Abstract
It has been reported that cellular prion protein (PrPc) is enriched in caveolae or caveolae-like domains with caveolin-1 (Cav-1) participating to signal transduction events by Fyn kinase recruitment. By using the Glutathione-S-transferase (GST)-fusion proteins assay, we observed that PrPc strongly interacts in vitro with Cav-1. Thus, we ascertained the PrPc caveolar localization in a hypothalamic neuronal cell line (GN11), by confocal microscopy analysis, flotation on density gradient, and coimmunoprecipitation experiments. Following the anti-PrPc antibody-mediated stimulation of live GN11 cells, we observed that PrPc clustered on plasma membrane domains rich in Cav-1 in which Fyn kinase converged to be activated. After these events, a signaling cascade through p42/44 MAP kinase (Erk 1/2) was triggered, suggesting that following translocations from rafts to caveolae or caveolaelike domains PrPc could interact with Cav-1 and induce signal transduction events.
INTRODUCTION
Multiproteic caveolar complexes represent a sophisticated membrane organization involved in signal transduction. Their efficiency is linked to the insertion of proteins in a restricted membrane area (50–100 nm) where the generation of a signal has a vectorial and oriented characteristic and allows the recruitment of low-abundance proteins in order to activate signaling pathways which, in neural cells, seem to control differentiation and cell survival [1–4]. Caveolae are a subclass of membrane microdomains distinguishable by their shape (they are flask-like invaginations) and by the presence of membrane proteins of the caveolin family. Caveolin-1 (Cav-1) is a small 22 kDa highly versatile protein capable of organizing several caveolar functions. Lisanti and coworkers have precisely mapped the molecule defining two sites involved in the binding of caveolar constituents: a hydrophobic region (aa 82–101) called scaffolding domain (SD), and amore hydrophilic motif present in the C-terminal region indicated as CID motif [5].
Cellular prion protein (PrPc) is a secreted protein anchored to cell surface via a GPI anchor and believed to function as a cell surface receptor [6] or ligand [7, 8]. PrPc is characterized by an amino terminal unstructured highly flexible region characterized by the presence of multiple octapeptide repeats highly conserved during evolution that are binding sites for copper ions [9]. In neuroblastoma cells lacking caveolae, PrPc has been isolated in detergent-insoluble complexes denominated “caveolae-like domains” (CLDs) and it has been hypothesized that PrPc conversion in its pathological conformer PrP scrapie (PrPsc) occurs in this subcellular compartment [10, 11]. Recent data obtained by electron microscopy in CHO cells clearly confirmed that PrPc is internalized by caveolae [12]. Moreover, it has been observed that Cav-1 is coimmunoprecipitated by using PrPc antibody and that Cav-1 mediates the recruitment and the activation of Fyn kinase after anti-PrPc antibody-mediated stimulation [13, 14]. Evidence supporting a role of PrPc in regulating cell proliferation, differentiation, and survival has been collected [15].
Fyn kinase is a member of Src family kinase involved in signal transduction events. It has been reported that Fyn kinase during signal transduction events is noncovalently associated with glycosylphosphatidylinositol (GPI)-anchored proteins [16–18] and that Fyn kinase, after the palmitoylation of its Cys3, is included in caveolae [19].Moreover, it has been shown that, following antibody-mediated cross linking, GPI-anchored proteins lead to signal transduction events in T cells, B cells, monocytes, and granulocytes [20] and that are sequestered in caveolae [21].
Erk 1/2 has been intensively studied in neurons because of its participation to hippocampal mechanisms leading to learning and memory consolidation [22]. Caveolae play an important role in Erk 1/2 regulation. In fact, it has been reported that Erk 1/2 is compartmentalized within caveolae [23, 24] and that Cav-1 can inhibit Erk 1/2 activity [25–29]. Interestingly, it has been reported that a reciprocal relationship between Cav-1 and Erk 1/2 as activation of the p42/44 MAP kinase cascade causes the downregulation of Cav-1 expression [30].Moreover, the role of PrPc in Erk 1/2 activation has been analyzed [14, 31, 32].
Findings reported here demonstrate that PrPc and Cav-1 interact in vitro and colocalize in GN11 cells, a hypothalamic neuronal cell line that highly expresses Cav-1 gene.Moreover, we examined the role played by caveolae and PrPc in signal transduction by transfecting GN11 cells with a novel PrPcexpressing vector showing a high transfection efficiency, in order to compare Fyn and Erk 1/2 kinases activity in wildtype and PrPc-overexpressing cells. Our results highlight the key role of caveolae as sophisticated microenvironments in which PrPc clusters to generate signal transduction pathways.
MATERIAL AND METHODS
Antibodies used
Anti-PrPc monoclonal antibody (Mab) 3F4 (western Blot (WB) 1 : 3000; Immunofluorescence (IF) 1 : 50; DakoCytomation, Denmark); antimurine PrP-Nterminus polyclonal antiserum Ab · Tg (kindly provided by Dr T. Yokoyama, Japan [33]), anti-human PrP-C terminus goat polyclonal antibody (Pab) C-20 (Santa Cruz Biotechnology, USA); antihuman recombinant Doppel protein (hurDpl) Pab Q55 (WB 1 : 100); Mab Dpl 79 (kindly provided by Dr J. Grassi, Commissariat a l'Energie Atomique/Saclay, France); anti-Cav-1 Pab (WB 1 : 5000; IF 1 : 100; BD Biosciences, USA), anti- Cav-1 FITC-conjugated antibody (IF 1 : 50; Santa Cruz Biotechnology, USA), antihaemagglutinin (HA)-epitope Pab (BD Biosciences, USA); anti-Fyn kinase Pab (IF 1 : 100; Santa Cruz Biotechnology, USA), anti-phospho-Src family (Tyr416) Pab (IF 1 : 200; Cell Signaling, USA), anti-phospho Erk1/2 Mab (WB 1 : 1000; Cell Signaling, USA); anti--actin Pab (WB 1 : 1200; Sigma, USA), secondary anti-mouse FITC- or Cy3-conjugated antibodies (IF 1 : 50, Sigma, USA); anti-rabbit Cy5-conjugated donkey Ab F(ab')2 fragment (IF 1 : 50, Jackson, USA); anti-mouse and anti-rabbit secondary Horseradish Peroxidase (HRP)-conjugated antibodies (WB 1 : 1000, Santa Cruz Biotechnology, USA). Antibodies were diluted in PBS-BSA 3% or in 5% nonfat milk powder-TBS-TWEEN buffer, for IF or WB experiments, respectively. For live cells stimulation the antibodies were used at the dilution of 1 : 100 in DMEM supplemented with 10% FCS.
Plasmid construction
Haemagglutinin-tagged PrP (PrP-HA) plasmid was prepared as follows: the coding region (23–254) of mouse PrPc was prepared, as described in Negro et al [34], with two Mets at positions 108 and 111 (L108M, V111M) to provide the epitope specific for commercial Mab 3F4. Then, the coding region was cloned into the mammalian expression vector pRK7HA (a kind gift of Dr Elisabetta Ciani, Bologna, Italy), downstream of the HA-tag sequence, between BamHI and EcoRI restriction sites. The region coding for mouse PrP signal peptide (1–22) was then cloned into the recombinant vector, upstream of the HA epitope sequence, between AgeI and HindIII restriction sites. Expression vector for human Dpl was prepared as in [34].
GST-binding assay
Recombinant full-length forms ofmorPrP (23–231) and hur- Dpl (28–152) were generated in, and purified from, Escherichia coli as described in Negro et al [34]. To provide the epitope specific for Mab 3F4, morPrP carried two Mets at positions 108 and 111 (L108M, V111M). Murine Cav-1 forms were recombinantly obtained as GST-fusion proteins as described in [35]. For binding assays, 0.2 μM of Glutathione STransferase (GST) or GST-Cav fusion proteins (prebound to glutathione-Sepharose beads) were incubated overnight with equimolar amounts of morPrP or hurDpl (in a final volume of 0.25 ml), under continuous shaking at 4°C. Proteins bound to glutathione-Sepharose beads were eluted, washed, and immunoblotted to detect the presence of PrP or Dpl. In competition experiments, equimolar amounts of either Ab · Tg or C-20 antibodies were present during the entire incubation period.
Determination of the dissociation constant for the binding GST-Cav fusion proteins and morPrP
Different concentrations of morPrP (0.03–0.7 μM) were incubated with 0.2 μMGST-Cav fusion proteins (GST-Cav 61–101, GST-Cav 135–178, and GST-Cav FL 1–178) in the same conditions described above. Proteins bound to glutathione-Sepharose beads were eluted and exposed to western blotting assay. The blots, stained with anti-PrP Mab 3F4, were analyzed in transmitted light by ImageMaster VDS software (Pharmacia Biotech, Sweden) to measure the intensity of the bands. PrP concentration was plotted as a function of band intensity expressed in arbitrary units.
Western blot
For western blot analysis, denatured proteins were first separated by a 12% SDS-PAGE, electrophoretically transferred onto nitrocellulose and then, depending on the experiment, incubated with the specific antibodies. Immunolabeling was visualized by ECL procedure (Amersham, UK) and the band intensities were measured by ImageMaster VDS software (Pharmacia Biotech, Sweden).
Cell culture and transfection
Murine GN11 cells [36] were grown at 37°C in 5% CO2 atmosphere in high glucose-culture medium (DMEM, Sigma, USA), supplemented with 10% heat-inactivated fetal calf serum (FCS, Cambrex BioWhittaker, USA), 2mM L-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml), as described in [37].
Cells were transiently transfected by using Lipofectamine 2000 reagent (Invitrogen, USA), following manufacturer's instructions. For cell differentiation, phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) (20 nM, Sigma, USA) was added to cells soon after the transfection step, depending on the experiment. For Western blot experiments, cells were trypsinized 24 hours after transfection and reseeded in 25 cm2 flasks to obtain the same number of transfected cells in each flask. To assess the Erk 1/2 phosphorylation status, cells were serum-starved for 4 hours before antibody-mediated ligation of PrPc. Depending on the experiments, 30 μM PP2 (Calbiochem, USA) was added to the medium for 4 hours. In both immunoblotting and immunocytochemistry experiments, 48 hours after transfection, antibody-mediated stimulation was carried out first by incubating (10, 20, or 30 min, 37°C) intact live cells withMab 3F4, Mab Dpl 79, or anti--actin Pab and then with secondary anti-mouse or anti-rabbit antibodies for additional 10–120 min. (37°C), depending on the experiment. Then, cells were lyzed in lysis buffer (50mM Tris-HCL, pH 7.5, 2 mM EDTA, 100 mM NaCl, 1% Triton X100, 5 mM NaF, 1 mM Na3VO4, 10 mM β-glycerolphosphate, and proteases inhibitors) or fixed with 4% paraformaldehyde, for western blot or immunofluorescence analysis, respectively.
Immunocytochemistry
PrPc-overexpressing GN11 cells grown on coverslips were fixed with 4% paraformaldehyde and incubated with Mab 3F4 or were stimulated by antibody-mediated ligation and fixed. Cells were permeabilized with Triton X-100 (0.1% in phosphate buffer saline (PBS)) and saturated in PBS-BSA 3% for 30 min. Then, cells were incubated with anti-Cav- 1 or anti-phospho Fyn Pabs (for double fluorescence staining experiments) or with anti-Fyn kinase Pab (for triple fluorescence staining experiments). After washings, cells were incubated with anti-mouse FITC-conjugated and with antirabbit Cy5- (for double staining) or Cy3- (for triple staining) conjugated secondary antibodies. For triple immunofluorescence staining experiments, anti-Cav-1 FITC-conjugated antibody was added. Finally, slides were mounted in glycerol- PBS medium containing 30 mg/ml DABCO (Sigma, USA). Evaluation of antibody specificity was carried out either by omitting primary antibody or by using unspecific sera.
Confocal microscopy
The confocal imaging was performed on a Radiance 2000 confocal laser scanning microscope (BioRad), equipped with a Nikon 40x, 1.4 NA objective, and with a Krypton and a Red Diode lasers, to excite FITC (green) and Cy5 (red) fluorescence simultaneously. Colocalization was evaluated on medial optical sections using LaserPix software (BioRad) [38]. Briefly, the two-dimensional scatter plot diagram of each cell was analyzed to evaluate the spatial colocalization of the fluorochromes. For each scatter plot diagram, pixels with highly colocalized fluorochromes, that is, with intensity values greater than 150 gray levels (on a scale from 0 to 255) for both detectors were selected to calculate the colocalization maps and create a binary image.
Optiprep gradient flotation
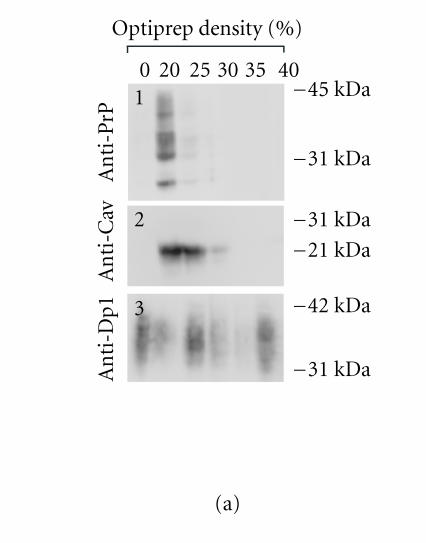
Optiprep density gradient flotation was performed as described in Harder et al [39]. Briefly, PrPc-overexpressing TPA-differentiated cells (one confluent 25 cm2 flask) were lyzed (20 min, 4°C) in 200 μl of a TNE/TX buffer (25mM Tris HCl (pH 7.4), 150mM NaCl, 5mM EDTA, 5mM DTT, 1% Triton X-100, and protease inhibitors),mixed with 400 μl of 60% Optiprep reagent (Sigma, USA), placed at the bottom of an ultracentrifuge tube, and overlaid with a 600 μl step of each of 35%, 30%, 25%, 20%, and 0% Optiprep (in TNE/TX). The gradient was spun at 200.000 xg (4 hours, 4°C), after which fractions were collected from the top of the gradient, acid-precipitated, and analyzed by western blot.
Coimmunoprecipitation assay
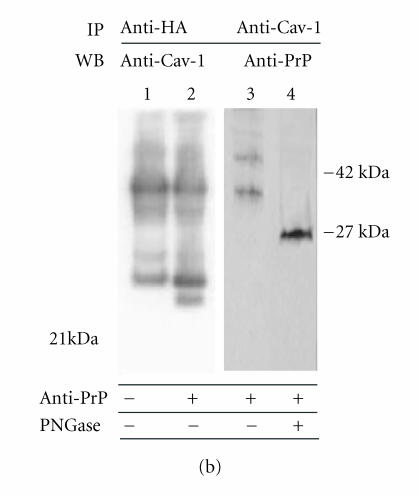
In Cav-1 immunoprecipitation experiments, cells transiently expressing the HA-tagged PrPc (one confluent 25 cm2 flask for each sample) were lyzed in 500 μl of a buffer containing 10mM Tris (pH 7.5), 150mM NaCl, 1mM EDTA, 1% Triton X-100, 0.5% NP40, 10% glycerol, and protease inhibitors. Precleared supernatants were immunoprecipitated using anti-Cav-l Pab (3 μg/ml), followed by incubation with protein A Sepharose [35]. After washings, immunoprecipitated samples were processed by western blot to detect PrPc. For sample deglycosylation, immunoprecipitates were treated (24 hours, 37°C) with PNGase-F (5 U) (Roche Molecular Biochemicals, Germany) [40]. For PrP immunoprecipitation, the same protocol described above was followed, except for the use of an anti-HA tag antibody to immunoprecipitate. The presence of Cav-1 in the immunoprecipitated samples was then assayed by immunoblotting with anti Cav-1 Pab.
RESULTS AND DISCUSSION
Interaction between PrP and Cav-1
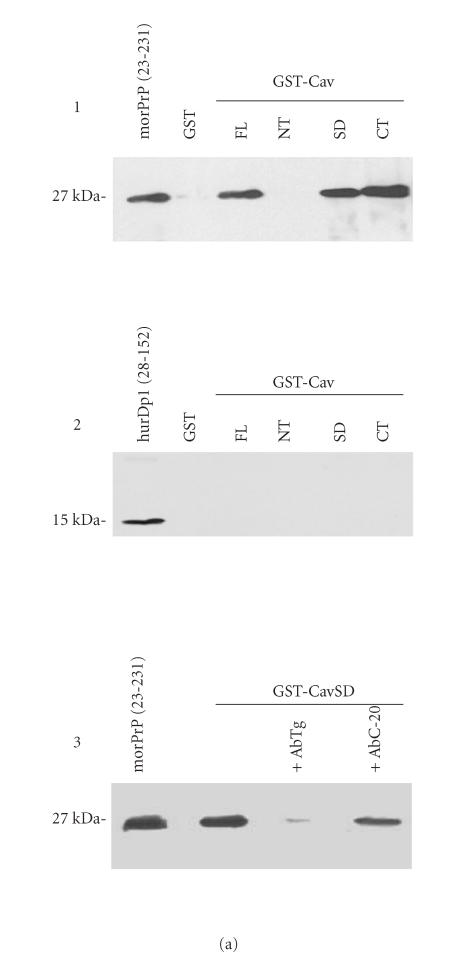
To evaluate the interaction between Cav-1 and PrPc, we carried out in vitro binding experiments as previously described in [35], by using four different GST-Cav1 fusion proteins and a control GST-nonfusion protein to challenge mor- PrP. Bound PrP molecules were then immunodetected by western blot analysis. Results clearly show that GST-CavFL (full lenght, aa 1–178), GST-CavSD (scaffolding domain, aa 61–101), and GST-CavCT (C-terminal region, aa 135–178) very efficiently bind morPrP. No morPrP was bound when GST alone and GST-CavNT (N-terminal region, aa 1–81) were challenged, indicating that the binding was specific (Figure 1(a), 1).
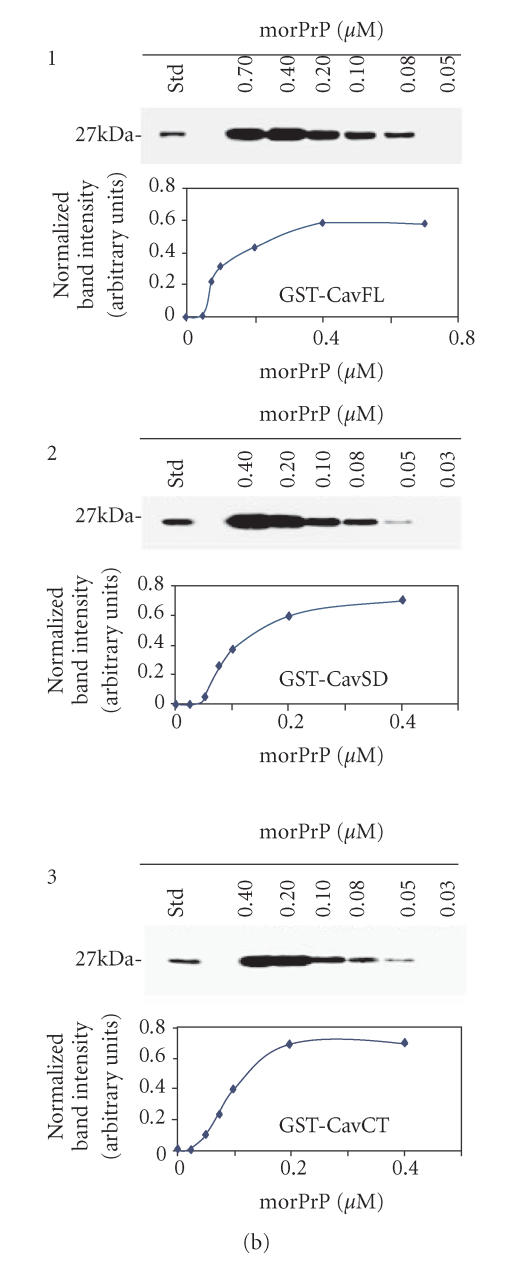
Figure 1.
(a) In vitro interaction between Cav-1 andmorPrP. To assess the binding of morPrP (23–231) and hurDpl (28–152) to different Cav-1 domains (CavFL: full-length form (1–178); CavNT: N-terminal domain (1–81); CavSD: scaffolding domain (61–101); CavCT: C-terminal domain (135–178)), prion proteins were incubated with equimolar amounts (0.2 μM) of GST-Cav fusions bound to glutathione-Sepharose beads. Bound morPrP (1, 3) and hurDpl (2) were then visualized by western blotting with Mab 3F4 and polyclonal antibody Q55, respectively. In 3, the binding assay was carried out in the presence of polyclonal antibodies (0.2 μM) to PrP, namely, Ab · Tg recognizing the octapeptide region, and C-20 directed against the C-terminus. In the first lane of (a) 1, 2, 3, 50 ng morPrP or hurDpl were added as standard, while in the second lane of (1) and (2) GST alone (ie, not fused to Cav-1) was used for the binding experiments as negative control. Data shown are representative of three independent experiments. (b) Evaluation of Kd values of morPrP/Cav-1 complexes. Increasing concentrations of morPrP (0.03–0.7 μM) were incubated with GST-Cav 1 fusion proteins: CavFL in 1, CavSD in 2, and CavCT in 3, as described under Materials and Methods. The upper parts of (b) 1, 2, 3 show the results of western blotting experiments performed with anti-PrPc antibody (3F4). Lane 1, morPrP used as standard; lanes 2–7, 50 pmoles of GST-Cav 1 fusion proteins, CavFL in 1, CavSD in 2, and CavCT in 3, were incubated with the indicated amounts of morPrP (final volume 250 μl). In lower parts of each panel, the concentration of morPrP used in binding assays was plotted as a function of band intensity, which was quantified using ImageMaster VDS Software (Pharmacia Biotech, Sweden). The dissociation constants calculated from each plot ranged from 8 to 9 × 108 M−1 The data are representative of two independent experiments.
We hypothesized that the unstructured PrP N-terminal region [41] could represent an important binding site for Cav-1. This region is characterized by the presence of an octapeptide sequence PHGGGWGQ repeated five times in tandem and involved in the binding of copper ions [9]. To demonstrate our hypothesis, we first evaluated whether Dpl protein binds to Cav-1. Dpl is a protein which shows high sequence homology with the C-terminus of PrPc but lacks the N-terminal octapeptide region [42, 43]. To this end, we probed the hurDpl under the same conditions used for mor- PrP binding to Cav-1. However, contrary to morPrP, hurDpl showed no interaction with Cav-1 (Figure 1(a), 2).
More details about the PrPc N-terminal sites involved in the binding to Cav-1 were obtained using a polyclonal antibody Ab · Tg. This antibody specifically recognized two nonadjacent octapeptide repeats (57–65 and 81–89) since a substitution G/S was sufficient to abrogate recognition. The use of Ab · Tg antibody almost completely blocked the morPrP binding to GST-Cav SD (61–101), while an antibody directed against a PrP C-terminal epitope had a much lower effect (Figure 1(a), 3). Taken together, these results indicate that the octapeptide repeats could represent a PrP binding site for Cav-1.
The results of experiments aimed at evaluating the PrP/Cav-1 binding affinity constants are shown in Figure 1(b). Amounts of morPrP ranging from 0.03 μM to 0.7 μM were incubated with different GST-caveolin fusion proteins, as described under Material and Methods. Bands intensity in western blots was quantified by densitometry. Plots representing the binding of GST-CavFL (1–178), GSTCavSD (61–101), and GST-CavCT (135–178) are shown in panels B1, B2, and B3, respectively. Evaluation of dissociation constant (Kd) by reciprocal plots showed that Kd ranged from 8 to 9 × 108 M−1 for all the three fusion proteins. Since no binding was observed using GST alone or GST CavNT fusion protein, it appears that unspecific binding is not occurring in our experimental conditions.
To determine if the interaction between PrP and Cav-1 observed in vitro occurred also in a physiological environment and if this interaction had any effects on signal transduction, we used, as cellular model, a cell line of immortalized luteinizing hormone-releasing hormone (LHRH) neurons that naturally expresses high levels of Cav-1 [44] and that was prone to be transiently transfected with high efficiency with a plasmid encoding the HA tagged morPrP protein.
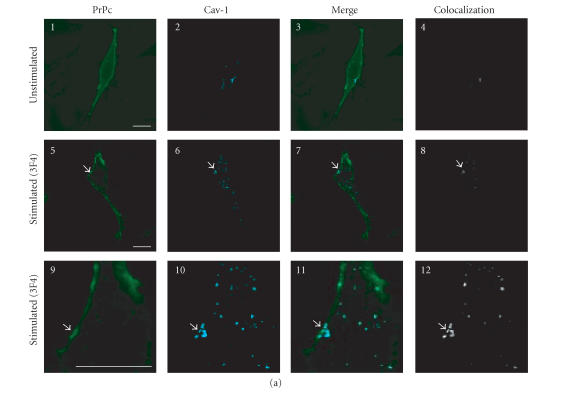
In PrPc-overexpressing GN11 cells, PrPc appears uniformly distributed on the plasma membrane (Figure 2(a), 1) and shows a low colocalization level with Cav-1 (Figure 2(a), 3 and 4). Given that it has been shown that GPI-anchored proteins residing in rafts translocate to caveolae following clustering induced by antibody-mediated cross-linki [21, 39], we stimulated live PrPc-overexpressing GN11 cells with anti-PrPc antibody (30 min) and with FITC-conjugated secondary antibody (30 min), observing that PrPc clusters in plasma membrane microdomains (in Figure 2(a), 5 and 9, arrowheads indicate the clusters of PrPc) rich in Cav-1 (Figure 2(a), 6 and 10). In these clusters we detected a high level of colocalization between PrPc and Cav-1 (Figure 2(a), 8 and 12).
Figure 2.
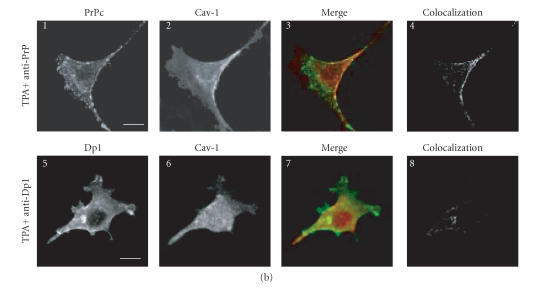
Confocal microscopy analysis of PrPc, Dpl, and Cav-1 distribution in GN11 cells transfected with morPrP or hurDpl. (a) PrP coverexpressing GN11 cells were fixed (1–4) and incubated with anti-PrP Mab 3F4 and with secondary FITC-conjugated antibody to allow the visualization of PrPc on the plasma membrane of unstimulated cells (1). Then cells were permeabilized and incubated with anti-Cav- 1 Pab to visualize Cav-1 distribution (2). Live GN11 cells PrPc transiently transfected were incubated with anti-PrP Mab 3F4 (30 min), followed by secondary FITC-conjugated antibody (30 min) (5–8, corresponding to 9–12 after magnification) to allow antibody-mediated ligation (and visualization) of PrPc (5), (9). After fixing, cells were permeabilized and incubated with anti-Cav-1 Pab and secondary CY5- conjugated antibody to visualize Cav-1 distribution ((6), (10)). Superposition of green PrPc ((1), (5), and (9)) and cyan-blue Cav-1 ((2), (6), and (10)) signals provided the images shown in 3, 7, and 11. The binary maps presented in 4, 8, and 12 are a more precise means to evaluate colocalization, whereby only those regions, in which PrPc and Cav-1 are copresent above a defined threshold of fluorescence intensity, appear.Micrographs show a single confocal plane. Arrowheads indicate PrPc clusters where Cav-1 is also present. Bar = 40 μm. (b) The same protocol used to analyze PrPc and Cav-1 distribution in GN11 cells in (a) was applied to GN11 cells stimulated by TPA (20 nM, 48 h) (1–4). Note that TPA treatment increases surface colocalization of PrPc (green) and Cav-1 (red) ((3), (4)). When the same methods were used to analyze the distribution of Dpl (green) and Cav-1 (red) in Dpl-transfected cells activated by anti-DplMab 79 antibody (5–8), no evident proteins colocalization was found ((7), (8)). For other experimental details see Material and Methods. Images result fromintegrating different optical sections. Bar = 5 μm.
In order to obtain a neuronal environment closer to that in which PrPc operates physiologically, we treated GN11 cells with TPA, an inducer of cell differentiation (Figure 2(b), 1–4). Analysis of PrPc-overexpressing GN11 cells treated with TPA has shown an increase in the co-localization level between PrPc and Cav-1 (Figure 2(b), 3 and 4). Since in vitro experiments (Figure 1(a), 2) have shown an incapacity of Cav-1 to bind Dpl and since Dpl is a surface protein exposed to the exoplasmic space through a GPI-anchor like PrPc [45], confocal microscopy analyses were performed on GN11 cells overexpressing the Dpl protein differentiated with TPA (Figure 2(b), 5–8). When Dpl was cross-linked using a specific antibody, colocalization with Cav-1 at plasma membrane level was undetectable (Figure 2(b), 7–8) confirming a different behavior between PrPc and Dpl and suggesting that Doppel, although located through a GPI anchor in the same microdomains of PrPc, could be unable to participate in the transduction of survival signals, thus favoring conditions slowly leading to apoptosis. For example, failure of Doppel to bind Cav-1 could impair Fyn kinase recruitment due to PrPc activation [13], thus altering the normal fate of cell differentiation occurring during brain development. Our results may also help to explain why the reintroduction of a wildtype Prnp gene, in PrPc 0/0 mice expressing Doppel protein in the brain, rescues them from neurodegeneration [46]. In fact, PrPc, having a higher affinity for transducing machinery, may displace Doppel protein and trigger survival signals. Evidence supporting a role of PrPc in regulating cell proliferation, differentiation, and survival has been recently collected by Satoh and coworkers [47], who compared the gene expression profile in fibroblast cell lines derived from PrPc-deficient mice (Prnp 0/0) and fromPrP wild-typemice. They observed that the disruption of Prnp gene resulted in an aberrant regulation of a battery of genes important for cell proliferation, differentiation, and survival, including those located in the Ras and Rac signaling pathways. Moreover, Kuwahara and coworkers [48] observed that the reintroduction of wild-type Prnp gene prevented apoptosis in hippocampal cells derived from Prnp 0/0 mice and cultured under serum-free conditions.
To further confirm the PrPc/Cav-1 colocalization, experiments of flotation in density gradient were performed. Extracting cells with Triton X-100 at 4°C and fractionating the insoluble material by OptiPrep or sucrose-gradient centrifugation are considered a stringent criterion to establish whether a protein is present in detergent-insoluble glycolipid-rich domains (DIGs) and colocalizes with Cav-1 [49]. Nevertheless, it is difficult to obtain a separation between rafts and caveolae.We applied this procedure to GN11 cells, transiently transfected with Prnp or Prnd, and evaluated PrPc, Cav-1, and Dpl distribution by western blot. Results obtained (Figure 3(a)) show a marked colocalization between PrPc and Cav-1 in low-density gradient fractions (Figure 3(a), 1 and 2, resp.). Interestingly, Dpl protein (Figure 3(a), 3), even if it is a GPI-linked protein too, shows a wide distribution in most fractions of the gradient that does not correlate with Cav-1 distribution.
Figure 3.
(a) Optiprep density gradient flotation of PrPc, Dpl, and Cav-1 proteins from GN11 cell lysates. Differentiated (TPA-treated) GN11 cells, transiently transfected with morPrP or hurDpl, were lyzed in 1% Triton X-100 at 4°C and subjected to flotation on discontinuous Optiprep gradient (0–40%). Gradient fractions were collected and examined by immunoblotting with anti-PrP (1), anti-Cav-1 (2), and anti- Dpl (3) antibodies. Optiprep concentration in each fraction is indicated above each lane. The data are representative of three independent experiments. Bar = 40 μm. (b) PrP and Cav-1 coimmunoprecipitation assay. Live GN11 cells transfected with HA tagged-PrP plasmid were untreated (lane 1) or activated by anti-PrP antibody-mediated ligation (lanes 2–4). Cell lysates were collected and immunoprecipitated with either anti-HA (lanes 1 and 2) or anti-Cav-1 (lanes 3 and 4) antibodies. The immunoprecipitates were immunoblotted and blots were incubated with anti-Cav-1 (lanes 1 and 2) or anti-PrP antibodies (lanes 3 and 4), respectively. In lane 2, anti-Cav-1 antibody detects a band of the apparent molecular weight of 22 kDa typical of Cav-1. Anti-PrP antibody reveals the presence of bands with apparent mass typical of PrPc glycoforms (lane 3). When cell immunoprecipitate was subjected to deglycosylation by PNGase-F, a unique band (at 27 kDa), having the expected electrophoretic mobility of unglycosylated PrPc, appeared (lane 4). Data are representative of two independent experiments
Mouillet-Richard et al [13] detected the presence of Cav- 1 in immunoprecipitated samples obtained by using an anti- PrPc antibody, particularly when neuronal cells were induced to differentiate. We proposed to confirm these results in GN11 neuronal cells transiently transfected with murine Prnpgene. Reciprocal coimmunoprecipitation experiments in HA tagged-PrPc-overexpressing GN11 cells were performed to further confirm PrPc-Cav1 interaction (Figure 3(b)). We found that anti-HA-tag antibodies coimmunoprecipitate Cav-1 (Figure 3(b), lane 2) and, reciprocally, that anti-Cav-1 antibodies coimmunoprecipitate PrP (Figure 3(b), lanes 3 and 4) in cells stimulated by anti-PrPc antibody- (30 min) and by secondary antibody- (30 min) mediated ligation. After deglycosylation treatment (Figure 3(b), lane 4), the smear typical of PrPc was resolved in a single band of 27 kDa. Interestingly, the sensitivity of western blot detection did not permit to reveal Cav-1 in coimmunoprecipitates of unstimulated cells (lane 1) confirming the importance of antibody stimulation in PrPc/Cav- 1 interaction.
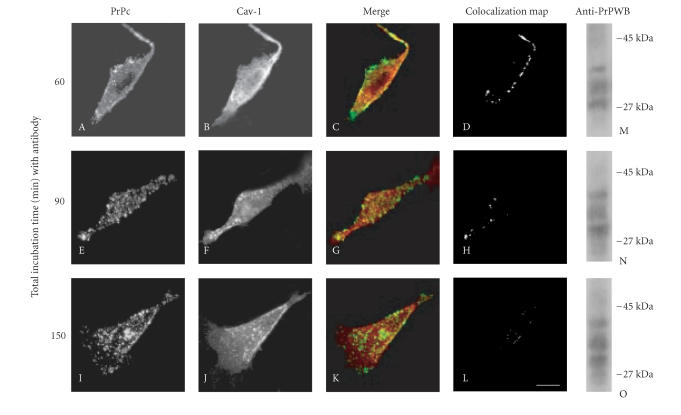
To follow the fate of PrPc/Cav-1 complexes, a timecourse of PrPc and Cav-1 distributions after anti-PrPc antibody-mediated stimulation was analyzed by confocal microscopy in living GN11 cells overexpressing PrPc. In Figure 4 it is shown that the PrPc/Cav-1 colocalized signal, at plasma membrane level, was evident after 60 min of anti-PrPc antibody-mediated stimulation (A–D). However, after 90 min, PrPc was prominent in the intracellular space and only a few spots of colocalized signals were present at plasma membrane level (E–H). After 150 min, PrPc became almost exclusively intracellular (I) while Cav-1 distribution appeared unchanged (J), and colocalization signals were undetectable (K), (L). It is likely that when caveolae are detached from surface membrane the dissociation of PrPc/Cav- 1 complexes takes place. However, western blotting analysis of PrPc at different times (M, N, and O) showed that no degradation of PrPc, even after 2 h, was evident. Moreover, the extent of PrPc glycosylation was unchanged.
Figure 4.
Time course of antibody-mediated PrPc internalization. As detailed above (Figure 2), the anti-PrPc antibody-mediated stimulation was achieved by incubating live GN11 cells with Mab 3F4 for 30 min, followed by FITC-conjugated secondary antibody, for a total incubation time of either 60 min (A–D), (M), 90 min (E–H), (N), or 150 min (I–L), (O). After fixing and permeabilizing, cells were probed with anti-Cav-1 Pab and Cy5-conjugated anti-rabbit secondary antibody. The distribution of PrPc (A, E, and I) and Cav-1 (B, F, and J) at the different incubation periods, the superposition (C, G, and K) of the green (PrPc) and red (Cav-1) signals, and the corresponding binary maps (D, H, and L) are shown. Note that after 150 min, the major internalization of PrPc is quite independent from Cav-1 movements (I–L). Panels M–O represent western blot (WB) analysis of cell lysates for the presence of PrPc glycoforms under the different conditions described. On the right margin, standard molecular weights are reported. For further experimental details see Material and Methods. Bar = 7.5 μm.
Involvement of PrPc in signal transduction
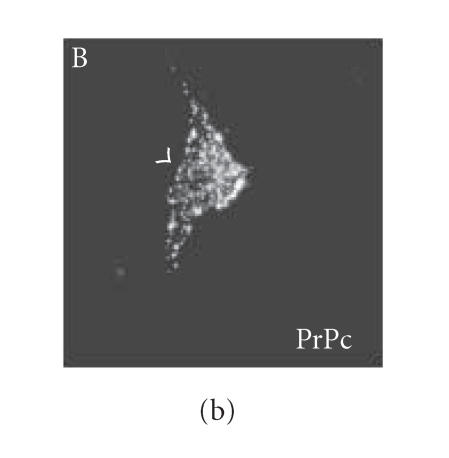
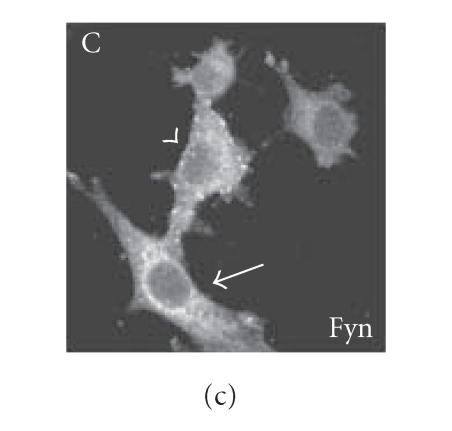
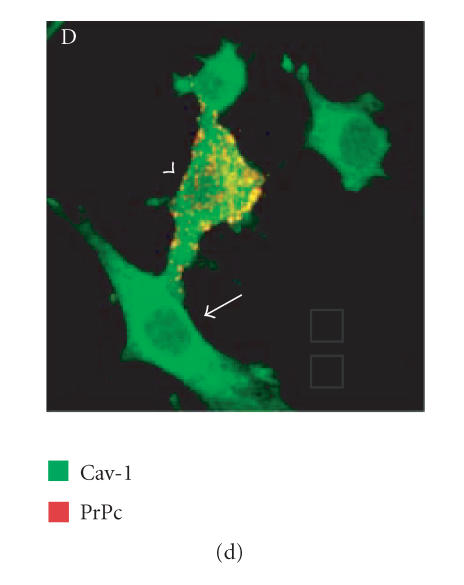
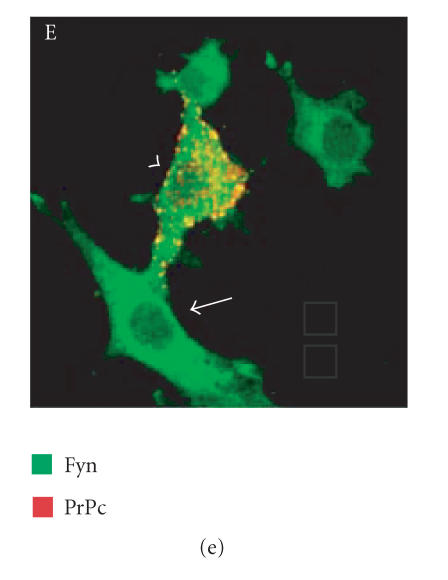
Since we hypothesized a role of PrPc clustering and of PrPc- Cv 1 colocalization in signal transduction, we performed confocal microscopy experiments to assess if Fyn kinase responds, in someway, to an anti-PrP antibody-mediated stimulation (Figure 4). Biedi and collaborators demonstrated the recruitment of Fyn kinase from cytoplasm to caveolae in fibroblasts overexpressing wild-type IGF-IR stimulated by IGF-I [50]. In a similar way, after the stimulation of PrPc-overexpressing GN11 cells with anti-PrPc antibody (30 min) and with secondary antibody (30 min) to induce PrPc patching, we observed the shift of Fyn kinase from the cytoplasm towards the membrane (in Figure 5(c), compare the Fyn distribution in a transfected cell subjected to the stimulation (arrowhead) with an untransfected cell not subjected to the stimulation (arrow)). After the antibody-mediated stimulation, we revealed a high colocalization between PrP and Cav-1 (panel D) and between PrP and Fyn kinase (panel E) on the plasma membrane. It was previously reported that PrPc and Fyn kinase do not colocalize in neurons; however in those experiments cells were not stimulated by anti-PrPc antibody [51].
Figure 5.
Fyn kinase shift from cytoplasm into membrane caveolae. Live GN11 cells PrPc transiently transfected were incubated with anti- PrP Mab 3F4 (30 min), followed by secondary Cy3-conjugated antibody (30 min), to allow antibody-mediated ligation (and visualization) of PrPc B. After fixing and permeabilizing cells, anti-Fyn kinase polyclonal antibody and secondary Cy5-conjugated antibody were added to visualize Fyn kinase distribution (C). Finally, anti-caveolin1 FITC-conjugated antibody was added to visualize Cav-1 distribution (A). The superposition of green (Cav-1 in D; Fyn kinase in E) and red (PrPc in D and E) signals provided the images shown in D and E. Arrowheads show the PrPc-overexpressing cells and arrows do not indicate transfected cells. Images (A–E) result from integrating different optical sections.. Bar = 7.5 μm.
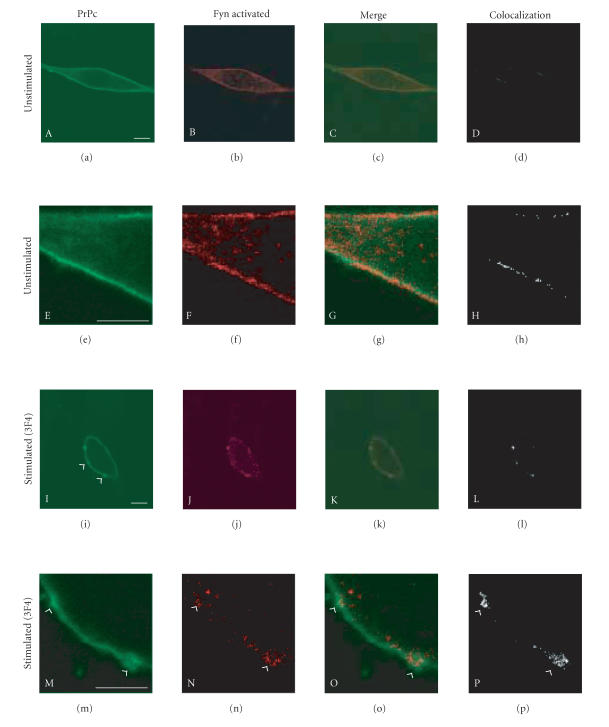
To evaluate if the Fyn kinase recruited in the clusters of PrPc/Cav-1 was activated, we performed confocal immunofluorescence experiments by using a specific antibody that recognizes the active form of Fyn kinase phosphorylated on Tyr 416 (Figure 6). In GN11 cells that do not express PrPc, the signal of active Fyn kinase is low and wide dispersed in the cytoplasm (data not shown). On the contrary, in PrP-overexpressing GN11 cells we detected a low activation of Fyn kinase on plasma membrane (Figure 6, B and F). Following anti-PrPc antibody-mediated stimulation (20 min), an increased level of active Fyn kinase located in the correspondence of PrPc clusters was detected (J, N, arrowheads indicate PrPc clusters). After 40 min of antibody stimulation, the signal of active Fyn kinase was weaker and after 60 min it was no more detectable (data not shown). Taken together, these data show that following anti-PrPc antibody mediated stimulation, PrPc patches on the plasma membrane in clusters in which Fyn kinase is recruited and activated.
Figure 6.
Fyn kinase is activated in PrPc/Cav-1 clusters on plasma membrane. PrPc-overexpressing GN11 cells were fixed and incubated with anti-PrP Mab 3F4 and with secondary FITC-conjugated antibody to allow PrPc visualization (A, E). Live GN11 cells PrPc transiently transfected were incubated with 3F4 Mab (30 min), followed by secondary FITC-conjugated antibody (30 min) (I–P), to allow antibody-mediated ligation (and visualization) of PrPc (I, M). After fixing, cells were permeabilized and incubated with anti-phospho Fyn kinase (Tyr 416) Pab and secondary Cy5-conjugated antibody to visualize the distribution of active Fyn kinase (B, F, J, and N). Superposition of the green PrPc (A, I corresponding to E, M after magnification) and red phospho Fyn (B, J corresponding to F, N after magnification) signals provided the images shown in C, G, K, O. The binary maps presented in D, H, L, P are a more precise means to evaluate colocalization, whereby only those regions, in which PrPc and phospho Fyn are copresent above a defined threshold of fluorescence intensity, appear. Arrowheads show the PrPc clusters on the plasma membrane. Micrographs show a single confocal plane. Bar = 40 μm.
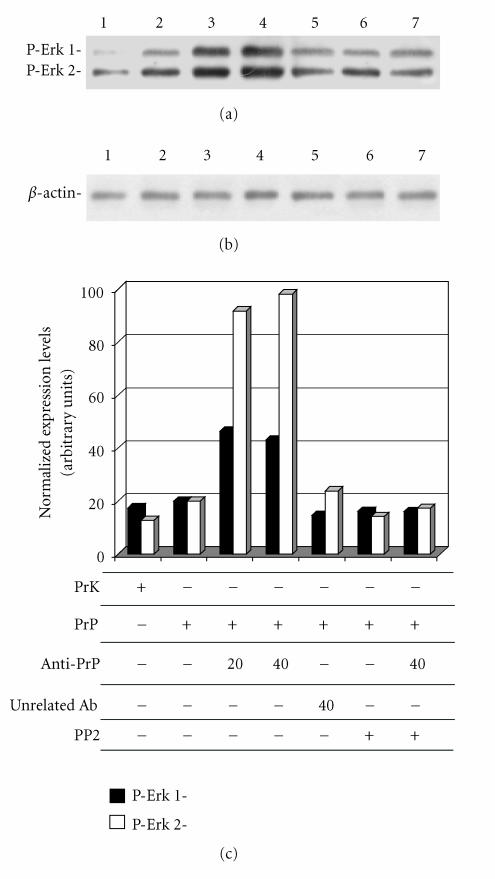
It has been reported that activated Fyn kinase can transduce a signal cascade through Erk 1/2 kinase [52]. To evaluate if in GN11 cells the activation of PrPc by antibody-mediated ligation triggers a signal transduction pathway through Erk 1/2, we perform the experiments reported in Figure 7. The incubation of PrP-overexpressing GN11 cells with anti-PrPc (3F4) and secondary antibody for a total time of 20 and 40 min caused the activation of of Fyn kinase following anti-PrPc antibody-mediated stimulation, observed by confocal microscopy, may trigger a downstream activation of Erk 1/2 kinase by inducing their phosphorylation (lanes 3 and 4), while the incubation with unrelated antibodies had no effects (lane 5). Erk1/2 phosphorylation levels did not increase if cells were previously treated with the Fyn kinase inhibitor PP2 (lane 6). This result suggests that the activation of Erk 1/2 signaling pathway. Our data, obtained in a neuronal model expressing Cav-1, confirm that PrPc triggers a Fyn kinase and Erk 1/2 signaling pathway as previously reported [15, 31, 32, 53].
Figure 7.
Erk 1/2 phosphorylation in GN11 cells stimulated with anti- PrPc antibody. GN11 cells were transfected with empty HA-plasmid Prk7HA (lane 1) or PrP-HA plasmid (lanes 2–7) and incubated with anti-PrP-antibody (3F4) for 20 (lane 3) and 40 (lanes 4 and 7) min. In lane 5, cells were incubated with an unspecific antibody (anti-β-actin) for 40 min. In lanes 6 and 7, cells were treated with the Fyn kinase inhibitor PP2 (30 μm, 4 h). Cell lysates were collected and analyzed by western blot using anti-phospho-p44/42 MAP kinase antibody (a) and anti-β-actin antibody (b). Band intensities were evaluated by ImageMaster software (Pharmacia Biotech, Sweden). In (c), we reported the intensity values of the bands corresponding to phosphorylated Erk 1 and Erk 2 (black and white bars, resp.) normalized with respect to β-actin band intensity values. Data are representative of three independent experiments.
To obtain this activation, PrPc and Fyn kinase need to co-localize in caveolae: PrPc clusters on the plasma membrane following the antibody (or other putative physiological ligands) binding, and Fyn kinase moves from cytoplasm to caveolae.
In order to demonstrate the essential role of Cav-1 in the PrPc-Fyn-Erk 1/2 signaling pathway, we down-regulated or misallocated caveolin 1 by using anti-Cav-1 antisense oligonucleotides and Filipin III, respectively. The aim was to obtain an experimental condition in which the absence of Cav-1 could prevent the activation of the transduction cascade triggered by PrPc clustering. However, these treatments in GN11 cells did not permit to obtain clear results as they caused a strong increase of Erk 1/2 phosphorylation level (data not shown). The ability of Cav-1 to directly inhibits Erk 1/2 activation has been reported in other experimental models [25, 26]. Moreover, the hyperactivation of Erk 1/2 signalingwas reported in Cav-1 (−/−) null mouse cells and tissues [28, 29, 54], and in NIH-3T3 cells in which Cav-1 was downregulated by antisense [27]. The inhibitory effect of Cav-1 on Erk 1/2 may explain why, in our cellularmodel, it is so hard to demonstrate the direct involvement of Cav-1 in signal transduction events triggered by PrPc stimulation.
The data reported here confirm the involvement of PrPc in signal transduction events through Fyn and Erk 1/2 kinase and demonstrate caveolae to be the physical place in which this signal cascade switches on. In vitro binding experiments's results suggest that a physical interaction between PrPc and Cav-1 could be involved in these signaling events, even if this is unlikely considering the nowadays-accepted location of PrPc and Cav-1 on the opposite side of the plasma membrane.
ACKNOWLEDGMENTS
We thank Dr R. Maggi and Prof F. Piva (Department of Endocrinology, University of Milan, Italy) for providing GN11 cells. Dr Romina D'Angelo (CNRS/Curie Institute, Paris, France) helped with the PrP-HA plasmid construction. The present work was supported by grants from MIUR (PRIN 2004) to VT.
References
- 1.Massimino ML, Griffoni C, Spisni E, Toni M, Tomasi V. Involvement of caveolae and caveolae-like domains in signalling, cell survival and angiogenesis. Cellular Signalling. 2002;14(2):93–98. doi: 10.1016/s0898-6568(01)00232-7. [DOI] [PubMed] [Google Scholar]
- 2.Shaul PW, Anderson RGW. Role of plasmalemmal caveolae in signal transduction. The American Journal of Physiology. 1998;275(5 pt 1):L843–L851. doi: 10.1152/ajplung.1998.275.5.L843. [DOI] [PubMed] [Google Scholar]
- 3.Simons K, Toomre D. Lipid rafts and signal transduction. Nature Reviews Molecular Cell Biology. 2000;1(1):31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 4.Teruel MN, Meyer T. Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell. 2000;103(2):181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 5.Schlegel A, Lisanti MP. A molecular dissection of caveolin-1 membrane attachment and oligomerization. Two separate regions of the caveolin-1 C-terminal domain mediate membrane binding and oligomer/oligomer interactions in vivo. Journal of Biological Chemistry. 2000;275(28):21605–21617. doi: 10.1074/jbc.M002558200. [DOI] [PubMed] [Google Scholar]
- 6.Shyng S-L, Huber MT, Harris DA. A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. Journal of Biological Chemistry. 1993;268(21):15922–15928. [PubMed] [Google Scholar]
- 7.Gauczynski S, Hundt C, Leucht C, Weiss S. Interaction of prion proteins with cell surface receptors, molecular chaperones, and other molecules. Advances in Protein Chemistry. 2001;57:229–272. doi: 10.1016/s0065-3233(01)57024-2. [DOI] [PubMed] [Google Scholar]
- 8.Hundt C, Peyrin J-M, Haik S, et al. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO Journal. 2001;20(21):5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffoni C, Toni M, Spisni E, et al. The cellular prion protein: biochemistry, topology, and physiologic functions. Cell Biochemistry and Biophysics. 2003;38(3):287–304. doi: 10.1385/cbb:38:3:287. [DOI] [PubMed] [Google Scholar]
- 10.Vey M, Pilkuhn S, Wille H, et al. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(25):14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. Journal of Biological Chemistry. 1997;272(10):6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- 12.Peters PJ, Mironov A, Jr, Peretz D, et al. Trafficking of prion proteins through a caveolae-mediated endosomal pathway. Journal of Cell Biology. 2003;162(4):703–717. doi: 10.1083/jcb.200304140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouillet-Richard S, Ermonval M, Chebassier C, et al. Signal transduction through prion protein. Science. 2000;289(5486):1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 14.Schneider B, Mutel V, Pietri M, Ermonval M, Mouillet-Richard S, Kellermann O. NADPH oxidase and extracellular regulated kinases 1/2 are targets of prion protein signaling in neuronal and nonneuronal cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(23):13326–13331. doi: 10.1073/pnas.2235648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satoh T, Nakatsuka D, Watanabe Y, Nagata I, Kikuchi H, Namura S. Neuroprotection by MAPK/ERK kinase inhibition with U0126 against oxidative stress in a mouse neuronal cell line and rat primary cultured cortical neurons. Neuroscience Letters. 2000;288(2):163–166. doi: 10.1016/s0304-3940(00)01229-5. [DOI] [PubMed] [Google Scholar]
- 16.Shenoy-Scaria AM, Gauen LKT, Kwong J, Shaw AS, Lublin DM. Palmitylation of an amino-terminal cysteine motif of protein tyrosine kinases p56lck and p59fyn mediates interaction with glycosyl-phosphatidylinositol-anchored proteins. Molecular and Cellular Biology. 1993;13(10):6385–6392. doi: 10.1128/mcb.13.10.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shenoy-Scaria AM, Kwong J, Fujita T, Olszowy MW, Shaw AS, Lublin DM. Signal transduction through decay-accelerating factor. Interaction of glycosyl-phosphatidylinositol anchor and protein tyrosine kinases p56lck and p59fyn 1. Journal of Immunology. 1992;149(11):3535–3541. [PubMed] [Google Scholar]
- 18.Stefanova I, Horejsi V. Association of the CD59 and CD55 cell surface glycoproteins with other membrane molecules. Journal of Immunology. 1991;147(5):1587–1592. [PubMed] [Google Scholar]
- 19.Shenoy-Scaria AM, Dietzen DJ, Kwong J, Link DC, Lublin DM. Cysteine3 of Src family protein tyrosine kinases determines palmitoylation and localization in caveolae. Journal of Cell Biology. 1994;126(2):353–363. doi: 10.1083/jcb.126.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lublin DM. Glycosyl-phosphatidylinositol anchoring of membranes proteins. Current Topics in Microbiology and Immunology. 1992;178:141–162. doi: 10.1007/978-3-642-77014-2_9. [DOI] [PubMed] [Google Scholar]
- 21.Mayor S, Rothberg KG, Maxfield FR. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 1994;264(5167):1948–1951. doi: 10.1126/science.7516582. [DOI] [PubMed] [Google Scholar]
- 22.Adams JP, Sweatt JD. Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annual Review of Pharmacology and Toxicology. 2002;42:135–163. doi: 10.1146/annurev.pharmtox.42.082701.145401. [DOI] [PubMed] [Google Scholar]
- 23.Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. American Journal of Physiology - Cell Physiology. 2005;288(3):C494–C506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Ying Y-S, Anderson RGW. Platelet-derived growth factor activates mitogen-activated protein kinase in isolated caveolae. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(25):13666–13670. doi: 10.1073/pnas.94.25.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelman JA, Chu C, Lin A, et al. Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Letters. 1998;428(3):205–211. doi: 10.1016/s0014-5793(98)00470-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, Razani B, Altschuler Y, et al. Caveolin-1 inhibits epidermal growth factor-stimulated lamellipod extension and cell migration in metastatic mammary adenocarcinoma cells (MTLn3). Transformation suppressor effects of adenovirus-mediated gene delivery of caveolin-1. Journal of Biological Chemistry. 2000;275(27):20717–20725. doi: 10.1074/jbc.M909895199. [DOI] [PubMed] [Google Scholar]
- 27.Galbiati F, Volonte D, Engelman JA, et al. Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO Journal. 1998;17(22):6633–6648. doi: 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capozza F, Combs TP, Cohen AW, et al. Caveolin-3 knockout mice show increased adiposity and whole body insulin resistance, with ligand-induced insulin receptor instability in skeletal muscle. American Journal of Physiology - Cell Physiology. 2005;288(6):C1317–C1331. doi: 10.1152/ajpcell.00489.2004. [DOI] [PubMed] [Google Scholar]
- 29.Cohen AW, Park DS, Woodman SE, et al. Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. American Journal of Physiology - Cell Physiology. 2003;284(2):C457–C474. doi: 10.1152/ajpcell.00380.2002. [DOI] [PubMed] [Google Scholar]
- 30.Engelman JA, Zhang XL, Razani B, Pestell RG, Lisanti MP. p42/44 MAP kinase-dependent and -independent signaling pathways regulate caveolin-1 gene expression. Activation of Ras-MAP kinase and protein kinase A signaling cascades transcriptionally down-regulates caveolin-1 promoter activity. Journal of Biological Chemistry. 1999;274(45):32333–32341. doi: 10.1074/jbc.274.45.32333. [DOI] [PubMed] [Google Scholar]
- 31.Monnet C, Gavard J, Mege R-M, Sobel A. Clustering of cellular prion protein induces ERK1/2 and stathmin phosphorylation in GT1-7 neuronal cells. FEBS Letters. 2004;576(1-2):114–118. doi: 10.1016/j.febslet.2004.08.076. [DOI] [PubMed] [Google Scholar]
- 32.Chiarini LB, Freitas ARO, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO Journal. 2002;21(13):3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoyama T, Kimura KM, Ushiki Y, et al. In vivo conversion of cellular prion protein to pathogenic isoforms, as monitored by conformation-specific antibodies. Journal of Biological Chemistry. 2001;276(14):11265–11271. doi: 10.1074/jbc.M008734200. [DOI] [PubMed] [Google Scholar]
- 34.Negro A, Meggio F, Bertoli A, Battistutta R, Sorgato MC, Pinna LA. Susceptibility of the prion protein to enzymic phosphorylation. Biochemical and Biophysical Research Communications. 2000;271(2):337–341. doi: 10.1006/bbrc.2000.2628. [DOI] [PubMed] [Google Scholar]
- 35.Spisni E, Griffoni C, Santi S, et al. Colocalization prostacyclin (PGI2) synthase-caveolin-1 in endothelial cells and new roles for PGI2 in angiogenesis. Experimental Cell Research. 2001;266(1):31–43. doi: 10.1006/excr.2001.5198. [DOI] [PubMed] [Google Scholar]
- 36.Radovick S, Wray S, Lee E, et al. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(8):3402–3406. doi: 10.1073/pnas.88.8.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pimpinelli F, Rovati GE, Capra V, Piva F, Martini L, Maggi R. Expression of prostacyclin receptors in luteinizing hormone-releasing hormone immortalized neurons: role in the control of hormone secretion. Endocrinology. 1999;140(1):171–177. doi: 10.1210/endo.140.1.6445. [DOI] [PubMed] [Google Scholar]
- 38.Riccio M, Di Giaimo R, Pianetti S, Palmieri PP, Melli M, Santi S. Nuclear localization of cystatin B, the cathepsin inhibitor implicated in myoclonus epilepsy (EPM1) Experimental Cell Research. 2001;262(2):84–94. doi: 10.1006/excr.2000.5085. [DOI] [PubMed] [Google Scholar]
- 39.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. Journal of Cell Biology. 1998;141(4):929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Negro A, Ballarin C, Bertoli A, Massimino ML, Sorgato MC. The metabolism and imaging in live cells of the bovine prion protein in its native form or carrying single amino acid substitutions. Molecular and Cellular Neuroscience. 2001;17(3):521–538. doi: 10.1006/mcne.2000.0953. [DOI] [PubMed] [Google Scholar]
- 41.Riek R, Hornemann S, Wider G, Glockshuber R, Wuthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231) FEBS Letters. 1997;413(2):282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 42.Moore RC, Lee IY, Silverman GL, et al. Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. Journal of Molecular Biology. 1999;292(4):797–817. doi: 10.1006/jmbi.1999.3108. [DOI] [PubMed] [Google Scholar]
- 43.Behrens A, Aguzzi A. Small is not beautiful: antagonizing functions for the prion protein PrPC and its homologue Dpl. Trends in Neurosciences. 2002;25(3):150–154. doi: 10.1016/s0166-2236(00)02089-0. [DOI] [PubMed] [Google Scholar]
- 44.Prioni S, Loberto N, Prinetti A, et al. Sphingolipid metabolism and caveolin expression in gonadotropin-releasing hormone-expressing GN11 and gonadotropin-releasing hormone-secreting GT1-7 neuronal cells. Neurochemical Research. 2002;27(7-8):831–840. doi: 10.1023/a:1020217309987. [DOI] [PubMed] [Google Scholar]
- 45.Silverman GL, Qin K, Moore RC, et al. Doppel is an N-glycosylated, glycosylphosphatidylinositol-anchored protein: expression in testis and ectopic production in the brains of Prnp(0/0) mice predisposed to purkinje cell loss. Journal of Biological Chemistry. 2000;275(35):26834–26841. doi: 10.1074/jbc.M003888200. [DOI] [PubMed] [Google Scholar]
- 46.Nishida N, Tremblay P, Sugimoto T, et al. A mouse prion protein transgene rescues mice deficient for the prion protein gene from Purkinje cell degeneration and demyelination. Laboratory Investigation. 1999;79(6):689–697. [PubMed] [Google Scholar]
- 47.Satoh J-I, Kuroda Y, Katamine S. Gene expression profile in prion protein-deficient fibroblasts in culture. American Journal of Pathology. 2000;157(1):59–68. doi: 10.1016/s0002-9440(10)64517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuwahara C, Takeuchi AM, Nishimura T, et al. Prions prevent neuronal cell-line death. Nature. 1999;400(6741):225–226. doi: 10.1038/22241. [DOI] [PubMed] [Google Scholar]
- 49.Kurzchalia TV, Parton RG. Membrane microdomains and caveolae. Current Opinion in Cell Biology. 1999;11(4):424–431. doi: 10.1016/s0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 50.Biedi C, Panetta D, Segat D, Cordera R, Maggi D. Specificity of insulin-like growth factor I and insulin on Shc phosphorylation and Grb2 recruitment in caveolae. Endocrinology. 2003;144(12):5497–5503. doi: 10.1210/en.2003-0417. [DOI] [PubMed] [Google Scholar]
- 51.Botto L, Masserini M, Cassetti A, Palestini P. Immunoseparation of Prion protein-enriched domains from other detergent-resistant membrane fractions, isolated from neuronal cells. FEBS Letters. 2004;557(1–3):143–147. doi: 10.1016/s0014-5793(03)01463-7. [DOI] [PubMed] [Google Scholar]
- 52.Maulon L, Mari B, Bertolotto C, et al. Differential requirements for ERK1/2 and P38 MAPK activation by thrombin in T cells. Role of P59Fyn and PKCε. Oncogene. 2001;20(16):1964–1972. doi: 10.1038/sj.onc.1204266. [DOI] [PubMed] [Google Scholar]
- 53.Krebs B, Dorner-Ciossek C, Schmalzbauer R, Vassallo N, Herms J, Kretzschmar HA. Prion protein induced signaling cascades in monocytes. Biochemical and Biophysical Research Communications. 2006;340(1):13–22. doi: 10.1016/j.bbrc.2005.11.158. [DOI] [PubMed] [Google Scholar]
- 54.Williams TM, Lee H, Cheung MW-C, et al. Combined loss of INK4a and caveolin-1 synergistically enhances cell proliferation and oncogene-induced tumorigenesis. Role of INK4a/CAV-1 in mammary epithelial cell hyperplasia. Journal of Biological Chemistry. 2004;279(23):24745–24756. doi: 10.1074/jbc.M402064200. [DOI] [PubMed] [Google Scholar]