Abstract
In Arabidopsis, a complex of Polycomb-group (PcG) proteins functions in the female gametophyte to control the initiation of seed development. Mutations in the PcG genes, including MEDEA (MEA) and FERTILIZATION-INDEPENDENT SEED 2 (FIS2), produce autonomous seeds where endosperm proliferation occurs in the absence of fertilization. By using a yeast two-hybrid screen, we identified MEA and a related protein, SWINGER (SWN), as SET-domain partners of FIS2. Localization data indicated that all three proteins are present in the female gametophyte. Although single-mutant swn plants did not show any defects, swn mutations enhanced the mea mutant phenotype in producing autonomous seeds. Thus, MEA and SWN perform partially redundant functions in controlling the initiation of endosperm development before fertilization in Arabidopsis.
Keywords: chromatin, endosperm, female gametophyte, fertilization, gene silencing
In angiosperms, seed development is initiated by a double-fertilization event within the female gametophyte (FG). The pollen tube delivers two sperm cells that fuse with the haploid egg cell and the typically homodiploid central cell to initiate the development of two distinct organs, an embryo and an endosperm, respectively (1). The diploid embryo forms the next generation, whereas the typically triploid endosperm is a short-lived nutritive tissue that supports embryogenesis or seedling development. In most angiosperms, including Arabidopsis, the endosperm undergoes a nuclear-type development that includes an initial period of coenocytic development in which the primary endosperm nucleus undergoes mitosis without cytokinesis followed by cellularization. In Arabidopsis, the cellularized endosperm is gradually absorbed by the developing embryo, and only an aleurone layer remains at seed maturity (2, 3).
Recent evidence indicates that the FG controls the initiation of seed development in Arabidopsis. Mutations in genes encoding the Polycomb-group (PcG) proteins FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), MEDEA (MEA), FERTILIZATION-INDEPENDENT SEED 2 (FIS2), and the Arabidopsis homologue of MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) cause the development of autonomous seeds in the absence of fertilization due mainly to proliferation of the diploid central cell nucleus. Molecular and genetic evidence indicates that these proteins are expressed in the FG and function to repress downstream genes required for normal endosperm proliferation before fertilization (4–11). The FIE, MEA, FIS2, and MSI1 genes encode proteins that are highly similar to components of the Polycomb repressive complex 2/3 (PRC2/3) that include the Drosophila Esc, E(z), Su(z)12, and p55 and the mammalian EED, EZH2, SUZ12, and RbAp48/46, respectively (12). PRC2/3 and the larger PRC1 complex function to silence and maintain epigenetic silencing of target genes involved in regulation of early development, cell growth, and proliferation. At least part of the gene-silencing function of PRC2/3 is likely mediated through its intrinsic histone H3 methylation activity (13, 14).
Plants presumably have evolved multiple PRC2/3-like complexes that function in various stages of the life cycle (12, 15). Except for FIE, the known plant PcG proteins are encoded by multigene families (www.chromdb.org). Genetic analysis in Arabidopsis indicates that, in addition to controlling seed initiation, PcG proteins control flower organ development, the switch from vegetative to floral development and vernalization (16–22). In all cases, at least one specific Arabidopsis homologue of E(z) (SET-domain PcG) or Su(z)12 (zinc-finger PcG) has been implicated genetically in the process (16, 17, 21). Protein-interaction studies have supported the presence of multiple evolutionarily conserved pairings of E(z)-Su(z)12 homologues in Arabidopsis (16, 23). However, it is not clear whether such pairings form the basis for the functional specificity of plant PcG complexes.
Here we report that, by using yeast two-hybrid (Y2H) interaction studies, we have identified MEA and SWINGER/EZA1 (SWN; refs. 8 and 16), a closely related SET-domain PcG protein, as partners of FIS2, a Su(z)12 homologue. Protein-reporter studies indicated that FIS2, MEA, and SWN are expressed during late FG development and fertilization, consistent with their role in the control of seed initiation. Finally, genetic-interaction studies indicated that MEA and SWN perform overlapping roles in this process.
Results
FIS2 Interacts with the SET-Domain PcG Protein MEA.
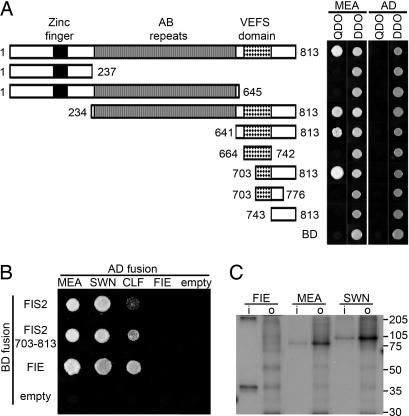
To identify FIS2-interacting proteins, we isolated a full-length FIS2 cDNA clone using RT-PCR and 5′-RACE. FIS2 is composed of a 5′ untranslated region of 159 bp, an ORF of 2,442 bp, and a 3′ untranslated region of 198 bp (Fig. 4, which is published as supporting information on the PNAS web site). Our data indicate that FIS2 should encode a protein of 813 aa, which is 121 aa longer than was reported as a full-length FIS2 product (9). In earlier studies, FIS2 failed to interact with both MEA and FIE in Y2H assays (8, 10, 24). We initially performed directed Y2H assays with our full-length FIS2, FIE, and MEA to test whether the reported lack of interaction between FIS2 and MEA could be due to the absence of the extra 121 aa of FIS2 in the previous assays. As shown in Fig. 1A and B, FIS2 interacted with MEA but not with FIE. However, FIE and MEA showed interactions as described (8, 10, 24), indicating that all of our clones produced functional proteins. Further analyses (see below) indicated that the extended N-terminal amino acids of FIS2 are not required for interaction with MEA. The lack of interaction between FIS2 and MEA reported previously (8, 10, 24) might have been due to instability, improper folding, or reduced accumulation of the FIS2 fusion protein. Nevertheless, we identified the SET-domain PcG MEA as a likely interacting partner of FIS2.
Fig. 1.
Interaction of FIS2 with SET-domain PcG proteins in yeast and in vitro. (A) Mapping MEA-interacting domains of FIS2 by using Y2H assays. The deletion constructs of FIS2 are shown schematically with numbers indicating the position of amino acids included in each construct. Diploid yeast harboring both pGBK (BD) and pGAD (AD) constructs were spotted on QDO (synthetic complete medium minus leucine, tryptophan, adenine, and histidine) and DDO (synthetic complete medium minus leucine and tryptophan). (B) Interaction of FIS2 and FIE with three SET-domain PcG proteins using Y2H assays is indicated by the growth of yeast on QDO medium. empty, empty pGBK, or pGAD vector. (C) In vitro binding of myc-tagged FIS2 (641–813) polypeptide with HA-tagged full-length MEA, SWN, and FIE labeled with [35S]methionine. The input lane (i) corresponds to 5% of the protein used for in vitro coimmunoprecipitation assays in output lanes (o).
FIS2 Interacts with the SET-Domain PcG Protein SWN.
To identify additional FIS2-interacting proteins, we screened a Y2H library constructed from reproductive organs of Arabidopsis using a FIS2-GAL4BD bait plasmid. One of the clones identified encoded the C-terminal region (amino acids 202–689) of the MEA protein, indicating that this screen identified relevant binding partners.
A second class of clones encoded full-length or partial SWN polypeptides (8, 16). To determine whether these interactions are specific or whether they represent a general affinity between FIS2 and all SET-domain PcG proteins, we used directed Y2H assays to test for FIS2 interactions with MEA, SWN, and CURLY LEAF (CLF), a SET-domain PcG protein closely related to MEA and SWN (16, 18). As shown in Fig. 1B, FIS2 showed significantly weaker interaction with CLF than with the other SET-PcG proteins. The reduced interaction was not due to differences in fusion protein stability or expression level, because all three SET-PcG proteins interacted with FIE (Fig. 1B). Together, these data indicate that the FIS2–MEA and FIS2–SWN interactions are specific and that FIS2 does not interact with SET-domain PcG proteins indiscriminately.
The C-Terminal Portion of FIS2 Is Sufficient for Interaction with MEA.
To determine the regions of the FIS2 protein required for interaction with SET-domain PcG proteins, we generated a series of FIS2 truncations and examined their interactions with MEA using Y2H assays. As shown in Fig. 1A, a 110-aa C-terminal portion of FIS2 (FIS2703–813) was necessary and sufficient for interaction with MEA. This region contains part of the Zn-finger-PcG-specific VRN2-EMF2-FIS2-Su(z)12 (VEFS) domain (25) and the nonconserved C-terminal region of FIS2 (9). Y2H assays indicated that the more N-terminal regions of FIS2, including the Zn-finger motif and the FIS2-specific AB repeats (9), were not required for interactions with MEA (Fig. 1A).
To identify the FIS2 regions required for specificity of protein–protein interaction, we tested interactions between truncated FIS2 polypeptides and CLF using Y2H. An N-terminal-truncated FIS2 protein containing the AB repeats and the C terminus (FIS2234–813) also did not show any significant interaction with a full-length CLF protein (data not shown). However, as compared with the full-length FIS2 protein, the C-terminal portion of FIS2 (FIS2703–813), shown to be sufficient for interaction with MEA (Fig. 1B), displayed an increased level of interaction with CLF (Fig. 1B). The data indicate that the FIS2 C terminus is necessary and sufficient for interaction with SET-domain PcG proteins, and that the specificity of interaction (MEA and SWN vs. CLF) is likely conferred by the adjacent AB repeat region.
To confirm the FIS2–MEA and FIS2–SWN Y2H interactions, we performed a series of coimmunoprecipitation assays using in vitro-transcribed and translated versions of FIS2, MEA, and SWN. As shown in Fig. 1C, an anti-myc-epitope antibody was able to precipitate the myc-FIS2 polypeptide (amino acids 641–813) in association with an HA-tagged MEA or SWN but not an HA-tagged FIE. We did not detect an association between an N-terminal FIS2 polypeptide (FIS21–237) and MEA or SWN (data not shown). Therefore, results of the in vitro coimmunoprecipitation experiments support the Y2H interaction studies. Together, the data indicate that FIS2 is capable of interacting with MEA and SWN SET-domain PcG proteins specifically and that the C-terminal region of FIS2 is sufficient for this interaction.
SWN, MEA, and FIS2 Localization Patterns Overlap During Late Megagametogenesis.
The protein-interaction studies discussed above suggest that FIS2 interacts with MEA and SWN during plant development. To identify the stages of plant development in which FIS2–MEA and FIS2–SWN interactions may take place, we analyzed MEA, FIS2, and SWN expression throughout the plant using RT-PCR and through the analysis of protein fusion constructs. As shown in Fig. 5, which is published as supporting information on the PNAS web site, FIS2 and MEA RNAs were mainly detected in reproductive structures, including closed buds, open flowers, and siliques. Unlike MEA and FIS2, SWN transcripts were present at high levels and detectable in both reproductive and vegetative structures (Fig. 5). The data indicate that MEA and SWN gene expression patterns are distinct and suggest that the two genes may perform divergent roles during plant development.
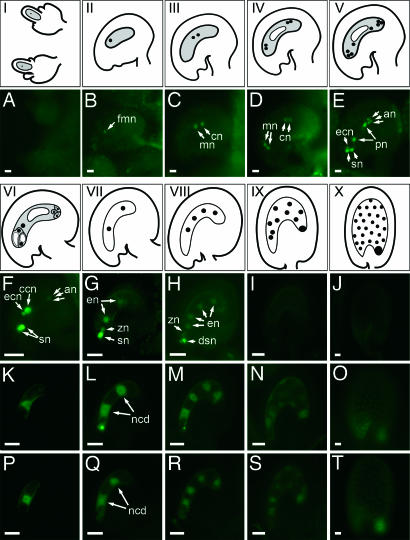
To determine the patterns of localization for all three proteins, we generated protein fusion constructs and introduced these into Arabidopsis plants. SWN-GFP/yellow fluorescent protein (YFP) fusion proteins were detected in the nucleus throughout most of the plant life cycle including ovule and FG development (Figs. 2I–VI and A–F and 3A; see also Fig. 6, which is published as supporting information on the PNAS web site), embryo development (Fig. 3B and C), and vegetative development (Fig. 3 D–F). After fertilization, SWN-GFP was down-regulated rapidly within developing endosperm (Fig. 2 G–J) and was not observed beyond the first or second rounds of endosperm mitosis (Fig. 2 G and H). However, expression persisted throughout embryogenesis and within vegetative structures after seed germination (Fig. 3). No SWN-GFP/YFP signal was observed in developing and mature pollen grains (data not shown). The data indicate that SWN is nuclear-localized and has a broad pattern of accumulation during the Arabidopsis life cycle.
Fig. 2.
Expression of SWN-GFP, FIS2-YFP, and MEA-YFP during FG and early seed development. (I–X) Schematic representations of stages of FG and early endosperm development in Arabidopsis. The FG is formed from a surviving megaspore after the megaspore mother cell undergoes meiosis (I). The shaded areas in II–VI represent the developing FGs (26). Dividing endosperm nuclei during early seed development are depicted in VII–X. (A–T) Epifluorescence images of GFP/YFP activity corresponding to stages shown in I–V (A–E) and VI–X (F–J, K–O, and P–T). Images were obtained with the focal planes through the megaspore mother cell (A), developing FG (B–F, K, and P), or developing endosperm (G–J, L–O, and Q–T). (A–J) SWN-GFP expression. In most transgenic plants, SWN-GFP expression was not detectable in the endosperm (G–H). (K–O) FIS2-YFP expression. (P–T) MEA-YFP expression. an, antipodal cell nucleus; cn, chalazal nucleus; ccn, central cell nucleus; dsn, degenerating synergid nucleus; ecn, egg cell nucleus; en, endosperm nucleus; fmn, functional megaspore nucleus; mn, micropylar nucleus; ncd, nuclear cytoplasmic domain; pn, polar nucleus; sn, synergid cell nucleus; zn, zygote nucleus. [Scale bars: 10 μm (A–E); 30 μm (F–T).]
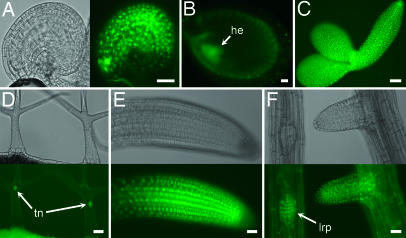
Fig. 3.
Expression of SWN-GFP in ovule, seed, and vegetative structures. (A) Bright-field (Left) and the corresponding epifluorescence (Right) images of a flower stage 13 (44) ovule showing SWN-GFP in all nuclei of the integument cells. The focal plane is through the outer integument. (B) Epifluorescence image of a developing seed containing a heart-stage embryo. (C) Epifluorescence image of a bent cotyledon-stage embryo dissected away from the endosperm and seed coat. (D) Bright-field (Upper) and the corresponding epifluorescence (Lower) images of a trichome on the surface of a rosette leaf. (E) Bright-field (Upper) and the corresponding epifluorescence (Lower) images of a root tip. (F) Bright-field (Upper) and the corresponding epifluorescence (Lower) images of a lateral root primordium and a lateral root. he, heart stage embryo; lrp, lateral root primordium; tn, trichome cell nucleus. (Scale bars: 30 μm.)
To compare the expression of SWN with FIS2 and MEA, we analyzed the expression of FIS2-YFP (Fig. 2 K–O) and MEA-YFP (Fig. 2 P–T) during similar stages of development, as outlined above. As shown in Fig. 2 K–T, FIS2-YFP and MEA-YFP fusion proteins were detectable only in the late-developing central cell [stages FG6 and FG7 (26); Fig. 2 K and P] and early-developing endosperm. Within the central cell, FIS2-YFP and MEA-YFP were detected in both the nucleus and the cytoplasm (Fig. 2 K and P). After fertilization, FIS2- and MEA-YFP showed a similar pattern of expression and were associated with the dividing nuclear cytoplasmic domains (27). Unlike SWN-GFP/YFP, FIS2-YFP and MEA-YFP activities were detectable during much of the nuclear phase of endosperm development (Fig. 2 L–O, Q–T) until the early heart stage of embryo development (Fig. 2 O and T). The FIS2 and MEA protein localization patterns during development are consistent with their role in repressing proliferation of the central cell nucleus before fertilization and control of early endosperm development. Taken together, the data identify overlapping patterns of expression for SWN, FIS2, and MEA and suggest a role for SWN in the regulation of proliferation of the central cell nucleus.
SWN Function Is Partially Redundant with MEA in Controlling Seed Initiation.
Protein interaction (Fig. 1) and expression (Fig. 2) studies suggest that SWN plays a role in repressing endosperm development before fertilization. To test this, we obtained three swn T-DNA alleles (28) and analyzed these for defects in FG and seed development. With all three alleles, heterozygous and homozygous plants did not exhibit morphological abnormalities in vegetative or reproductive development, including seed-set defects. Heterozygous and homozygous swn plants did not show consistent autonomous seed development or silique elongation in the absence of fertilization (Table 1; see also Fig. 7 A–C and G–I, which is published as supporting information on the PNAS web site).
Table 1.
Fertilization-independent development in swn, mea, and swn;mea double-mutant plants
| Genotype | Experiment 1 |
Experiment 2 |
||
|---|---|---|---|---|
| Length, mm | Seed, % | Length, mm | Seed, % | |
| swn-3/swn-3;mea-3/+ | 11.50§ | 49.31§ | 10.48§ | 37.10§ |
| swn-3/+;mea-3/+ | 7.92‡ | 24.46‡ | 9.23‡ | 24.19‡ |
| mea-3/+ | 4.53† | 10.49† | 4.93† | 5.64† |
| swn-3/swn-3 | 3.79* | 0.00* | 3.65* | 0.00* |
| swn-3/+ | 4.58† | 0.89* | 3.88* | 0.00* |
| Col-0 WT | 3.65* | 0.00* | 3.48* | 0.00* |
| swn-4/swn-4;mea-3/+ | 10.82¶ | 44.53§ | 10.66¶ | 44.63§ |
| swn-4/+;mea-3/+ | 8.31§ | 18.80‡ | 8.56§ | 21.13‡ |
| mea-3/+ | 3.87† | 3.98† | 3.99† | 3.09† |
| swn-4/swn-4 | 3.65*† | 0.00* | 4.48‡ | 0.00* |
| swn-4/+ | 4.43‡ | 0.00* | 3.34* | 0.00*† |
| Col-0 WT | 3.21* | 0.00*† | 3.79† | 0.00*† |
| swn-2/swn-2;mea-3/+ | 10.72§ | 45.85§ | ||
| swn-2/+;mea-3/+ | 8.50‡ | 27.24‡ | ||
| mea-3/+ | 5.54† | 11.82† | ||
| swn-2/swn-2 | 4.02* | 0.00* | ||
| swn-2/+ | 4.28* | 0.00* | ||
| Col-0 WT | 4.28* | 0.00* | ||
The six genotypes were obtained among the progenies by selfing a single swn/+;mea-3/+ double-heterozygous plant for each swn allele. Flowers for two to three individual plants (9–12 siliques per plant) for each genotype were emasculated. Silique lengths (in millimeters) and the number of autonomous seed-like structures were determined 7 days after emasculation for each genotype separately, and the data were analyzed by using PROC GLM in SAS as described in Materials and Methods. Average values with the same superscript symbol do not differ significantly. Seed, % refers to the occurrence of seed-like structures as a proportion of total number of ovules per silique multiplied by 100.
The absence of a phenotype for swn single mutants may be due to functional redundancy or compensation between SWN and MEA. To test this, we generated swn;mea-3 double-mutant plants for all three swn alleles and analyzed these for autonomous seed development and the associated autonomous silique elongation. As shown in Table 1 and Fig. 7J, 3–12% of ovules gave rise to seed-like structures in mea-3/+ plants. By contrast, in swn-3/+;mea-3/+ and swn-3/swn-3;mea-3/+ double-mutant plants, 24% and 37–49% of ovules, respectively, developed into seed-like structures in the absence of fertilization (Table 1 and Fig. 7 K and L). Similar results showing a more-than-additive increase in the number of autonomous seeds were obtained for swn-2 and -4 alleles in combinations with mea-3 (Table 1). These data indicate that most of the FGs inheriting both the swn-3 and mea-3 mutant alleles developed autonomously. In addition, all double-mutant combinations of the swn alleles and mea-3 (including double heterozygotes) exhibited an enhanced silique elongation phenotype compared with mea-3/+ single-mutant plants (Table 1). Therefore, swn mutations enhanced the number of autonomous seeds forming in mea plants (and consequently the length of autonomous siliques) indicating functional overlap between MEA and SWN in the control of seed initiation before fertilization.
To determine the extent of autonomous endosperm development on a seed basis, we measured the size of autonomous seeds formed in swn;mea-3 double mutants and compared them with mea single-mutant plants. The average sizes of autonomous seeds were significantly different (P < 0.001, two-sample Student’s t test) between mea-3/+ single-mutant (0.043 mm3, n = 47) and swn-3/swn-3;mea-3/+ double-mutant (0.124 mm3, n = 55) plants. Microscopic analysis indicated that the autonomous seeds obtained from emasculated swn-4/swn-4;mea-3/+ double-mutant plants produced more free-nuclear autonomous endosperm than mea-3/+ single-mutant plants (Fig. 8, which is published as supporting information on the PNAS web site). Thus, the swn;mea-3 mutant plants exhibit an enhanced penetrance for development of the autonomous seeds as compared to mea single-mutant plants; they show an increase in the frequency of individual mutant FGs capable of producing autonomous seeds and an increase in the quantity of autonomous endosperm proliferation in each mutant FG. Together, our data indicate that SWN performs a partially redundant role to MEA in controlling seed initiation by helping to suppress central cell nucleus/endosperm proliferation within the FG.
Discussion
We have found that FIS2 can interact specifically with two SET-domain PcG proteins, MEA and SWN, in Y2H assays and in vitro (Fig. 1). All three proteins are expressed in overlapping patterns in planta, particularly during late FG development (Fig. 2). In addition, swn mutations enhance the autonomous seed development phenotype of a mea mutant (Table 1 and Fig. 7), indicating functional overlap between SWN and MEA in control of endosperm proliferation and initiation of seed development.
Results from our protein-interaction studies indicating FIS2–MEA and FIS2–SWN interactions are supported by the recent finding that SUZ12, the mammalian counterpart of the Drosophila Su(z)12, directly interacts with the SET-domain PcG protein EZH2 (23). The SUZ12/Su(z)12 genes are homologous to the Arabidopsis family of Zn-finger PcG genes that include VERNALIZATION2 (VRN2), EMBRYONIC FLOWER2 (EMF2), and FIS2 (9, 17, 22, 25). Drosophila Su(z)12 is required to maintain the repressed state of Hox genes throughout development. In addition, Su(z)12 functions in germ-cell development and heterochromatin-mediated silencing (25). The SUZ12 protein is a component of the Drosophila and mammalian PRC2/3 complex that functions in initiating transcriptional repression and is associated with methylation of histone H3 at Lys-27 (H3K27; refs. 29–32). In fact, this histone methyltransferase activity in mammalian cells requires a minimum of three proteins that include EZH2, EED, and SUZ12 (14). Depletion of SUZ12 activity can lead to severe developmental and proliferative defects in knockout mice and in cell lines, presumably because of a loss of H3K27 marks and subsequent up-regulation of target genes (33, 34). Therefore, EED, EZH2, and SUZ12 constitute the core of a functional PRC2/3 complex in animals.
Our data and previous studies by others (12) support the notion that PRC2/3-like complexes control key transitional events during the Arabidopsis life cycle. Genetic evidence in Arabidopsis indicates that EZH2- and SUZ12-like proteins, encoded by multigene families, have specific roles in control of seed initiation, flower organ development, and vernalization-mediated initiation of flowering (4, 6, 9, 17, 18, 22). However, recent data indicate that EZH2-like genes may have redundant roles during plant development. Chanvivattana et al. (16) have shown that SWN is responsible for partially masking the clf mutant phenotype. Homozygous swn/swn plants do not exhibit any obvious morphological defects. However, swn mutant alleles were shown to enhance the clf mutant phenotypes in swn;clf double-mutant combinations, producing very small, early-flowering plants. Our data indicate a similar role for SWN in the control of seed initiation. Null alleles of MEA do not exhibit a fully penetrant phenotype in the production of autonomous seeds (5, 6). Here, swn mutations enhanced the mea mutant phenotype in double-mutant combinations, increasing the penetrance of autonomous seed development observed in mea/+ single-mutant plants to nearly 100%. Thus, SWN has a role in at least two phases of the plant life cycle, although it is seemingly essential for neither (see below).
Consistent with the genetic data, protein–protein interaction studies indicate that SWN and MEA are capable of interacting with SUZ12 homologues and thus may form distinct PcG complexes. Full-length SWN and MEA proteins can interact directly with FIS2 in Y2H and in vitro pull-down assays (Fig. 1B). Moreover, these interactions are mediated through the VEFS domain of FIS2. We show that a C-terminal portion of FIS2 containing part of the VEFS domain is necessary for binding to both SWN and MEA (Fig. 1A; data not shown). Similarly, the SUZ12 VEFS domain has been shown to be required for the interaction between SUZ12 and EZH2 (23). In Drosophila, a C-terminal truncation of Su(z)12 causes homeotic transformations and ectopic activation of Hox genes (25), indicating a conserved role for the VEFS domain. Interestingly, we did not observe any detectable interaction between full-length CLF and FIS2 proteins (Fig. 1B). This result is in contrast to a published report showing an extensive repertoire of interactions between partial SWN/CLF proteins and VEFS domains of EMF2/VRN2/FIS2 proteins using Y2H assays (16). In our hands, the C-terminal VEFS domain is in fact sufficient for interaction with full-length SWN, MEA, and CLF proteins (Fig. 1 A and B), whereas a longer FIS2 peptide (FIS2234–813) does not interact with CLF in Y2H assays (data not shown). This result indicates that the more central AB-repeat sequences that are specific to FIS2 (9) may control the specificity of interaction between FIS2 and its SET-domain partner. Thus, our protein-interaction data indicate that the specificity of the plant PRC2/3 complexes may be regulated by restricted interactions between SET-domain and Zn-finger PcG proteins.
The specificity of the plant PcG function may also be conferred through the regulation of specific components of the complex (15, 16). Our data indicate that MEA and FIS2 proteins are highly restricted in their pattern of accumulation during development (Fig. 2 K–T). Recent data indicate that various transcriptional-regulatory mechanisms, including PcG-mediated mechanisms, contribute to establishing this pattern of expression (35–39). In contrast, SWN is localized in the nucleus at all stages of development except in pollen grains and during endosperm development (Figs. 2 A–J and 3), suggesting that the SWN gene is not under the same tight regulatory control. Phylogenetic analysis indicates that SWN and CLF are more closely related to each other than to MEA, and that the SWN/CLF clade is the more ancestral one as compared with the MEA clade (16). Therefore, the MEA gene may have gained its specific role in the control of initiation of seed development, in part through acquiring both positive and negative regulatory sequences that confer FG- and early-endosperm-specific expression.
Our data support the notion that the functional specificity of the individual PcG complexes may be determined, at least in part, by the pairing of E(z)-Su(z)12 homologues, itself controlled by the availability of the specific protein components and their inherent ability to interact with each other. How this specificity would translate to the recruitment of individual plant PcG complexes to their specific target genes remains to be determined. In Drosophila, recruitment of PcG complexes is initiated by proteins such as Pleiohomeotic and Zeste, bound at Polycomb Response Elements (PRE) in the regulatory regions of the target genes (40, 41). To date, there is no direct evidence in plants for the existence of a PRE-like sequence or a PRE-binding protein that would recruit plant PcG proteins.
What is the function of SWN during plant development? The SWN, MEA, and CLF proteins may represent discrete PcG regulatory complexes or modules that regulate distinct and overlapping sets of target genes. Although it is expressed widely, the SWN module may regulate only a subset of a larger group of target genes that are controlled by the CLF and MEA modules during flowering and early seed development, respectively. Consequently, the function of SWN would be completely masked by CLF and MEA, whereas SWN would partially mask the function of CLF and MEA. Such a scenario could drive rapid accumulation of mutations in SWN and is therefore inconsistent with the current phylogenetic data (16). However, other components of the SWN module (including its direct protein partners) may impose constraints on its rapid divergence (42). Alternatively, SWN may not have an equivalent role to CLF or MEA in forming independent PcG complexes. For example, it may play a more fundamental role in chromatin regulation, a function that is masked by yet other unknown proteins. A full understanding of the SWN function will require a detailed analysis of all of its partners and target genes.
Materials and Methods
Plant Materials and Growth Conditions.
The swn-2 and -3 alleles described in ref. 16 and the swn-4 allele (SALK_109121) were obtained from the Salk Institute Genomic Analysis Laboratory (28) through the Arabidopsis Biological Resource Center. The mea-3 mutant allele was originally obtained in Ler (6). Details on the single- and double-mutant genotypes used for the analysis of phenotypes and the relevant growth conditions are provided as Supporting Text, which is published as supporting information on the PNAS web site.
Analysis of FIS2 Gene Structure and mRNA Accumulation.
Standard procedures were used to clone a full-length FIS2 cDNA and to analyze mRNA accumulation patterns using RT-PCR (see Supporting Text).
Directed Y2H–Interaction Studies, Y2H cDNA Library Construction, and Screen.
A detailed description of constructs used for directed Y2H studies, Y2H library construction, and the bait for the Y2H screen is provided as Supporting Text.
In Vitro Coimmunoprecipitation Assays.
The Matchmaker Co-IP kit (BD Biosciences, Heidelberg, Germany) was used according to the manufacturer’s protocol. All proteins were synthesized by using the TnT T7 quick-coupled transcription/translation system (Promega, Madison, WI) in the presence of [35S]methionine (Amersham Pharmacia, Uppsala, Sweden). The cDNA clones of FIE, MEA, SWN, and FIS2 in pGADT7 or pGBKT7 vectors were used as templates for in vitro protein synthesis, as described in Supporting Text. Because of the difficulty in expressing a full-length version of FIS2 in vitro, we used a myc-tagged FIS2 C-terminal fragment (amino acids 641–813) in combination with full-length MEA and SWN proteins tagged with an HA epitope.
Reporter Constructs and Plant Transformation.
Construction and transformation of Arabidopsis with the SWN-GFP/YFP, FIS2-GFP/YFP, and MEA-YFP transgenes were carried out by using standard methods (see Supporting Text).
Thirty-one, 10, 18, 21, and 12 independent Kanamycin-resistant T1 plants were generated for SWN-GFP, SWN-YFP, FIS2-GFP, FIS2-YFP, and MEA-YFP constructs, respectively. FIS2-GFP/YFP and MEA-YFP showed consistent patterns of transgene expression. All SWN-GFP/YFP lines (31 and 10) showed nuclear localization of GFP/YFP within the FG (Figs. 2 B–F and 6). The majority of SWN-GFP (27 of 31) and SWN-YFP (8 of 10) lines showed high levels of GFP/YFP signal in integument cells (Figs. 3A and 6). The remaining SWN-GFP (Fig. 2 A–J) and SWN-YFP (data not shown) lines displayed low levels of GFP/YFP signal in integument cells.
Light and Epifluorescence Microscopy.
Ovules, seeds, roots, and leaves were excised and imaged as described (24). Autonomous seeds were fixed, cleared, and imaged following a published protocol (43). Bright-field and epifluorescence images were obtained by using an Axiophot compound epifluorescence microscope (Zeiss, Jena, Germany) equipped with a GFP bandpass filter (exciter 450–490, dichroic 495, emitter 500–550; Chroma Technology, Brattleboro, VT), YFP bandpass filter (exciter 490–510, dichroic 515, emitter 520–550; Chroma Technology), and FITC longpass filter (exciter 450–490, dichroic 510, emitter 515; Zeiss). Intact and dissected siliques were analyzed and imaged by using an SZX stereomicroscope (Olympus, Melville, NY). All microscopic images were captured by using an Optronics MicroFire (Goleta, CA) CCD camera and processed with Photoshop CS (Adobe Systems, San Jose, CA).
Analysis of Fertilization-Independent Phenotypes.
Flowers corresponding to stages 12B–12C (44) were emasculated from two to three individual plants for each genotype. After 7 days, siliques and autonomous seeds were imaged, and the numbers of autonomous seeds developing per silique were recorded. Methods for measuring silique length and autonomous seed size are provided as Supporting Text. Data for silique length and percentage of autonomous seed were analyzed by using PROC GLM in SAS (45), assuming a mixed-effects model (see Supporting Text). Data for autonomous seed size were analyzed by using Student’s t test (see Supporting Text). Statistical significance was assigned at P = 0.05 throughout.
Supplementary Material
Acknowledgments
We thank Gary Drews and Karen Schumaker for critical reading of the manuscript; Sasha Schlicher and Amanda Durbak for the initial characterizations of the swn alleles; Gary Drews (University of Utah, Salt Lake City, UT) for providing the fis2-8 allele; David Jackson (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for providing the Citrine-YFP reporter gene and linker sequences; Byeong-ha Lee for creating the GFP/YFP binary vectors; the Arabidopsis Biological Resource Center for providing the swn alleles; and Steven Smith (University of Arizona, Tucson, AZ) for help with the statistical analysis of mutant phenotypes. M.D.T. and S.S.J. were supported in part through the Undergraduate Biology Research Program, which is funded by Grant 52003749 from the Howard Hughes Medical Institute to the University of Arizona. This work was supported by research setup funds from the Department of Plant Sciences, University of Arizona; Southwest Consortium on Plant Genetics and Water Resources Grant 2004-34186-14533; and U.S. Department of Energy Basic Energy Sciences Grant DE-FG02-03ER15438 (to R.Y.).
Abbreviations
- PcG
polycomb group
- FG
female gametophyte
- PRC2/3
polycomb repressive complex 2/3
- Y2H
yeast two-hybrid
- YFP
yellow fluorescent protein
- VEFS domain
VRN2-EMF2-FIS2-Su(z)12 domain.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ275158).
References
- 1.Yadegari R., Drews G. N. Plant Cell. 2004;16:Suppl. S133–S141. doi: 10.1105/tpc.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa L. M., Gutierrez-Marcos J. F., Dickinson H. G. Trends Plant Sci. 2004;9:507–514. doi: 10.1016/j.tplants.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Olsen O. A. Plant Cell. 2004;16:Suppl. S214–S227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossniklaus U., Vielle-Calzada J. P., Hoeppner M. A., Gagliano W. B. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 5.Guitton A. E., Page D. R., Chambrier P., Lionnet C., Faure J. E., Grossniklaus U., Berger F. Development (Cambridge, U.K.) 2004;131:2971–2981. doi: 10.1242/dev.01168. [DOI] [PubMed] [Google Scholar]
- 6.Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., Katz A., Margossian L., Harada J. J., Goldberg R. B., et al. Proc. Natl. Acad. Sci. USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. EMBO J. 2003;22:4804–4814. doi: 10.1093/emboj/cdg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo M., Bilodeau P., Dennis E. S., Peacock W. J., Chaudhury A. Proc. Natl. Acad. Sci. USA. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo M., Bilodeau P., Koltunow A., Dennis E. S., Peacock W. J., Chaudhury A. M. Proc. Natl. Acad. Sci. USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spillane C., MacDougall C., Stock C., Kohler C., Vielle-Calzada J. P., Nunes S. M., Grossniklaus U., Goodrich J. Curr. Biol. 2000;10:1535–1538. doi: 10.1016/s0960-9822(00)00839-3. [DOI] [PubMed] [Google Scholar]
- 11.Vielle-Calzada J. P., Thomas J., Spillane C., Coluccio A., Hoeppner M. A., Grossniklaus U. Genes Dev. 1999;13:2971–2982. doi: 10.1101/gad.13.22.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert D., Clarenz O., Goodrich J. Curr. Opin. Plant Biol. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Levine S. S., King I. F., Kingston R. E. Trends Biochem. Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Cao R., Zhang Y. Curr. Opin. Genet. Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Kohler C., Grossniklaus U. Curr. Opin. Cell Biol. 2002;14:773–779. doi: 10.1016/s0955-0674(02)00394-0. [DOI] [PubMed] [Google Scholar]
- 16.Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y. H., Sung Z. R., Goodrich J. Development (Cambridge, U.K.) 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 17.Gendall A. R., Levy Y. Y., Wilson A., Dean C. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- 18.Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E. M., Coupland G. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 19.Hennig L., Taranto P., Walser M., Schonrock N., Gruissem W. Development (Cambridge, U.K.) 2003;130:2555–2565. doi: 10.1242/dev.00470. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita T., Harada J. J., Goldberg R. B., Fischer R. L. Proc. Natl. Acad. Sci. USA. 2001;98:14156–14161. doi: 10.1073/pnas.241507798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon Y. H., Chen L., Pan R. L., Chang H. S., Zhu T., Maffeo D. M., Sung Z. R. Plant Cell. 2003;15:681–693. doi: 10.1105/tpc.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida N., Yanai Y., Chen L., Kato Y., Hiratsuka J., Miwa T., Sung Z. R., Takahashi S. Plant Cell. 2001;13:2471–2481. doi: 10.1105/tpc.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto K., Sonoda M., Inokuchi J., Shirasawa S., Sasazuki T. J. Biol. Chem. 2004;279:401–406. doi: 10.1074/jbc.M307344200. [DOI] [PubMed] [Google Scholar]
- 24.Yadegari R., Kinoshita T., Lotan O., Cohen G., Katz A., Choi Y., Nakashima K., Harada J. J., Goldberg R. B., Fischer R. L., Ohad N. Plant Cell. 2000;12:2367–2381. doi: 10.1105/tpc.12.12.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birve A., Sengupta A. K., Beuchle D., Larsson J., Kennison J. A., Rasmuson-Lestander A., Muller J. Development (Cambridge, U.K.) 2001;128:3371–3379. doi: 10.1242/dev.128.17.3371. [DOI] [PubMed] [Google Scholar]
- 26.Christensen C. A., King E. J., Jordan J. R., Drews G. N. Sex. Plant Reprod. 1997;10:49–64. [Google Scholar]
- 27.Brown R. C., Lemmon B. E., Nguyen H., Olsen O.-A. Sex. Plant Reprod. 1999;12:32–42. [Google Scholar]
- 28.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 29.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 30.Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 31.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., O’Connor M. B., Kingston R. E., Simon J. A. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 33.Cao R., Zhang Y. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 34.Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi Y., Gehring M., Johnson L., Hannon M., Harada J. J., Goldberg R. B., Jacobsen S. E., Fischer R. L. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 36.Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., Goldberg R. B., Fischer R. L. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao W., Gehring M., Choi Y., Margossian L., Pu H., Harada J. J., Goldberg R. B., Pennell R. I., Fischer R. L. Dev. Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- 38.Baroux C., Gagliardini V., Page D. R., Grossniklaus U. Genes Dev. 2006;20:1081–1086. doi: 10.1101/gad.378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jullien P. E., Katz A., Oliva M., Ohad N., Berger F. Curr. Biol. 2006;16:486–492. doi: 10.1016/j.cub.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 40.Ringrose L., Paro R. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz Y. B., Kahn T. G., Dellino G. I., Pirrotta V. Cold Spring Harbor Symp. Quant. Biol. 2004;69:301–308. doi: 10.1101/sqb.2004.69.301. [DOI] [PubMed] [Google Scholar]
- 42.Pereira-Leal J. B., Levy E. D., Teichmann S. A. Philos. Trans. R. Soc. London Ser. B. 2006;361:507–517. doi: 10.1098/rstb.2005.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohad N., Margossian L., Hsu Y. C., Williams C., Repetti P., Fischer R. L. Proc. Natl. Acad. Sci. USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smyth D. R., Bowman J. L., Meyerowitz E. M. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Littell R. C., Milliken G. A., Stroup W. W., Wolfinger R. D. SAS System for Mixed Models. Cary, NC: SAS Institute; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.