Google Scholar]
- Langeveld SMJ, van Wijk R, Stuurman N, Kijne JW, de Pater S. B-type granule containing protrusions and interconnections between amyloplasts in developing wheat endosperm revealed by transmission electron microscopy and GFP expression. J Exp Bot. 2000;51:1357–1361. [PubMed] [Google Scholar]
- Lavintman N, Cardini CE. Particulate UDP-glucose:protein transglucosylase from potato tuber. FEBS Lett. 1973;29:43–46. doi: 10.1016/0014-5793(73)80011-0. [DOI] [PubMed] [Google Scholar]
- Lomako J, Lomako WM, Whelan WJ. A self-glucosylating protein is the primer for rabbit muscle glycogen biosynthesis. FASEB J. 1988;2:3097–3103. doi: 10.1096/fasebj.2.15.2973423. ; erratum Lomako J, Lomako WM, Whelan WJ (1989) FASEB J 3: 1873. [DOI] [PubMed] [Google Scholar]
- Meijer AH, Scarpella E, van Dijk EL, Qin L, Taal AJ, Rueb S, Harrington SE, McCouch SR, Schilperoort RA, Hoge JH. Transcriptional repression by Oshox1, a novel homeodomain leucine zipper protein from rice. Plant J. 1997;11:263–276. doi: 10.1046/j.1365-313x.1997.11020263.x. [DOI] [PubMed] [Google Scholar]
- Memelink J. Two yeast/Escherichia coli Lambda/plasmid vectors designed for yeast one-and two-hybrid screens that allow directional cDNA cloning. Technical Tips Online. 1997;1:TO1111. [Google Scholar]
- Memelink J, Swords KMM, Staehelin LA, Hoge JHC. Southern, northern and western blot analysis. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. F1–F23. [Google Scholar]
- Moreno S, Cardini CE, Tandecarz JS. α-glucan synthesis on a protein primer. A reconstituted system for the formation of protein-bound α-glucan. Eur J Biochem. 1987;162:609–614. doi: 10.1111/j.1432-1033.1987.tb10682.x. [DOI] [PubMed] [Google Scholar]
- Nijtmans LGJ, de Jong L, Sanz MA, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma-Haarsma AD, Verwoert II, Stronk OP, Koster J, Lamers GE, Hoge JH, Meijer AH. Characterization of the KNOX class homeobox genes Oskn2 and Oskn3 identified in a collection of cDNA libraries covering the early stages of rice embryogenesis. Plant Mol Biol. 1999;39:257–271. doi: 10.1023/a:1006153506868. [DOI] [PubMed] [Google Scholar]
- Rothschild A, Wald FA, Bocca SN, Tandecarz JS. Inhibition of UDP-glucose: protein transglucosylase by a maize endosperm protein factor. Cell Mol Biol. 1996;42:645–651. [PubMed] [Google Scholar]
- Saxena IM, Brown RM., Jr Are the reversibly glycosylated polypeptides implicated in plant cell wall biosynthesis non-processive β-glycosyltransferases? Trends Plant Sci. 1999;4:6–7. doi: 10.1016/s1360-1385(98)01358-2. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM, Jr, Fevre M, Geremia RA, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DG, Lomako J, Lomako WM, Whelan WJ, Meyer HE, Serwe M, Metzger JW. β-Glucosylarginine: a new glucose-protein bond in a self-glucosylating protein from sweet corn. FEBS Lett. 1995;376:61–64. doi: 10.1016/0014-5793(95)01247-6. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Moore I. The plant Golgi apparatus: structure, functional organization and trafficking mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:261–288. [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. ; erratum Tatusova TA, Madden TL (1999) FEMS Microbiol Lett 177:187–188. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. Comparison of the activities of CaMV 35S and FMV 34S promoter derivatives in Catharanthus roseus cells transiently and stably transformed by particle bombardment. Plant Mol Biol. 1997;33:943–946. doi: 10.1023/a:1005763605355. [DOI] [PubMed] [Google Scholar]
- Von Heijne G. Membrane proteins: from sequence to structure. Annu Rev Biophys Biomol Struct. 1994;23:167–192. doi: 10.1146/annurev.bb.23.060194.001123. [DOI] [PubMed] [Google Scholar]
- Wald FA, Rothschild A, Moreno S, Tandecarz JS. Identification of a UPTG inhibitor protein from maize endosperm: high homology with sucrose synthase protein. Cell Mol Biol. 1998;44:397–406. [PubMed] [Google Scholar]
- Wiggins CA, Munro S. Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]