Abstract
Previous studies showed that exposure of C3H10T1/2 stem cells to bone morphogenetic protein-4 (BMP-4) produced cells that convert into adipocytes at high frequency when treated with differentiation inducers. In the present investigation, an independent approach shows that BMP-4 is required for stable commitment of pluripotent stem cells to the adipocyte lineage. Exposure of proliferating 10T1/2 stem cells to 5-azacytidine, a potent DNA methylation inhibitor, gave rise to a subpopulation of cells that can be cloned and that have the capacity to undergo conversion into adipocytes upon treatment with terminal differentiation inducers. Detailed studies performed with a cloned committed subline, the A33 line, verified stable adipocyte lineage determination in the absence of exogenous BMP-4. Remarkably, this cell line expresses and secretes BMP-4 during proliferation in the same time window that exogenous BMP-4 must be added to naïve 10T1/2 cells to induce maximal adipocyte commitment. Furthermore, exposure of A33 cells to noggin, a naturally occurring BMP-4–binding antagonist, during this critical time window blocks subsequent differentiation. The role of BMP-4 in adipocyte lineage commitment is further strengthened by gene expression profiling of proliferating 10T1/2 stem cells and A33 preadipocytes. These findings revealed changes in the molecular circuitry, specifically coordinated changes in the expression of members of the BMP-4 signaling pathway, that distinguish A33 preadipocytes from uncommitted parental 10T1/2 stem cells. Together, these studies provide compelling evidence for the participation of BMP-4 in adipocyte lineage determination.
Keywords: 5-azacytidine, adipogenesis, determination, mesenchymal stem cell, preadipocyte
Obesity results from increases in both fat cell size and number (1, 2). During enlargement of adipose tissue depots, adipocytes develop from preadipocytes that are recruited from pluripotent mesenchymal stem cells (MSCs) in the vascular stroma of the tissue. MSCs also have the potential to develop into other mesodermal cell types, including myoblasts, chondroblasts, and osteoblasts. An understanding of the mechanisms that govern stem cell lineage determination should aid efforts to direct or prevent the development of stem cells into specific cell types for therapeutic uses.
The development of mesenchymal stem cells into terminally differentiated adipocytes can be divided into four stages: lineage commitment, preadipocyte proliferation, growth arrest, and terminal differentiation. Adipose lineage commitment results from a combinatorial signal that recruits stem cells to become preadipocytes. In this process, asymmetric cell division occurs, in which one daughter cell becomes a lineage-restricted preadipocyte, whereas the other daughter cell remains pluripotent to perpetuate the stem cell population. Proliferation of committed preadipocytes amplifies the preadipocyte population until space constraints result in growth arrest. Finally, terminal differentiation of growth-arrested preadipocytes into mature adipocytes is induced by a second combinatorial signal, and mitotic clonal expansion further amplifies the population of incipient adipocytes. Thus, commitment of a single stem cell can ultimately result in the production of large numbers of mature adipocytes. Further, angiogenesis and neurogenesis are coordinated with adipogenesis to provide a blood supply and innervation to allow communication with the central nervous system.
Although 3T3 preadipocyte cell lines (notably the 3T3-L1 and 3T3-F442A lines) have provided an excellent model cell system with which to elucidate the terminal adipocyte differentiation program (3–9), a more primitive cell line should be useful in unraveling earlier steps in adipocyte development. The C3H10T1/2 stem cell line derived from C3H mouse embryos (3) has characteristics that suit it well for studies on the stem cell commitment program. When dividing 10T1/2 cells are exposed to 5-azacytidine (5-azaC), a potent DNA methylation inhibitor, they undergo commitment to new cell phenotypes, specifically chondrocytes, myocytes, and adipocytes (10). Blocking DNA methylation with 5-azaC is thought to induce stable expression of one or a few genes responsible for lineage commitment (10–12). Several lines of evidence support this hypothesis (12). By transfecting cDNA from 5-azaC-committed myoblasts or preadipocytes into uncommitted stem cells, it was shown that myocyte (13) or adipocyte (14) commitment could be induced. Indeed, the first transcription factor shown to be crucial for myoblast lineage determination, i.e., MyoD, was identified in such a genetic screen using 10T1/2 cells and myoblasts (13). Although analogous preadipocyte cell lines have been isolated after 5-azaC treatment of 10T1/2 cells (11, 15), to date no analogous “preadipocyte commitment genes” have been identified.
Bone morphogenetic proteins (BMPs) are members of the TGF-β superfamily. Previously, administration of exogenous BMP-4 to 10T1/2 cells was shown to induce commitment to the adipocyte lineage (16, 17). Specifically, when 10T1/2 cells were treated with recombinant BMP-4 during proliferation and upon reaching confluence, subsequent treatment with differentiation inducers resulted in terminal adipocyte differentiation (17). Although levels of 50 or 100 ng/ml BMP-4 resulted in the differentiation of virtually all 10T1/2 cells into adipocytes, a lower level of BMP-4 (10 ng/ml) was completely ineffective (17). Thus, a threshold of BMP-4 must be surmounted to induce adipocyte lineage commitment of 10T1/2 cells.
During dorsoventral patterning in both invertebrates and vertebrates, a gradient of BMP-4 signaling encodes positional information. This morphogen gradient is established in the extracellular space through the combined actions of several secreted proteins. Both noggin and chordin bind to BMP-4 with high affinities and prevent BMP-4 from binding to its cognate cell surface receptors. However, the inhibition of BMP signaling by chordin, but not noggin, can be reversed by xolloid/tolloid proteases. Further, twisted gastrulation (twsg) can have a negative or a positive effect on BMP signaling. Initially, twsg promotes the formation of inactive BMP-4–chordin–twsg ternary complexes. However, in the presence of twsg, chordin becomes a better substrate for BMP-1 (tolloid homologue), and as a result, twsg positively affects BMP signaling by promoting the degradation of an inhibitor (18). Thus, BMP-1 functions as a molecular switch that converts inactive BMP complexes into active complexes (19). Although BMP-4 was shown to induce adipocyte commitment of pluripotent stem cells, the other extracellular components of the BMP-4 network have not previously been examined in the context of adipogenesis, several of which are investigated in this paper.
In the present study, 10T1/2 cells and a committed preadipocyte cell line, the A33 line, derived from 10T1/2 cells after 5-azaC treatment were used to further elucidate the role of BMP-4 in the commitment of pluripotent 10T1/2 stem cells to the adipocyte lineage. Here we show that proliferating A33 cells express BMP-4 mRNA and secrete BMP-4, whereas parental 10T1/2 cells express little BMP-4 during proliferation. This finding is remarkable, given that A33 cells had not previously been exposed to exogenous BMP-4.
Results and Discussion
Derivation of the Lineage-Committed A33 Cell Line from 10T1/2 Stem Cells.
To investigate the genetic events underlying adipocyte commitment, 10T1/2 stem cells were treated with 5-azaC at low/cloning density, and foci were selected for their ability to produce adipocytes when treated with differentiation inducers. Of the clonal lines of preadipocytes generated, the A33 line exhibited the greatest propensity to differentiate into adipocytes and was further characterized. In the experiments described below, preadipocyte characteristics of A33 cells were compared to those of the parental 10T1/2 cells.
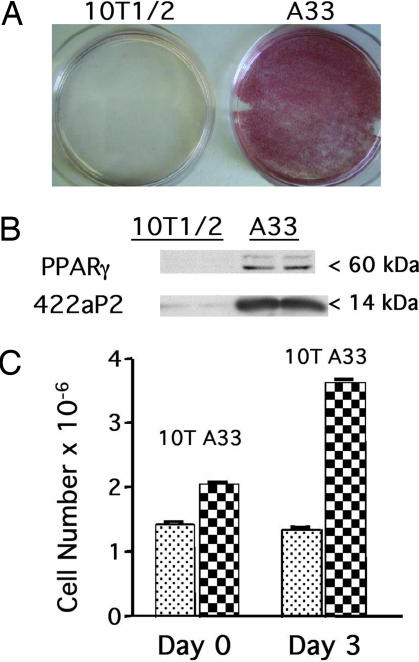
To verify that A33 cells are bona fide preadipocytes, A33 and 10T1/2 stem cells were plated at low density, allowed to proliferate until confluent/growth arrested, and then induced to differentiate using our standard differentiation protocol (20, 21). Staining with Oil red O 7 days later revealed that A33 cells had accumulated cytoplasmic fat, whereas the parental 10T1/2 stem cells had not (Fig. 1A). Immunoblotting of cell extracts prepared 6 days after exposure to differentiation inducers showed that markers of terminal adipocyte differentiation, i.e., peroxisome proliferator-activated receptor (PPARγ) and 422/aP2, were expressed by A33 but not by 10T1/2 cells (Fig. 1B). Further, A33 but not 10T1/2 cells underwent mitotic clonal expansion, an established prerequisite for adipogenesis (7), exhibiting an ≈2-fold increase in cell number after induction of differentiation (Fig. 1C).
Fig. 1.
Adipocyte characteristics of A33 cells. (A) Oil red O staining of cytoplasmic triacylglycerols in 10T1/2 and A33 cells 7 days after addition of differentiation inducers. (B) Immunoblotting of the adipocyte markers PPARγ and 422/aP2 from whole-cell extracts of 10T1/2 and A33 cells 6 days after induction. (C) Cell number of 10T1/2 and A33 cells before and 3 days after induction.
Additional evidence that A33 cells are preadipocyte derivatives of 10T1/2 cells was provided by quantitative real-time PCR (qPCR) analysis of the adipocyte marker PPARγ2 (Table 1). Although PPARγ is known to be expressed by terminally differentiated adipocytes, it is also expressed, albeit at a very low level by A33 but not 10T1/2 cells before induction of terminal differentiation. After induction of differentiation, however, A33 cells express a much higher level of PPARγ2 mRNA. The low level of expression of PPARγ2 mRNA by 10T1/2 cells appears to reflect the low level (≤1%) of spontaneous differentiation that normally occurs with parental 10T1/2 cells treated with differentiation inducers. Thus, the expression of PPARγ by A33 preadipocytes is initially low, but increases dramatically upon induction of differentiation. In contrast, uncommitted 10T1/2 stem cells, neither differentiate (Fig. 1 A–C) nor express significant levels of PPARγ, even when treated with differentiation inducers (Table 1 and Fig. 1C).
Table 1.
PPARγ2 and Pref-1 mRNA by qPCR in 10T1/2 and A33 cells
| Day | Inducers | PPARγ2 mRNA |
Pref-1 mRNA |
||
|---|---|---|---|---|---|
| 10T1/2 | A33 | 10T1/2 | A33 | ||
| 3 | − | 0 | 0 | 0 | 1 ± 0.1 |
| 5 | − | 0 | 0.19 ± 0.02 | 2 ± 0.2 | 20 ± 4.1 |
| 7 | − | 0 | 0.21 ± 0.03 | 36 ± 1.3 | 22 ± 0.8 |
| 10 | + | 0.76 ± 0.6 | 66 ± 4.7 | 23 ± 3.8 | 2 ± 0.03 |
Cells were plated such that confluence was reached on day 5, and differentiation inducers were added on day 7. Values are arbitrary units normalized to 18S rRNA (mean ± SD).
The expression pattern of Pref-1, a preadipocyte marker (22), was also assessed during commitment and differentiation. Replicate dishes of 10T1/2 stem cells and A33 cells were plated at low density, allowed to proliferate and on day 7 (2 days after reaching confluence), and treated with differentiation inducers. As illustrated in Table 1, 10T1/2 stem cells, which lack the capacity to differentiate, express Pref-1 mRNA whether induced to differentiate or not. In contrast, committed A33 initially express Pref-1, but upon entry of the terminal differentiation program, Pref-1 mRNA levels are markedly reduced. Together, these findings demonstrate that A33 cells are determined preadipocytes, whereas 10T1/2 stem cells are not.
Committed A33 Preadipocytes Express BMP-4 During Proliferation.
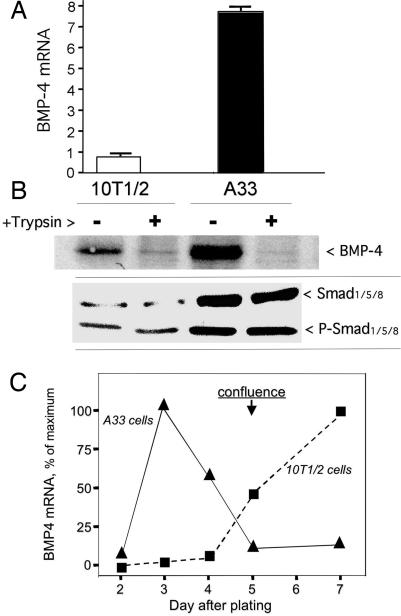
Because a single gene encoding MyoD activated by 5-azaC was sufficient to commit 10T1/2 stem cells to the muscle lineage (13), we speculated that a single gene (or perhaps a few genes) in A33 cells might have been responsible for commitment to the adipocyte lineage. The time window during which we attempted to identify differentially expressed mRNAs with commitment potential was the period of rapid cell division preceding growth arrest. Because growth arrest is required for the induction of terminal differentiation (23), it seemed likely that the expression of adipocyte commitment genes had already occurred by this point in the development program. Initially therefore, 10T1/2 stem cells were plated at low density, and mRNA was analyzed 72 h later, long before growth arrest at confluence was achieved. Because BMP-4 was known to induce adipocyte lineage commitment in 10T1/2 cells (17), qPCR was initially performed 72 h after plating to compare mRNA levels of this gene in 10T1/2 cells and in the A33 preadipocyte subline. At this point, cell density is ≈15% of the total number of cells at confluence. Although initial cell density and growth time were identical by 3 days after plating, the number of A33 cells per dish was ≈35% greater than that of 10T1/2 cells, indicating a higher growth rate for A33 cells. Remarkably, qPCR indicated a ≥10-fold increase in BMP-4 mRNA in A33 cells compared with 10T1/2 cells at this time in the program (Fig. 2A). This increase in BMP-4 mRNA is reflected in increased BMP-4 protein, as demonstrated by immunoblotting of whole cell extracts prepared at this time (Fig. 2B). Little free BMP-4 was detected in the culture medium (results not shown), but when the intact A33 cells were treated with trypsin before lysis, the BMP-4 protein was digested, demonstrating that virtually all of the BMP-4 present in the whole-cell extracts is extracellular, as would be expected for secreted BMP-4. Thus, most if not all of the BMP-4 was cell-associated, i.e., bound to the cell surface or extracellular matrix, as reported (24). In addition, an antibody that recognizes Smads -1, -5, and -8 shows that the levels of these BMP transducers are increased (Fig. 2B). Moreover, an antibody specific to the active phosphorylated forms of these Smads indicates increased BMP-4 activity in proliferating A33 cells (Fig. 2B).
Fig. 2.
Expression of BMP-4 mRNA and protein during proliferation of 10T1/2 and A33 cells. (A) Real-time PCR determination of BMP-4 mRNA expression from total RNA prepared from 10T1/2 and A33 cells 3 days after plating 22,000 cells per 6-cm dish. (B) BMP-4 protein expression in whole-cell extracts from 10T1/2 and A-33 cells treated or not with trypsin and corresponding to day 3 in A. (C) Time course of expression of BMP-4 mRNA by 10T1/2 and A33 cells after plating as in A.
Next, to assess the timing of BMP-4 expression, BMP-4 mRNA was followed for 7 days after plating at low density, and confluence was reached after 5 days. As shown in Fig. 2C, actively proliferating A33 cells express high levels of BMP-4 mRNA, reaching a maximum 3 days after plating and then declining as growth arrest at confluence occurs. Although both cell lines are growth-arrested/confluent by day 5, their patterns of BMP-4 per cell expression are markedly different. During active proliferation, 10T1/2 cells express an extremely low level of BMP-4 mRNA, which is dramatically up-regulated at growth arrest/confluence. These findings correlate well with our previous observation (17) with naïve 10T1/2 cells that exogenous BMP-4 must be administered during proliferation and growth arrest to achieve maximal adipocyte determination. Taken together, the present findings suggest that BMP-4 provides a signal for adipocyte determination in A33 cells. To more firmly establish the importance of BMP-4 in adipocyte commitment, additional functional tests described below were performed.
Disruption of BMP-4 Signaling by Noggin Blocks the Preadipocyte Phenotype Exhibited by A33 Cells.
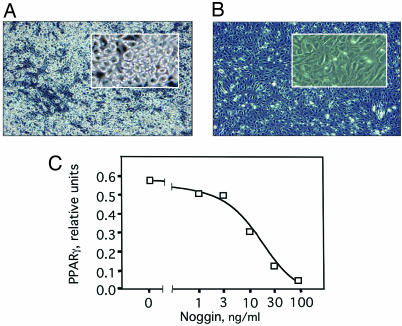
To verify that the expression of BMP-4 by proliferating A33 cells is required for commitment, the BMP-4-binding protein, noggin, was used to interrupt signaling. Noggin is known to antagonize BMP-4 action by binding BMP-4 with high affinity and preventing it from binding to its cognate receptors. A33 cells were plated in replicate and were treated with recombinant murine noggin 2 and 4 days after plating. The 2-day time point precedes the spike in BMP-4 mRNA expression that occurs in proliferating A33 cells on day 3 after plating (Fig. 2C). Treatment with noggin throughout proliferation and postconfluence completely blocked subsequent terminal adipocyte differentiation after exposure to differentiation inducers, as indicated by a lack of cytoplasmic triacylglycerol accumulation and PPARγ2 expression (Fig. 3). Further, progressively smaller doses of noggin were progressively less efficacious, as evidenced by a stepwise decline in PPARγ2 mRNA, a marker of terminal adipocyte differentiation (Fig. 3C). These results provide functional evidence that BMP-4 signaling is required for the acquisition of the preadipocyte phenotype by A33 cells.
Fig. 3.
Effect of the BMP-4 antagonist noggin on the acquisition of adipocyte characteristics by A33 cells. One day after plating replicate dishes (22,000 cells per 6-cm dish) of A33 cells, cells were exposed or not to recombinant mouse noggin 0–100 ng/ml. Noggin treatment was continued until postconfluent dishes were treated with differentiation inducers; photomicrographs were taken after 4 days and RNA harvested after 6 days. (A and B) Photomicrographs of incipient adipocytes in untreated control A33 cells (A) or A33 cells treated with 100 ng/ml noggin (B). (C) Effect of noggin on the expression of PPARγ2 mRNA.
Importantly, the specificity of noggin for BMP-4 in this experiment was illustrated by adding 100 ng/ml rBMP-4 to A33 cells that were exposed to 100 ng/ml noggin. Because BMP-4 and noggin have similar molecular masses (≈13 kDa), these concentrations produced an ≈1:1 molar ratio. Under such conditions, exogenous BMP-4 completely reversed the inhibitory effect of noggin (data not shown). These findings demonstrate that BMP-4 is required for A33 cells to exhibit their determined preadipocyte phenotype.
Switch in Promoter Usage During Commitment of 10T1/2 Stem Cells to A33 Preadipocytes.
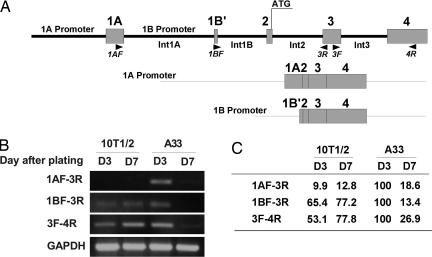
The murine BMP-4 gene is reported to have at least two promoters that are differentially regulated during development (25, 26). Fig. 4A shows a map of the BMP-4 locus that identifies promoter 1A and 1B and the locations of primer sets for RT-PCRs to distinguish transcripts produced by these promoters. RNA from committed A33 cells and 10T1/2 stem cells was isolated on day 3 after plating at the point of maximal BMP-4 expression (Fig. 2C) and on day 7 after both cell types had undergone growth arrest at confluence. Semiquantitative RT-PCR of RNA from day 3 cells was analyzed by using primer set 1AF-3R and showed that promoter 1A is used to drive expression of BMP-4 in A33 cells, but not 10T1/2 cells (Fig. 4 B and C). On day 7, use of promoter 1A and expression of BMP-4 cease. In contrast, RT-PCR of BMP-4 mRNA from uncommitted 10T1/2 cells with primer set 3BF-3R showed that promoter 1B was used exclusively to drive expression of the BMP-4 gene at both time points (Fig. 4 B and C). These results suggest that commitment to the adipocyte lineage induced by 5-azaC may have altered the methylation status of the BMP-4 gene, rendering promoter 1A more accessible for transcriptional activation. Preliminary findings in our laboratory indicate that a region of the BMP-4 locus, rich in CpG islands, has fewer methylated Cs in genomic DNA from A33 cells than from 10T1/2 cells.
Fig. 4.
Analyses of BMP-4 mRNA transcripts by semiquantitative RT-PCR. (A) Map of BMP-4 showing location of the primers 1AF (specific for promoter 1A) and 1BF (specific for promoter 1B) as well as the reverse primer (3R) for these transcripts. The amplicon from primers 3F and 4R is common to all BMP-4 transcripts. (B) RT-PCR data show promoter 1A is only used in proliferating A33 cells. (C) Quantification of the data presented in B.
Cell Density and BMP-4 Alter Expression of BMP-4 mRNA by A33 Cells.
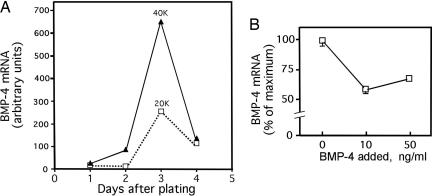
The expression of BMP-4 mRNA is markedly affected by the initial plating density of A33 cells. As shown in Fig. 5A, increasing the initial plating density from 20,000 to 40,000 cells per 6-cm culture dish caused a 2- to 3-fold increase in the expression of BMP-4 mRNA (normalized to 18S rRNA) by day 3 after plating (Fig. 5A). Microscopic examination of the cells at this stage of proliferation revealed many more foci and cell–cell contacts between with A33 cells than with 10T1/2 cells at comparable cell densities. Two possible causal factors could be involved: (i) A factor(s) secreted by committed A33 preadipocytes could activate the expression or decrease the turnover of BMP-4 mRNA, or (ii) increased cell–cell contact at higher cell densities could affect BMP-4 synthesis or turnover.
Fig. 5.
Effects of cell density and exogenous BMP-4 on BMP-4 mRNA expression by A33 cells. (A) Effect of initial plating density (20,000 or 40,000 cells per 6-cm dish) on BMP-4 mRNA expression. (B) Effect of exogenous BMP-4 (10 or 50 ng/ml) on BMP-4 mRNA expression by A33 cells.
The expression of BMP-4 also appears to be affected by the ambient concentration of BMP-4. It was repeatedly observed that, after reaching a maximal level at ≈3 days after plating, there is a rapid “fall off” in the expression of BMP-4 over the next several days (Figs. 5A and 2C). Because BMP-4 is secreted by A33 cells, it was considered that BMP-4 itself may cause this decline. As shown in Fig. 5B, the addition of BMP-4 to the culture medium at a level of 10 or 50 ng/ml caused an ≈50% decrease in the expression of BMP-4 mRNA. This finding suggests that BMP-4 may inhibit its own expression by a negative autocrine feedback loop.
mRNA Expression Profiling of 10T1/2 Stem Cells and A33 Preadipocytes Implicates BMP-4 Signaling Pathway Components in Adipocyte Lineage Commitment.
High-throughput mRNA expression profiling with a customized PCR array, as described in Methods, was used. The gene list focused on those known to be involved in BMP-2/4 signaling, but a few other relevant genes were included (Table 2, which is published as supporting information on the PNAS web site). 10T1/2 and A33 cells were plated at low density, and total RNA was isolated 3 days later, corresponding to the spike in BMP-4 mRNA expression by A33 cells in Figs. 2C and 5B.
Consistent with results presented in Fig. 2, the PCR array data show BMP-4 was up-regulated 7-fold in A33 compared to 10T1/2 cells (Table 2). BMP-2 and -7 mRNAs were not detected in 10T1/2 or A33 cells (data not shown). Of note, A33 cells expressed ≈2-fold more BMPr2 and BMPr1a mRNA than 10T1/2 cells, whereas 10T1/2 cells expressed a low level of BMPr1b that was not detected in A33 cells (Table 2). These observations are particularly interesting in light of a study showing the BMPr1a subtype is coupled to adipogenesis, whereas the BMPr1b subtype is coupled to chondrogenesis (27).
SMADs are well known intracellular signal transducers of the BMP pathway, and remarkably all four SMADs (SMADs -1, -4, -5, and -8) that are downstream of BMP receptors are up-regulated in A33 cells, Smad-8 increasing 9-fold (Table 2). These findings agree with the immunoblots of Smad-1, -5, and -8 and the phosphorylated forms of these Smads presented in Fig. 2B.
The transcriptional profile of several known BMP target genes including inhibitor of DNA-binding (Id) -1, -2, and -3 were up-regulated in A33 cells, providing further evidence for increased BMP-4 signaling pathway activity in proliferating A33 cells vs. their undetermined progenitors (Table 2). Ids play important roles in cell fate determination, so their increase in expression is likely to be an important component of adipocyte determination in A33 cells. The Msh-like 2 homeobox gene (Msx2) is a BMP target gene that plays a role in osteoblast determination, and Msx2 is reported to be a BMP-2 target in 10T1/2 cells (28, 29). Msx2 is markedly (≈10-fold) down-regulated in A33 cells. These changes in BMP targets are likely to play a role in the determined preadipocyte phenotype exhibited by A33 cells.
Genes classified as “extracellular modulators” exhibited the most dynamic differential expression patterns. Noggin mRNA decreases in A33 cells to a level approximately one-third that in 10T1/2 cells, consistent with its inhibitory effect on adipocyte differentiation (Fig. 3). Similarly, the BMP-4 antagonist gremlin is down-regulated in A33 cells (30). In contrast, chordin mRNA is increased by ≈8-fold. Because chordin binds BMP-4 and antagonizes its activity (31), this observation appears counterintuitive. It must be recognized, however, that BMP-1 can cleave and thereby inactivate chordin, and BMP-1 expression is increased 80% in A33 cells. Further, the murine twsg homologue (twsg1) increases 2-fold. twsg1 promotes BMP-4 signaling in vivo by forming a BMP-4–chordin–twsg complex, which makes chordin a better substrate for BMP-1 (18). Thus, expression of BMP-1 and twsg1 by A33 cells would be expected to render the cells insensitive to chordin. Indeed unlike exogenous noggin, exogenous chordin is unable to block adipogenesis by A33 cells (data not shown). Although the basis for up-regulation of chordin by A33 cells is uncertain, it may serve to ensure that any BMP-4 that escapes the immediate vicinity of BMP-4 expression is sequestered by chordin and thus affects only cells that express BMP-1. In this connection, it should be noted that in Drosophila chordin is required for maximal BMP-4 signaling.
The most differentially expressed genes identified were the serine peptidase Htra1 that increased ≈29-fold in A33 cells and chordin-like 1 (chrdl1) that increased ≈56-fold. Although there is biochemical evidence for Htra1 binding to BMP-4, and overexpression of this protein can antagonize BMP-4 activity (32), the physiological role of Htra1 is not clear. Htra1 binds to collagens and may play a role in extracellular matrix remodeling. Further investigations are necessary to determine the role of Htra1 in lineage determination. The rat homologue of chrdl1, neurogenesin-1, is 97% identical at the amino acid level to mouse Chrdl1 and is implicated in adult neurogenesis (33). This observation and the fact that BMP-4 is implicated in sympathetic nervous system development raise the intriguing possibility that differentiating preadipocytes may recruit/attract sympathetic neurons to innervate them. Additional experiments to address this hypothesis are underway. Together, these PCR array findings indicate coordinated changes in the BMP pathway in A33 preadipocytes vs. their pluripotent progenitors that shed light on their preadipocyte phenotype.
Mesenchymal Cell Fate Determination.
Mounting evidence indicates mesenchymal lineage determination is regulated by a network of extracellular signaling factors that ultimately impinge on the promoters of lineage-specific transcription factors. In addition, cell–cell and cell–extracellular matrix interactions affect lineage determination. Further, as opposed to simple linear pathways leading to specific cell types, the different paths of stem cell differentiation are interconnected. It is the balance of key signaling molecules that determines the developmental pathway, and lineage-specific factors often simultaneously promote one pathway while inhibiting another. Many key components, extracellular and intracellular, of the developmental network that controls lineage determination have a bipolar nature. For example, Wnt 10b promotes myogenesis while inhibiting adipogenesis (34), and BMP-4 promotes adipogenesis while inhibiting myogenesis (35). In addition, PPARγ inhibits chondrogenesis while stimulating adipogenesis, and Msx2 stimulates osteogenesis while inhibiting adipogenesis by inhibiting the transcriptional activity of PPARγ. These observations illustrate the interconnected nature of adipogenesis and chondro/osteogenesis, the understanding of which has important implications for the treatment of osteoporosis as well as obesity and diabetes. The present findings of decreased Msx2 expression in A33 preadipocytes and detectable PPARγ2 mRNA in these determined cells complement previous studies and provide leads for further investigations. These studies provide evidence that BMP-4 signaling is sufficient and necessary to induce adipocyte lineage determination in a mesenchymal stem cell model. Further, coordinated changes in BMP pathway components characterize determined A33 preadipocytes vs. their pluripotent progenitors. Although the key targets of BMP-4 have not yet been identified, the A33 cell line provides an excellent model to identify BMP-4 targets involved in adipocyte determination. In addition, regulatory elements of the BMP-4 gene are expected to bind transcription factors that play more upstream roles in adipocyte determination. Studies to identify activators of BMP-4 as well as its targets are needed.
Methods
Cell Culture and Derivation of A33 Cells.
A33 and C3H10T1/2 stem cells (American Type Culture Collection, Bethesda, MD) were propagated and differentiated as described (20, 21). Recombinant murine noggin and chordin and recombinant human BMP-4 were from R&D Systems (Minneapolis, MN). The A33 cell line was isolated essentially by the method of Constantinides et al. (36), as described by Konieczny and Emerson (11). 10T1/2 cells were plated at cloning density (1,000 cells per 6-cm dish) and treated with 3 μM 5-azaC (Sigma, St. Louis, MO) for 24 h. 5-azaC exposure began 24 h after plating as with previous studies (10, 11, 15, 36). After ≈20 days, the cells had formed foci, and at this time they were treated with differentiation inducers. Some foci had cells that responded as preadipocytes, i.e., rounded up and began accumulating fat. Cells from these foci were picked with cloning cylinders and replated. After subcloning of one of these cell lines two times, the clone of cells named A33 was isolated. It should be noted that A33 cells maintained the characteristics of committed preadipocytes through numerous (>20) passages.
RNA Isolation, Semiquantitative RT-PCR, and qPCR.
Total RNA and was isolated and DNase I treated by using the RNeasy mini kit according to the protocol of the manufacturer (Qiagen, Valencia, CA). All qPCR experiments were conducted with a Mx3000P Quantitative PCR System (Stratagene, La Jolla, CA), essentially as described (37). One-step RT-PCR was used for examining 18S rRNA, BMP-4, PPARγ2, and Pref-l mRNA levels. Primers were as follows: 18S rRNA (5′-tggttgatcctgccagta; 3′-cgaccaaaggaaccataact), PPARγ2 (5′-ccagagcatggtgccttcgct; 3′-cagcaaccattgggtcagctc), and Pref-1 (ctgtgtcaatggagtctgcaag; ctacgatctcacagaagttgc). BMP-4 primers were obtained from SuperArray Bioscience (Frederick, MD). Standard curves were used to calculate mRNA levels, and values were normalized to 18S rRNA amounts. Semiquantitative RT-PCR was used to distinguish among BMP-4 transcripts as described (25). For PCR array experiments, an RT2 Profiler Custom PCR Array was used to simultaneously examine the mRNA levels of 89 genes, including five “housekeeping genes” in 96-well plates according to the protocol of the manufacturer (SuperArray Bioscience). Each reaction included 40 ng of total RNA and the proper negative controls (no reverse transcription, no template). 10T1/2 and A33 cells were plated at low density, and total RNA was isolated 3.3 (10T1/2) or 3.0 (A33) days later. The 10T1/2 cells were allowed to grow slightly longer, so that at the time of RNA isolation, the cell density was indistinguishable from that of the more rapidly dividing A33 cells. RNA of both 10T1/2 and A33 cells was analyzed in triplicate, and data were normalized for GAPDH levels by the ΔΔCt method.
Immunoblotting.
Western blots of cells were performed as described (38) with antibodies against BMP-4 (39), C/EBPα (38), or 422/aP2 (38).
Supplementary Material
Acknowledgments
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health Research Grant DK66627 (to M.D.L.) and National Research Service Award Grant DK074294 (to R.R.B.).
Abbreviations
- 5-azaC
5-azacytidine
- BMP-4
bone morphogenetic protein-4
- qPCR
quantitative real-time PCR
- twsg
twisted gastrulation
- PPARγ
peroxisome proliferator-activated receptor γ
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Faust I. M. In: Eating and Its Disorders. Stunkard A. J., Stellar E., editors. New York: Raven; 1984. pp. 97–107. [Google Scholar]
- 2.Shepherd P. R., Gnudi L., Tozzo E., Yang H., Leach F., Kahn B. B. J. Biol. Chem. 1993;268:22243–22246. [PubMed] [Google Scholar]
- 3.Reznikoff C. A., Brankow D. W., Heidelberger C. Cancer Res. 1973;33:3231–3238. [PubMed] [Google Scholar]
- 4.Green H., Kehinde O. Cell. 1976;7:105–113. doi: 10.1016/0092-8674(76)90260-9. [DOI] [PubMed] [Google Scholar]
- 5.MacDougald O. A., Lane M. D. Annu. Rev. Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 6.Loftus T. M., Lane M. D. Curr. Opin. Genet Dev. 1997;7:603–608. doi: 10.1016/s0959-437x(97)80006-8. [DOI] [PubMed] [Google Scholar]
- 7.Tang Q. Q., Otto T. C., Lane M. D. Proc. Natl. Acad. Sci. USA. 2003;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Q. Q., Otto T. C., Lane M. D. Proc. Natl. Acad. Sci. USA. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otto T. C., Lane M. D. Crit. Rev. Biochem. Mol. Biol. 2005;40:229–242. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- 10.Taylor S. M., Jones P. A. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 11.Konieczny S. F., Emerson C. P., Jr. Cell. 1984;38:791–800. doi: 10.1016/0092-8674(84)90274-5. [DOI] [PubMed] [Google Scholar]
- 12.Pinney D. F., Emerson C. P., Jr. Environ. Health Perspect. 1989;80:221–227. doi: 10.1289/ehp.8980221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis R. L., Weintraub H., Lassar A. B. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 14.Chen S., Teicher L. C., Kazim D., Pollack R. E., Wise L. S. Science. 1989;244:582–585. doi: 10.1126/science.2470149. [DOI] [PubMed] [Google Scholar]
- 15.Chapman A. B., Knight D. M., Dieckmann B. S., Ringold G. M. J. Biol. Chem. 1984;259:15548–15555. [PubMed] [Google Scholar]
- 16.Ahrens M., Ankenbauer T., Schroder D., Hollnagel A., Mayer H., Gross G. DNA Cell Biol. 1993;12:871–880. doi: 10.1089/dna.1993.12.871. [DOI] [PubMed] [Google Scholar]
- 17.Tang Q. Q., Otto T. C., Lane M. D. Proc. Natl. Acad. Sci. USA. 2004;101:9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oelgeschlager M., Larrain J., Geissert D., De Robertis E. M. Nature. 2000;405:757–763. doi: 10.1038/35015500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larrain J., Oelgeschlager M., Ketpura N. I., Reversade B., Zakin L., De Robertis E. M. Development. Vol. 128. Cambridge, U.K.: 2001. pp. 4439–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Student A. K., Hsu R. Y., Lane M. D. J. Biol. Chem. 1980;255:4745–4750. [PubMed] [Google Scholar]
- 21.Mandrup S., Loftus T. M., MacDougald O. A., Kuhajda F. P., Lane M. D. Proc. Natl. Acad. Sci. USA. 1997;94:4300–4305. doi: 10.1073/pnas.94.9.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smas C. M., Sul H. S. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 23.Gregoire F. M., Smas C. M., Sul H. S. Physiol. Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 24.Suzawa M., Takeuchi Y., Fukumoto S., Kato S., Ueno N., Miyazono K., Matsumoto T., Fujita T. Endocrinology. 1999;140:2125–2133. doi: 10.1210/endo.140.5.6704. [DOI] [PubMed] [Google Scholar]
- 25.Thompson D. L., Gerlach-Bank L. M., Barald K. F., Koenig R. J. Mol. Cell. Biol. 2003;23:2277–2286. doi: 10.1128/MCB.23.7.2277-2286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng J. Q., Chen D., Cooney A. J., Tsai M. J., Harris M. A., Tsai S. Y., Feng M., Mundy G. R., Harris S. E. J. Biol. Chem. 1995;270:28364–28373. doi: 10.1074/jbc.270.47.28364. [DOI] [PubMed] [Google Scholar]
- 27.Chen D., Ji X., Harris M. A., Feng J. Q., Karsenty G., Celeste A. J., Rosen V., Mundy G. R., Harris S. E. J. Cell Biol. 1998;142:295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichida F., Nishimura R., Hata K., Matsubara T., Ikeda F., Hisada K., Yatani H., Cao X., Komori T., Yamaguchi A., Yoneda T. J. Biol. Chem. 2004;279:34015–34022. doi: 10.1074/jbc.M403621200. [DOI] [PubMed] [Google Scholar]
- 29.Cheng S. L., Shao J. S., Charlton-Kachigian N., Loewy A. P., Towler D. A. J. Biol. Chem. 2003;278:45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 30.Michos O., Panman L., Vintersten K., Beier K., Zeller R., Zuniga A. Development. Vol. 131. Cambridge, U.K.: 2004. pp. 3401–3410. [DOI] [PubMed] [Google Scholar]
- 31.Larrain J., Bachiller D., Lu B., Agius E., Piccolo S., De Robertis E. M. Development. Vol. 127. Cambridge, U.K.: 2000. pp. 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oka C., Tsujimoto R., Kajikawa M., Koshiba-Takeuchi K., Ina J., Yano M., Tsuchiya A., Ueta Y., Soma A., Kanda H., et al. Development. Vol. 131. Cambridge, U.K.: 2004. pp. 1041–1053. [DOI] [PubMed] [Google Scholar]
- 33.Ueki T., Tanaka M., Yamashita K., Mikawa S., Qiu Z., Maragakis N. J., Hevner R. F., Miura N., Sugimura H., Sato K. J. Neurosci. 2003;23:11732–11740. doi: 10.1523/JNEUROSCI.23-37-11732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vertino A. M., Taylor-Jones J. M., Longo K. A., Bearden E. D., Lane T. F., McGehee R. E., Jr., MacDougald O. A., Peterson C. A. Mol. Biol. Cell. 2005;16:2039–2048. doi: 10.1091/mbc.E04-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerhart J., Neely C., Stewart B., Perlman J., Beckmann D., Wallon M., Knudsen K., George-Weinstein M. J. Cell Biol. 2004;164:739–746. doi: 10.1083/jcb.200309152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Constantinides P. G., Taylor S. M., Jones P. A. Dev. Biol. 1978;66:57–71. doi: 10.1016/0012-1606(78)90273-7. [DOI] [PubMed] [Google Scholar]
- 37.Cha S. H., Hu Z., Chohnan S., Lane M. D. Proc. Natl. Acad. Sci. USA. 2005;102:14557–14562. doi: 10.1073/pnas.0507300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang Q. Q., Lane M. D. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuhara K., Nakase T., Suzuki S., Takaoka K., Matsui M., Anderson H. C. Bone. 1995;16:91–96. doi: 10.1016/s8756-3282(94)00014-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.