Abstract
We present an overview of selected computational methods for microRNA prediction. It is especially aimed at viral miRNA detection. As the number of microRNAs increases and the range of genomes encoding miRNAs expands, it seems that these small regulators have a more important role than has been previously thought. Most microRNAs have been detected by cloning and Northern blotting, but experimental methods are biased towards abundant microRNAs as well as being time-consuming. Computational detection methods must therefore be refined to serve as a faster, better, and more affordable method for microRNA detection. We also present data from a small study investigating the problems of computational miRNA prediction. Our findings suggest that the prediction of microRNA precursor candidates is fairly easy, while excluding false positives as well as exact prediction of the mature microRNA is hard. Finally, we discuss possible improvements to computational microRNA detection.
INTRODUCTION
Since 2000 the interest in microRNAs (miRNAs) and their role as gene expression regulators has grown immensely. Lee et al were the first to identify such a small regulator: the lin-4 RNA in Caenorhabditis elegans [1]. It has been shown that the 21 nt lin-4 RNA represses mRNA and controls part of the C elegans larval development [1, 2]. The next small regulatory RNA to be discovered was the let-7, which controls a later stage in the development of C elegans [3, 4]. The lin-4 and let-7, previously known as small temporal RNAs (stRNAs), are today recognized as the first of a large class of small regulatory noncoding RNA molecules now called microRNAs [5]. This class of molecules is not limited to development but regulates a wide range of biological processes [6]. The microRNAs have been reported to be encoded within noncoding regions of genomes [5, 7, 8], and within protein coding genes [9] as well as noncoding genes [10].
Primary precursor miRNAs (pri-miRNAs) are long transcripts that contain one or more miRNA precursors (pre-miRNAs) [11]. Subsequently the pri-miRNA is cut by the Drosha enzyme into one or more ∼ 70 nt long pre-miRNA stem-loop (hairpin) structure(s) while still in the nucleus [12]. The pre-miRNAs are transported by exportin-5 to the cytoplasm [13–15], where they are cut by the RNase III Dicer enzyme into active ∼ 22 nt long miRNAs [16–18] (Figure 1). Usually only one side of the stem encodes a mature miRNA [5, 19], however the process of selecting the side and region of the pre-miRNA that becomes a mature miRNA is still not fully understood. The mature miRNAs are then incorporated as subunits of the micro-ribonucleoproteins (miRNPs) [20]. The miRNP is able to repress the transcription of target mRNAs by binding to or cleaving the mRNA. Thus the miRNA is capable of posttranscriptional regulation [1–4, 21–23]. Such a posttranscriptional silencing complex is often called an miRNA-initiated (or associated) RISC complex (RNA-induced silencing complex), and is very similar to the small interfering RNA-initiated RISC complexes [21, 24]. Detailed descriptions of the stepwise maturation of microRNAs are presented by Chen and Meister [25] and by Bartel [26].
Figure 1.
Sequence and structure of a pre-miRNA molecule encoding a miRNA detected by Pfeffer et al [33] from the Epstein-Barr virus. The mature 21 nt EBV mir-BART1 miRNA sequence is shown on a grey background.
Different miRNAs have been detected in a variety of organisms; including 114 C elegans miRNAs, 326 human miRNAs, and a total of 35 virus-encoded miRNAs (miRBase release 7.1, October 2005) [27, 28]. It is estimated that as much as 30% of human genes are regulated by miRNAs [29, 30].
COMPUTATIONAL DETECTION OF miRNAs IN SELECTED ORGANISMS
Until 2003 miRNAs were identified almost exclusively by experimental molecular biology [31] because there were few computational miRNA prediction tools available (except for homology searches).
According to Lai et al [32], three observations suggest that computational miRNA prediction approaches will be feasible. “First, miRNAs are generally derived from precursor transcripts of 70–100 nucleotides with extended stem-loop structure. Second, miRNAs are usually highly conserved between the genomes of related species. Third, miRNAs display a characteristic pattern of evolutionary divergence.”
Already in 2001 Lee and Ambros used both bioinformatics and cDNA cloning to identify potential C elegans miRNAs [7]. They searched the C elegans genome for sequences conserved in C briggsae that also had characteristic pre-miRNA features and a secondary structure similar to lin-4 and let-7, as computed by the mfold program [37]. They reported 15 novel miRNAs, of which two were the results of the computational screening, while the rest were derived from the cDNA cloning. Table 1 contains an overview of computational miRNA prediction studies.
Table 1.
Overview, in chronological order, of approaches and results of selected miRNA detection studies. Computational and experimental approaches used as well as the total number of predicted candidates and verified miRNAs are indicated for each study.
| Reference | Genome(s) | Stem-loop | Homology | Folding | Experimental | Novel candidates | Novel verified |
| (free energy) | (comp/exp) | (comp/tot) | |||||
| Lee and Ambros [7] | C elegans | X | X | X | X | 40/38 (only 53 tested) | 2/13 |
| Lim et al [31] | C elegans | X | X | — | X | 35/NA | 16/30 |
| Ambros et al [34] | C elegans | X | X | X | X | 407/NA | 9/21 |
| Lai et al [32] | D melanogaster | X | X | X | — | 166/0 (only 38 tested) | 24/24 |
| Pfeffer et al [35] | Epstein-barr virus (EBV) | — | — | — | X | 0/NA | 5/5 |
| Pfeffer et al [33] | Human cytomegalovirus | X | — | X | X | 11/NA | 5/9 |
| (HCMV)* | |||||||
| Grey et al [36] | Human cytomegalovirus | X | X | — | — | 10/0 | 2/2 |
| (HCMV) | |||||||
*One selected genome of a range of herpesviruses studied.
Another computational tool for miRNAs identification is MiRscan, described by Lim et al in 2003 [31]. MiRscan was designed to identify miRNA genes conserved between genomes, and was initially applied to C elegans and C briggsae. MiRscan was utilized together with extensive sequencing of clones, resulting in the detection of 30 additional miRNAs.
MiRscan starts out with two closely related genomes A and B. It scans genome A for sequences that could form hairpin structures and then checks if the sequences are conserved in genome B. This initial search aims at capturing most of the homologous pre-miRNAs in the two genomes. The program uses the captured miRNAs that are already experimentally verified as a training set, and then computes a score for all the initially recognized sequences.
Lim et al found 35 novel miRNA candidates in C elegans using MiRscan, of which 16 were experimentally validated. In addition, the program used a detection threshold that would have identified half (29) of the known (58) miRNAs. This implies that in the worst case, the MirScan program would have a sensitivity of 0.70 for miRNAs detection in this study.
Lim et al also showed that the accuracy of MirScan is lower than for programs designed to detect one special type of RNA, such as tRNAs [38], but on the other hand it is at least as good as general computer algorithms for detection of bacterial ncRNAs [39–41]. Due to the homology criterion of MiRscan, it may be questionable whether this program is suitable for the detection of viral miRNAs as there are reports on viral miRNAs not being conserved across species [33], as well as reports on the opposite [36]. MiRscan has proved itself able to detect a large number of miRNAs in vertebrate genomes with a detection sensitivity of 0.74 [42].
In May 2003, Ambros et al reported on the testing of different methods for the detection of miRNAs in C elegans [34]. This study was a follow up to their 2001 study, when only 10% of the C briggsae genome was available [7]. Two computational approaches were based on sequence similarities and stem-loop structure features, but used slightly different algorithms. The algorithms were complementary in the way that the methods uniquely identified miRNAs and in total these two approaches identified 9 new miRNAs. Combined with a third approach, cDNA cloning followed by Northern blots, they discovered a total of 21 novel miRNAs.
Others have also screened the C elegans genome for miRNAs using computational approaches based on hairpin structure searches, secondary structure predictions, and interspecies sequence conservation. Grad et al suggested 214 miRNA candidates of which 14 were confirmed by expression analysis [43].
In 2003 Lai and colleagues described a computational method for miRNA identification in Drosophilia [32]. The approach was named miRseeker, and the initial step was to search the euchromatic DNA sequences of D melanogaster and D pseudoobscura for transcripts potentially forming stem-loop structures and having a “pattern of nucleotide divergence characteristic of known miRNAs.” Subsequently they considered the conservation of this sequence in more distantly related insects. Lai et al started by aligning 24 pre-miRNA sequences from the two Drosophilia species and found the degree of conservation to be higher than in protein coding regions. The candidates were then subjected to a stricter selection procedure due to the many conserved possible pre-miRNA stem-loops found. Further analysis proved that most divergence in the orthologous Drosophila miRNAs originated in loop-mutations. In more diverged species only the 21–24 nt mature miRNAs were found to be preserved. The algorithm consists of three steps. Initially it aligns all D melanogaster and D pseudoobscura intronic and intergenic regions. It then slides a window along the conserved regions and uses mfold [37] to estimate the free energy of potential secondary structure formed by the sequence in the window. A minimum arm length of 23 nt was required as well as a free energy of at most −23.0 kcal/mol for one isolated miRNA precursor arm. Both strands of the DNA sequence in the sliding window were mfolded. Additional scoring of the stem-loops was also applied. Finally, miRseeker attempts to fit all the remaining miRNA-precursor candidates into one of six stem-loop pattern classes defined by the initial 24 pre-miRNA training set. This procedure left 208 miRNA candidates, including 18 (75%) from the training set among the 124 highest scoring candidates. Out of the 208 candidates 42 were also found to be conserved (by sequence and structure) in a third species. In a selection of 38 candidates, 24 were confirmed as novel miRNA genes (20/27 of those conserved in a third species and 4/11 of the Drosophila specific candidates). Lai and colleagues also estimated miRNAs to make up about 1% of the total amount of genes in the Drosophila genomes (94–124 miRNA genes), while Grad et al estimated C elegans to code for 140–300 miRNA genes [43]. As a concluding remark, Lai et al state that their algorithm excludes at least one known miRNA (miR-100).
Another study exploiting both characteristic miRNA features and sequence conservation was developed by Wang et al [44]. This approach was used in their search for Arabidopsis thaliana miRNAs. Their prediction identified 63% of known Arabidopsis miRNAs, and they claim identification of 83 novel miRNAs, of which 25 were verified. The computer algorithm evaluated possible miRNA precursors based on their stem-loop structure, the GC content of the mature miRNA, the loop length, mismatches in the stem containing the mature miRNA and the conservation of mature miRNA sequence in the Orysa sativa genome. Interestingly, 15 of the 19 already known unique Arabidopsis miRNAs have a loop ranging from 20–75 nt, which is much longer than in the known viral miRNAs [19, 33, 35, 36].
In plants, the alignment of the miRNA and its target mRNA contains few mismatches. This fact has been successfully exploited in combination with typical miRNA feature and conservation searches, as described above, in a search for Arabidopsis thaliana miRNA [45].
Yet another project combining bioinformatics and experimental biology in the quest for A thaliana and Nicotiana tabacum miRNA chose a “reverse” approach [46]. Billoud first created a cDNA library of all short N tabacum RNAs, then computational methods were used to identify potential miRNAs. Their pattern matching program, Patbank, was used for finding homologues and their MIRFOLD program was used to check for possible miRNA secondary structures.
In this context, the microHarvester should be mentioned as it is a useful web service designed to detect miRNA homologues in any set of sequences, given an miRNA precursor [47]. The microHarvester is filter based and uses the conservation patterns of the microRNAs combined with BLAST [48], Smith-Waterman [49], and RNAfold [50].
Wang et al presented a new computational tool in 2005 designed to search for homologues and paralogues of known miRNAs; miRAlign [51]. It is claimed that miRAlign outperforms all earlier programs of this kind, due to a less strict conservation search, the ability to take more structural properties into account, as well as its capability to create structural alignments based on a single miRNA. It should be noted that miRAlign is tested primarily on animal data. It was able to detect 59 miRNA candidates in Anopheles gambiae of which 37 has later been reported in the MicroRNA registry [27, 28].
COMPUTATIONAL DETECTION OF miRNAs IN VIRAL GENOMES
The first miRNAs detected in a viral genome were reported in Science 2004 [35]. Pfeffer and colleagues recorded the small RNA profile of Epstein-Barr virus (EBV) positive cells. They detected several expressed miRNA genes in EBV, and given the function of miRNAs they concluded that they had identified regulators of host and/or viral gene expression. The detection of these 5 novel miRNAs was made by cloning of small RNAs from EBV-infected cells. 4% of the small RNAs originated from EBV. The 5 novel EBV miRNAs were detected by Northern blotting. One miRNA was found in the 5′ UTR, one in the coding sequence, and one in the 3′ UTR of the same gene, BHRF-1. The last two miRNAs are from a cluster in the intronic regions of the BART gene. The miRANDA algorithm was used in their prediction of mRNA targets, a method developed for detecting miRNA targets in Drosophila [52]. Several host and/or EBV mRNA targets were found for every miRNA. The majority of the target mRNAs have more then one miRNA binding site.
In 2005 Pfeffer et al reported on the identification of several miRNAs in the herpesvirus family [33]. Their study combined a new computational method for miRNA prediction with a cloning approach similar to the one used in their initial discovery of viral miRNAs [35]. They were able to predict miRNAs in many large DNA viruses, but they were unable to predict or experimentally identify miRNAs in small RNA viruses or retroviruses. Another important finding in this study was that the EBV miRNAs neither have any significant sequence similarity with host miRNAs, nor do they seem to be conserved in the herpesvirus family. This observation indicated that methods depending on cross-species sequence conservation such as MiRscan and miRseeker, described above, are not well suited for prediction of viral miRNAs. The computational approach developed by Pfeffer and colleagues was based on defining a set of properties of known miRNA precursor stems and subsequently training a support vector machine (SVM) to separate known pre-miRNAs from stem-loops unlikely to code for miRNAs. The SVM was then applied on the set of all genomic regions potentially forming a stem-loop secondary structure. The SVM reported predictions based on a chosen threshold that resulted in the detection of 71% of the true pre-miRNAs from the training set with only 3% false positives. Their program also had a method for ranking the candidates with a score above the threshold; this method is independent of the SVM threshold score. Disregarding the direction of transcription, Pfeffer et al made 23 unique predictions of which 14 (61%) were experimentally verified. One should keep in mind that some of the predicted miRNAs can be very hard to detect as they may be expressed only under rare conditions.
Further studying the expression of the EBV BHRF-1 gene and its miRNAs, Pfeffer and colleagues suggest that viruses are able to simultaneously transcribe both miRNAs and mRNA from the same region. Pfeffer et al also suggest that their conclusions support the view of independent miRNA evolution in viruses, as viral miRNAs seem to lack sequence conservation. In addition, most miRNAs are transcribed by pol II [53], while viral miRNAs may also be transcribed by pol III [25, 33].
Almost at the same time as Pfeffer et al published their results [33], Cai et al published a paper on the detection of miRNAs in the human pathogenic Kaposi's sarcoma-associated herpesvirus (KSHV) [54]. They reported the detection of 11 distinct miRNAs, of which all were expressed in latent KSHV infected cells. These 11 miRNAs were detected by cloning small RNAs followed by RT-PCR and Northern blot analyses. MirBase (release 7.1, October 2005) [27, 28] lists 12 KSHV miRNAs, of which 10 were identified in both studies, while both Pfeffer et al and Cai et al report one additional unique miRNA.
Grey et al developed a computational method based on pre-miRNA stem-loop properties and combined it with stem-loop conservation [36], despite the findings by Pfeffer et al about lack of sequence conservation for viral miRNAs, but in line with the findings in primates [55]. Grey and colleagues studied the closely related human and chimpanzee cytomegaloviruses (HCMV and CCMV). First, all conserved stem-loop structures scoring better than a 60% similarity threshold were detected. The resulting 110 highly conserved stem-loop sequences were then run through the MiRscan program [31]. MiRscan then suggested 13 high-scoring candidates. Northern blot analysis was used on total RNA harvested at different time points for transcription verification. Five of the 13 candidates were expressed during infection, and three of these were among the ones detected by Pfeffer et al. All but one of the miRNAs found in the study by Pfeffer et al but not identified in the study by Grey et al were conserved in CCMV and had a MiRscan score above the threshold. The reason they were not detected was the initial stem-loop finder algorithm.
The miRNAs of the simian virus 40 (SV40) has also been studied [19]. Sullivan et al created a computer program called VirMir that identifies miRNA precursor candidates in small genomes (max 300 kbp). The VirMir program utilizes the RNAfold package [50]. Sullivan and colleagues ended up with two candidates out of which one region produced a suitably sized pre-miRNA that was detected by a Northern blot. The detected miRNA precursor was found to be a member of a seemingly small fraction of the miRNA precursor family, namely, those producing one mature miRNA from each stem of the precursor hairpin. Interestingly, they also discovered that both of these miRNAs are acting on the same target mRNA.
Bennasser et al argue that there are 5 likely miRNA candidates in the human immunodeficiency virus (HIV-1) [56]. Attempts to validate the candidates were in progress, but all of the miRNA candidates were found to have several cellular mRNA targets by a rule based target finder algorithm. As small-interfering RNAs (siRNAs) are somewhat related to miRNAs due to the fact that their pathways partially overlap and both become part of a RISC complex [21, 24], it is worth mentioning that the HIV-1 genome encodes an siRNA [57]. So there is evidence that viruses can encode both miRNAs and siRNA. The existence of both viral miRNAs and siRNAs was also suggested by Lu and Cullan in their paper on the adenovirus VA1 [58].
A COMPUTATIONAL SEARCH FOR EBV miRNA PRECURSORS
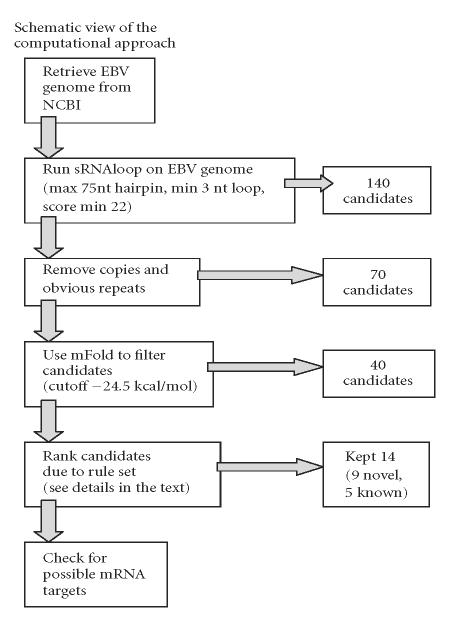
In 2004 we investigated the challenges in computational detection of miRNAs encoded in the EBV genome. The EBV genome sequence (NC_001345) was retrieved from NCBI, and then the sRNAloop program [43] (parameters: hairpin structure no more than 75 nt, loop longer than 3 nt, score threshold 22) was used to scan the entire genome for potential miRNA precursors. A total of 148 candidates were found, including all the five known EBV miRNAs. We kept only one copy of the candidates appearing more than once in the genome, narrowing down the number of candidates to 70. Potential miRNA precursors inside coding regions were not excluded. We then used mfold [37] to estimate the free energy of the entire precursors, using the web service (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). The free energy estimates for the five known EBV miRNAs ranged from −25 kcal/mol to −33.8 kcal/mol. We kept approximately 40 candidates having a free energy less than −24.5 kcal/mol, which is about the same threshold as used in the study by Lai et al [32].
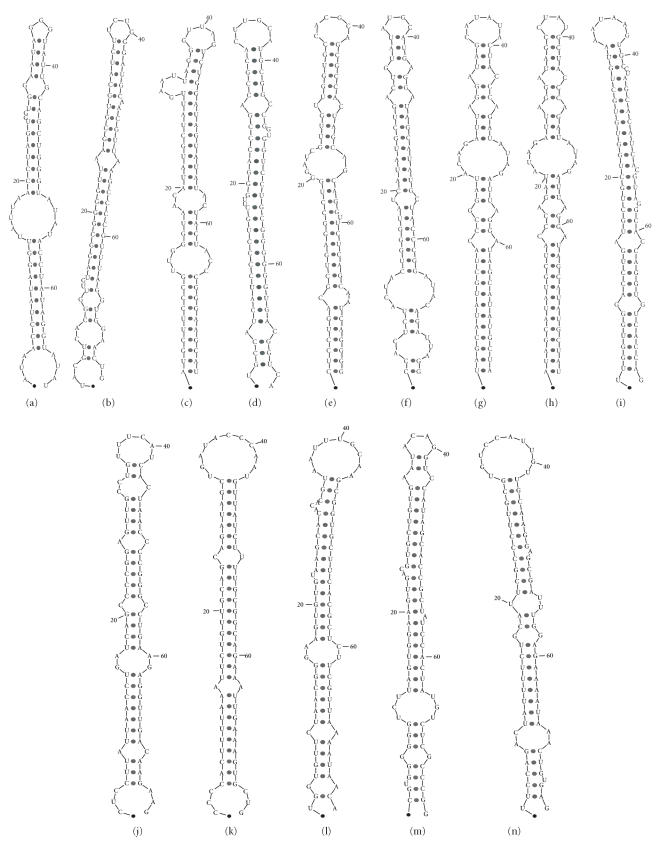
We then ranked the candidates as follows: the candidates from nonrepeat noncoding regions or hypothetical protein coding regions were ranked first, followed by candidates in known protein coding regions, and finally the remaining candidates. All of the five known pre-miRNAs were among the top ten candidates. Based on these criteria we selected the top 14 candidates for further studies, including the 5 known miRNAs. This leaves 9 novel predictions, as shown in Table 2, the according secondary structure predictions can be found in Figure 2. Attempts to experimentally verify either the 5 known miRNAs or the 9 new candidates were unsuccessful. Several possible human and EBV target mRNAs were predicted for the 9 novel pre-miRNA candidates (data not shown) using a ParAlign [59] sequence similarity search with the predicted stem sequences and a set of rules similar to the ones used by the miRANDA algorithm [52]. A schematic view of our approach can be found in Figure 3.
Table 2.
Computationally predicted miRNA precursor candidates from the Epstein-Barr virus (this study), ranked according to our criteria. The free-energy estimates were computed by the online mfold [37] version of December 2004. All predicted secondary structures can be found, according to the given letter, in Figure 2.
| Name,1 structure | Position | Direction | Length | Free energy2 | Sequence | Notes |
| PMRP 1, a | 53263–53332 | + | 69 | −25.4 kcal/mol | AUAACCUAUAGGUUAUUAACCUAGUGGUGGAAUAGGGUAUUGCAGCUGGGUAUAUACCUAUAGGUAUAU | Intergenic region, poly A signals upstream |
| PMRP 2, b | 6838–6912 | − | 74 | −32.7 kcal/mol | UACGUCACGGUUGUAGGCGGGGUUAAGCGUGCAUCUUCUGGGAUGCAACGUUAAGCCCCGUUUAGGUGGAACUG | Intergenic region |
| PMRP 3, c | 9041–9116 | + | 74 | −29.8 kcal/mol | AUGCUUCCCGUUGGGUAACAUAUGCUAUUGAAUUAGGGUUAGUCUGGAUAGUAUAUACUACUACCCGGGAAGCAU | Intergenic region, poly A signals upstream, promoter at 8573 |
| PMRP 4, d | 61262–61333 | + | 71 | −43.8 kcal/mol | UGCCAUCAUCCCCUGCUUGGGACCCGACCGCACUUGCAUGCGGCCGGUGGUCCUGCGGGGGGUGACGGUCA | Inside a hypothetical protein coding region |
| PMRP 5, e | 1898–1973 | − | 75 | −32.7 kcal/mol | CUCCUGACGCUGAGGCCUGGGAUCGUUGUUGGUGCCACGCAGCGCCACUAGCAGCAGGUUCUCAGCAAUCAGGGG | Inside a coding region |
| PMRP 6, f | 7408–7483 | + | 75 | −24.6 kcal/mol | CCACUCUACUACUGGGUAUCAUAUGCUGACUGUAUAUGCAUGAGGAUAGCAUAUGCUACCCGGAUACAGAUUAGG | Intergenic repeat region3 |
| PMRP 7, g | 7454–7526 | + | 72 | −25.4 kcal/mol | UAGCAUAUGCUACCCGGAUACAGAUUAGGAUAGCAUAUACUACCCAGAUAUAGAUUAGGAUAGCAUAUGCUA | Intergenic repeat region |
| PMRP 8, h | 7929–8003 | + | 74 | −29.4 kcal/mol | AUAGCAUAUGCUACCCAGAUAUAGAUUAGGAUAGCCUAUGCUACCCAGAUAUAGAUUAGGAUAGCAUAUGCUAU | Intergenic repeat region, promoter at 7888 |
| PMRP 9, i | 151510–151584 | + | 74 | −34.0 kcal/mol | UUGGUGGGACCUGAUGCUGCUGGUGUGCUGUAAAUAAGUGCCUAGCACAUCACGUAGGCACCAGGUGUCACCAG | Intergenic repeat region |
| BHRF 1-1, j | 53754–53829 | + | 75 | −27.9 kcal/mol | CUCCUUAUUAACCUGAUCAGCCCCGGAGUUGCCUGUUUCAUCACUAACCCCGGGCCUGAAGAGGUUGACAAGAAG | Holds known miRNA; BHRF 1-1 |
| BHRF 1–2, k | 55131–55206 | + | 75 | −32.1 kcal/mol | CCCCACUUUUAAAUUCUGUUGCAGCAGAUAGCUGAUACCCAAUGUUAUCUUUUGCGGCAGAAAUUGAAAGUGCUG | Holds known miRNA; BHRF 1–24 |
| BHRF 1–3, l | 55248–55323 | + | 75 | −25.0 kcal/mol | UGGUGUUCUAACGGGAAGUGUGUAAGCACACACGUAAUUUGCAAGCGGUGCUUCACGCUCUUCGUUAAAAUAACA | Holds known miRNA; BHRF 1–3 |
| BART 1, m | 151631–151706 | + | 75 | −33.8 kcal/mol | CGUGGGGGGUCUUAGUGGAAGUGACGUGCUGUGAAUACAGGUCCAUAGCACCGCUAUCCACUAUGUCUCGCCCGG | Holds known miRNA; BART 1 |
| BART 2, n | 153197–153272 | + | 75 | −30.8 kcal/mol | UUCCAGACUAUUUUCUGCAUUCGCCCUUGCGUGUCCAUUGUUGCAAGGAGCGAUUUGGAGAAAAUAAACUGUGAG | Holds known miRNA; BART 2 |
1The novel candidates are named PMRP (possible micro RNA precursor) 1 through 9.
2Energy calculations made using mfold [37].
3Mfold suggests two possible secondary structures for this sequence, only one structure is shown.
4Pfeffer et al [35] state that this hairpin structure gives two mature miRNAs, one from each stem-arm, the other is named BHRF 1–2*.
Figure 2.
(a)–(i) The predicted structure of the nine top scoring novel miRNA precursor candidates. (j)–(n) The predicted structure of the five known EBV miRNA precursors.
Figure 3.
Schematic view of the computational approach.
DISCUSSION
It is important to assess the significance of viral miRNA-induced posttranscriptional gene regulation in an infected cell. In C elegans, miRNAs play vital roles during development [3, 4], while such a critical role for miRNAs has not yet been discovered in viruses. Sullivan et al argue that the importance of the EBV miRNAs in viral mRNA regulation is uncertain, while claiming a more important role of the SV40 miRNA, which they have proven to reduce the cytotoxic T-lymphocyte susceptibility and also reduce local cytokine release [19]. The homology findings of Grey et al indicate that the viral miRNAs have not evolved independently [36], suggesting a more significant role than implied by theories of independent evolution.
The importance of further miRNA knowledge is illustrated by the successful use of miRNA expression profiles to classify human cancers [60], as well as data indicating that many human miRNAs are located in regions frequently associated with cancer [61].
Our study clearly indicates that predicting pre-miRNA structures seems reasonably easy apart from deciding on a score threshold for candidates. The most challenging task is to predict the accurate position of the mature miRNA within the precursor. The most promising strategy for predicting novel miRNAs in viruses appears to be a combination of the conserved stem-loop search by Grey et al and the precursor miRNA feature searches used in the Grey and Pfeffer studies. Grey et al suggest a refinement of the stem-loop finder to improve the search results as it excluded true positives that would have been accepted by the later stages of the algorithm. A broader search for stem-loop structures is also anticipated by the reports by Wang et al [44] of much longer loops (20–75 nt) in A thaliana than in the loops in the known HMCV miRNAs (4–12 nt) [33, 36].
Algorithms might also be improved by exploiting the findings of Berezikov et al [55]; while miRNAs stems show strong conservation and the loops vary in their degree of conservation, the miRNA precursors' flanking regions show a striking drop in conservation. This conservation profile can be used for phylogenetic shadowing [62], a technique for sequence comparison between closely related species. This approach was used to predict and identify several primate miRNAs [55].
Introducing a search for miRNA targets [29, 52, 63–67] at an earlier stage of the algorithm could also improve the results. In most miRNA detection approaches this is often a final separate part [44, 45]. We suggest that including an miRNA regulatory module (MRM) [68] search at an early stage could be a valuable improvement.
Concerning experimental approaches and verification it should be noted that miRNA candidates found to originate from within exons are often regarded as cloning artefacts and therefore discarded. However, as stated by Berezikov et al, there is no experimental evidence excluding miRNAs candidates in these regions [55]. Furthermore, there is evidence indicating that a region coding for both an miRNA and a protein can be used almost simultaneously for miRNA and protein production [54]. A large portion of the currently known miRNAs have emerged as a result of cloning, but cloning approaches are likely to be biased towards abundant miRNAs [43].
Current computational methods are useful tools for identifying miRNA candidates, however before better methods have been developed, we still need to verify candidates using Northern blots.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Molecular Cell. 2000;5(4):659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 6.Berezikov E, Plasterk RHA. Camels and zebrafish, viruses and cancer: a microRNA update. Human Molecular Genetics. 2005;14(suppl 2):R183–R190. doi: 10.1093/hmg/ddi271. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 8.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 9.Smalheiser NR. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biology. 2003;4(7):403. doi: 10.1186/gb-2003-4-7-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Research. 2004;14(10 A):1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO Journal. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes and Development. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohnsack MT, Czaplinski K, Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 16.Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 17.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint É, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nature Genetics. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 20.Mourelatos Z, Dostie J, Paushkin S, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes and Development. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 22.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589):2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(17):9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes and Development. 2004;18(14):1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen PY, Meister G. MicroRNA-guided posttranscriptional gene regulation. Biological Chemistry. 2005;386(12):1205–1218. doi: 10.1515/BC.2005.139. [DOI] [PubMed] [Google Scholar]
- 26.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S. The microRNA registry. Nucleic Acids Research. 2004;32(Database issue):D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright JA. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nature Genetics. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Lim LP, Lau NC, Weinstein EG, et al. The microRNAs of Caenorhabditis elegans. Genes and Development. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biology. 2003;4(7):R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeffer S, Sewer A, Lagos-Quintana M, et al. Identification of microRNAs of the herpesvirus family. Nature Methods. 2005;2(4):269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 34.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Current Biology. 2003;13(10):807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 35.Pfeffer S, Zavolan M, Grässer FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 36.Grey F, Antoniewicz A, Allen E, et al. Identification and characterization of human cytomegalovirus-encoded microRNAs. Journal of Virology. 2005;79(18):12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods in Molecular Biology. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 38.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Argaman L, Hershberg R, Vogel J, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Current Biology. 2001;11(12):941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 40.Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Current Biology. 2001;11(17):1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 41.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes and Development. 2001;15(13):1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 43.Grad Y, Aach J, Hayes GD, et al. Computational and experimental identification of C. elegans microRNAs. Molecular Cell. 2003;11(5):1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 44.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biology. 2004;5(9):R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 46.Billoud B, De Paepe R, Baulcombe D, Boccara M. Identification of new small non-coding RNAs from tobacco and Arabidopsis. Biochimie. 2005;87(9-10):905–910. doi: 10.1016/j.biochi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Dezulian T, Remmert M, Palatnik JF, Weigel D, Huson DH. Identification of plant microRNA homologs. Bioinformatics. 2006;22(3):359–360. doi: 10.1093/bioinformatics/bti802. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Smith TF, Waterman MS. Identification of common molecular subsequences. Journal of Molecular Biology. 1981;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 50.Hofacker IL, Fontana W, Stadler PF, Bonhöffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie. 1994;125(2):167–188. [Google Scholar]
- 51.Wang X, Zhang J, Li F, et al. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21(18):3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- 52.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biology. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO Journal. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(15):5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH-A, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 56.Bennasser Y, Le S-Y, Yeung ML, Jeang K-T. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1(1):43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennasser Y, Le S-Y, Benkirane M, Jeang K-T. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22(5):607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 58.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. Journal of Virology. 2004;78(23):12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rognes T. ParAlign: a parallel sequence alignment algorithm for rapid and sensitive database searches. Nucleic Acids Research. 2001;29(7):1647–1652. doi: 10.1093/nar/29.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 61.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boffelli D, McAuliffe J, Ovcharenko D, et al. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299(5611):1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 63.Grün D, Wang Y-L, Langenberger D, Gunsalus KC, Rajewsky N. MicroRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Computational Biology. 2005;1(1):e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiriakidou M, Nelson PT, Kouranov A, et al. A combined computational-experimental approach predicts human microRNA targets. Genes and Development. 2004;18(10):1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis BP, Shih I-H, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 66.Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(11):4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biology. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon S, De Micheli G. Prediction of regulatory modules comprising microRNAs and target genes. Bioinformatics. 2005;21(suppl 2):ii93–ii100. doi: 10.1093/bioinformatics/bti1116. [DOI] [PubMed] [Google Scholar]