Abstract
Transmissible spongiform encephalopathies (TSEs) are infectious, fatal neurodegenerative diseases characterized by aggregates of modified forms of the prion protein (PrP) in the central nervous system. Well known examples include variant Creutzfeldt-Jakob Disease (vCJD) in humans, BSE in cattle, chronic wasting disease in deer and scrapie in sheep and goats. In humans, sheep and deer, disease susceptibility is determined by host genotype at the prion protein gene (PRNP). Here I examine the molecular evolution of PRNP in ruminants and show that variation in sheep appears to have been maintained by balancing selection, a profoundly different process from that seen in other ruminants. Scrapie eradication programs such as those recently implemented in the UK, USA and elsewhere are based on the assumption that PRNP is under positive selection in response to scrapie. If, as these data suggest, that assumption is wrong, eradication programs will disrupt this balancing selection, and may have a negative impact on the fitness or scrapie resistance of national flocks.
Keywords: heterozygote advantage, Tajima's D, allele frequency, negative frequency-dependent selection
1. Introduction
It is well documented that variation in the coding sequence of PRNP plays a key role in susceptibility to prion disease in many mammals, including humans (Collinge et al. 1996; Mead et al. 2003), sheep (Goldmann et al. 1990; Hunter 1997) and deer (O'Rourke et al. 1999, 2004). For example, in humans virtually every vCJD patient has been homozygous for methionine at codon 129 (Peden et al. 2004), while heterozygotes at the same codon appear most resistant to kuru (Mead et al. 2003) (a prion disease of Papua New Guinean Highlanders transmitted during cannibalistic mortuary feasts). In sheep, three PRNP codons (136, 154 and 171) are largely responsible for determining scrapie susceptibility (Goldmann et al. 1990; Hunter 1997). Two alleles are observed at codons 136 (A and V) and 154 (R and H) and three are observed at codon 171 (Q, R and H). Of the five commonly observed haplotypes, A136R154R171 (hereafter ARR) is usually the most resistant, while haplotypes VRQ and ARQ are the most susceptible (Goldmann et al. 1990; Bossers et al. 2000), although the degree of susceptibility does vary between breeds. To date very few ARR homozygous sheep have tested positive for scrapie (Ikeda et al. 1995).
Despite the obvious importance of the effect of PRNP variation on transmissible spongiform encephalopathy (TSE) susceptibility, its coding sequence is highly conserved across mammalian orders (Wopfner et al. 1999; van Rheede et al. 2003), implying that the gene is under functional constraint and has historically been subject to purifying selection. This assumption is supported by a recent analysis of PRNP in cattle (Seabury et al. 2004). Paradoxically, sheep (DeSilva et al. 2003; Heaton et al. 2003) and goats (Billinis et al. 2002) appear to exhibit a high degree of PRNP variation including an excess of nonsynonymous (amino-acid changing) polymorphisms. This observation is consistent with either adaptive evolution or a relaxation of purifying selection in these species.
The relatively large amount of PRNP variation in sheep and goats is even more surprising when one examines the history of prion diseases. Scrapie was first documented in 1732 (Parry 1983; Woolhouse et al. 2001), indicating that it is older than many other TSEs. For example, BSE was first reported in 1986, kuru in the early 1900s and vCJD in 1996. Given that scrapie is an old disease it is perhaps surprising that susceptible genotypes remain at relatively high frequencies in modern sheep. Several possible explanations have been proposed for the persistence of susceptible genotypes (Woolhouse et al. 2001), including: (i) a selective advantage of susceptible genotypes when scrapie is absent; (ii) heterozygote advantage; (iii) negative frequency-dependent selection; and (iv) ongoing positive selection during a three-hundred-year epidemic that will result in the eventual eradication of susceptible genotypes, and ultimately of scrapie (Woolhouse et al. 2001). Note that the first three explanations are all forms of balancing selection that would help to maintain variation at PRNP, while positive selection would eventually lead to the loss of variation. It is uncertain which of these explanations is the most likely, but resolving this issue is key to eradicating and controlling scrapie epidemics.
Eradicating scrapie is important for two reasons. First, scrapie causes large economic loss to the sheep industry. Second, sheep may harbour concealed BSE (which is phenotypically indistinguishable from scrapie), which could, in theory, be consumed by humans, thereby exposing them to vCJD risk. In July 2001, the UK government implemented a National Scrapie Plan (NSP) with the aim of eradicating scrapie (and if present, BSE) from the national flock. The NSP has been used as a blueprint for similar schemes in USA, France and The Netherlands. Under the NSP, breeding rams must be carriers of the ARR haplotype, such that it is predicted to increase in frequency from its current value (∼0.5) to eventual fixation. In other words, scrapie eradication programs are accelerating a natural process only if PRNP has historically been under positive selection (Woolhouse et al. 2001). Conversely, if PRNP variation has been maintained by balancing selection this natural process will be disrupted. The aim of this study is to use a molecular evolution approach to understand what form of selection, if any, has been acting on PRNP during the course of ruminant evolution. Addressing this question in sheep is of particular importance given the large investment in scrapie eradication programs and the unusually large amount of genetic variation at this locus.
2. Material and methods
(a) Data collection
Sixty eight PRNP entire coding region sequences (length=771–819 bp) representing eight ruminant species were obtained from GenBank (http://www.ncbi.nlm.nih.gov/; see table 2 in the electronic supplementary material). Sequences were aligned with ClustalW (http://www.ebi.ac.uk/clustalw/), and where necessary, modified manually. An additional 16 sheep PRNP exon 3 genotypes (length ∼4.7 kb) were obtained from the literature (Hills et al. 2003) and their haplotypes were resolved using a Bayesian approach implemented in Haplotyper (Niu et al. 2002). These sequences were used for allele frequency-based tests (see below) and are unbiased with respect to PRNP genotype or allele frequency.
(b) Data analysis
Two fundamentally different approaches are available to detect the signature of selection in genomic data (Otto 2000; Ford 2002); both use Kimura's Theory of Neutral Evolution (Kimura 1983) as a null hypothesis against which various models of selection can be tested. One method examines the relative rates at which nonsynonymous (amino-acid changing) substitutions and synonymous substitutions occur. The other category of test is based on the frequency distribution of alleles within a genomic region of interest.
(i) Tests based on nonsynonymous and synonymous mutations
The ratio, ω, of nonsynonymous substitutions per nonsynonymous site (dN) to synonymous substitutions per synonymous sites (dS) was estimated for each lineage of the ruminant phylogeny in a maximum likelihood framework, using the codeml program of PAML (Yang 1997). PRNP evolution within each lineage of the ruminant phylogeny was initially examined by identifying one consensus (ancestral) haplotype from each species. Consensus sequences were identified by parsimony and were unambiguous. Three models were constructed: the one-ratio model estimates ω under the assumption that it is constant on each lineage of the phylogeny; the free-ratio model permits ω to vary between lineages; the fixed-ratio model constrains ω to a pre-specified value throughout the tree. Each estimate of ω on a lineage incorporates an additional parameter into the model. Specific hypotheses can be tested by likelihood ratio tests derived from the log-likelihood score of each model. Two null hypotheses were tested: (i) ω does not vary between lineages, tested by comparison of the one-ratio and free-ratio models; (ii) ω is not significantly different from 1 (the expectation under the neutral model), tested by comparison of the one-ratio and fixed-ratio models. A similar within-species analysis was conducted for those species for which >1 PRNP coding sequences were available. Median haplotype networks (Bandelt et al. 1995) were manually constructed, and within-species ω was estimated for each branch of the network. Putative recombinant haplotypes were excluded from the within-species analysis, although treating these haplotypes as nonrecombinant does not qualitatively alter any conclusion reached in this study because most mutations arose as singletons.
(ii) Tests based on allele frequency distributions
Test statistics designed to detect departures from neutrality including Tajima's D (Tajima 1989; hereafter DT), Fu and Li's D* and F* (Fu 1997; hereafter D*, F*) and Fay and Wu's H (Fay & Wu 2000) were calculated for 4.7 kb of sheep exon 3, a sample that was obtained at random with respect to PRNP genotype (Hills et al. 2003). Coalescent simulations (10 000 replicates) were used to generate the null distribution and statistical significance of each test statistic. Because the tests are sensitive to recombination, 4Ner (see below) was used in simulations rather than an assumption of zero recombination. All tests were implemented in DnaSP v. 4.00 (Rozas et al. 2003).
(iii) Recombination and linkage disequilibrium
Evidence for recombination within 4.7 kb of sheep PRNP exon 3 was examined using Ldhat (McVean et al. 2002), a composite likelihood estimator of the population recombination rate parameter, 4Ner, where 4Ne is the effective population size and r is the per gene per generation crossing over rate. Statistical significance was determined by the likelihood permutation test implemented within Ldhat.
Four of the 23 sheep exon 3 polymorphisms were within the coding region and all were nonsynonymous (one at codon 136, one at codon 154 and two at codon 171). Linkage disequilibrium (LD) between all pairs of exon 3 polymorphisms was estimated using the Hill & Robertson (1968) measure, and the relationship between LD and distance was examined by Mantel test (10 000 permutations). LD within this region was represented graphically with the GOLD software (Abecasis & Cookson 2000).
3. Results
(i) Tests based on nonsynonymous and synonymous mutations
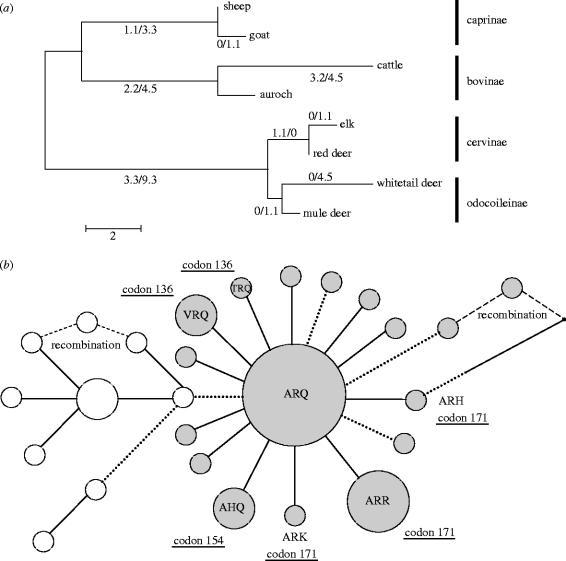
Consistent with the published literature, sheep and goats had a relatively greater proportion of nonsynonymous substitutions than other species (table 1). A free-ratio model of ω, the dN:dS ratio in the ruminant phylogeny (figure 1a), did not give a significantly better fit to the data than a one-ratio model (χ2(12)=7.64, ns), indicating that ω is not significantly variable among ruminant lineages. The one-ratio model estimate of ω=0.156 is significantly lower than 1.0 (χ2(1)=29.1; p<0.001). Thus, PRNP evolved by purifying selection during the course of ruminant speciation.
Table 1.
Summary of PRNP coding region variation in eight ruminants.
| species | N | θw | N/S | ω |
|---|---|---|---|---|
| sheep | 26 | 0.0062 | 14/5 | 1.66* |
| goats | 10 | 0.0046 | 6/2 | 1.25* |
| cattle | 16 | 0.0028 | 7/6 | 0.53 |
| auroch | 1 | – | – | – |
| elk | 6 | 0.0011 | 1/1 | 0.34 |
| red deer | 1 | – | – | – |
| mule deer | 3 | 0.0070 | 1/7 | 0.06 |
| whitetail deer | 5 | 0.0038 | 3/3 | 0.29 |
N, number of haplotypes; θw, Watterson's (Watterson 1975) estimator of the proportion of segregating sites (corrected for sample size); N/S, the number of nonsynonymous and synonymous polymorphic sites; ω, the ratio dN : dS. * indicates that ω is significantly (p<0.05) greater than 0.156 after Bonferroni correction (six independent tests).
Figure 1.
(a) PRNP coding region evolution within the ruminant phylogeny. Branches are drawn in proportion to their lengths, defined as the number of sequence differences per gene. The maximum likelihood estimates of the number of nonsynonymous and synonymous substitutions are shown for each branch (e.g. 1.1 nonsynonymous and 3.3 synonymous substitutions on the branch leading to the caprinae). The excess of synonymous substitutions is consistent with purifying selection. (b) PRNP coding region haplotype networks for sheep (filled circles) and goats (open circles). Haplotype frequency is proportional to the circle diameter. Substitutions affecting codons 136, 154 and 171 are indicated. Nonsynonymous changes are indicated by solid lines and synonymous substitutions by dotted lines. Note the excess of nonsynonymous substitutions. The number of substitutions that differentiate two haplotypes is indicated by branch length. Nearly all branches represent a single substitution.
Rates of nonsynonymous and synonymous mutations were estimated within each species to determine whether PRNP had continued to evolve by purifying selection. ω was significantly greater than 0.156 in both sheep (ω=1.66, χ2(1)=18.4, p<0.001, table 1, figure 1b) and goats (ω=1.25, χ2(1)=7.78, p<0.01, table 1, figure 1b). Therefore, the purifying selection that has typified ruminant PRNP evolution does not apply to sheep and goats. Because ω was not significantly greater than 1.0 in either species this test alone cannot distinguish between adaptive and neutral models of PRNP evolution, although testing whether ω>1.0 for an entire gene is a very conservative test of adaptive evolution, as many sites are expected to be under purifying selection to retain gene function (Nielsen 2001).
It has been suggested that tests of neutral evolution based on the relative rates of dN and dS in a phylogeny are sensitive to the effects of undetected recombination (Anisimova et al. 2003; Shriner et al. 2003). Only one sheep and one goat sequence analysed here can possibly be recombinant because most branches contain a single, unique mutation (figure 1b). When these possible recombinant sequences were excluded from the analysis the results from the codeml analysis were qualitatively identical. Additional evidence for the relaxation of purifying selection in sheep and goats is provided by McDonald–Kreitman tests (McDonald & Kreitman 1991; see table 3 in the electronic supplementary material.
(ii) Tests based on allele frequency distributions
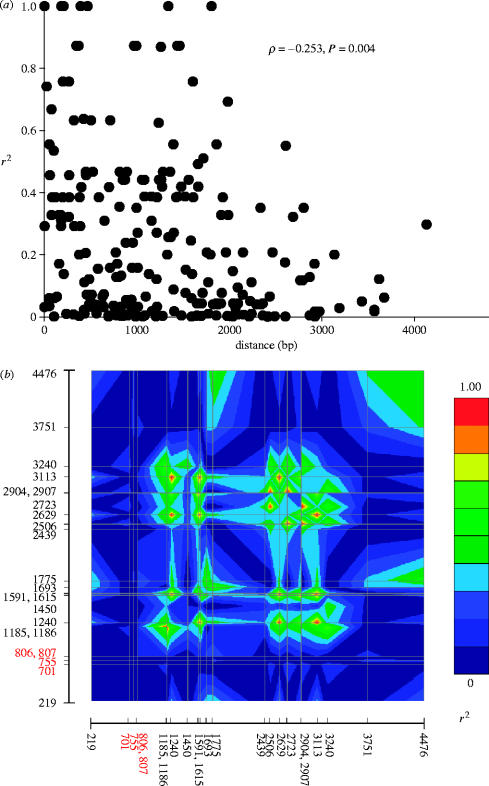
Prior to carrying out allele frequency-based tests, I investigated whether there was evidence of recombination in exon 3 of sheep PRNP. The recombination rate parameter was significantly greater than zero (4Ner=37, p=0.001), and this recombination appears to have caused LD to decline as a function of distance (figure 2a). Under the neutral model, tests based on allele frequency distributions should yield non-significant values close to zero. Here, DT, D* and F* were all significantly greater than zero (DT=1.71, p=0.0014; D*=1.68, p=0.0033; F*=1.99, p=0.0002). By contrast, H, a statistic designed to detect selective sweeps, was not significant (H=−2.54, p=0.123). Taken together, these findings (an ω>1, an excess of replacement polymorphisms and significantly positive values of DT, D* and F*) are consistent with balancing selection and not with purifying selection, positive selection or neutral evolution.
Figure 2.
(a) LD in sheep PRNP. LD was measured between all pairs of polymorphic sites within a 4.7 kb region of exon 3. Note the significant decline in LD with distance, consistent with recombination. (b) Graphical representation of LD between all pairs of polymorphic sites. The position (in bp) of each of the 23 polymorphisms is indicated on the scale bars with the 4 nonsynonymous sites shown in red font. Note that LD is very low between the nonsynonymous sites and every other site, suggesting that variation at these sites is not caused by recent mutation. Position 701 corresponds to the codon 136 A-V polymorphism, 755=codon 154 R-H, 806=codon 171Q-R, 807=codon 171Q-H.
4. Discussion
A molecular evolution analysis of PRNP in sheep suggests that this gene has evolved by balancing selection and not positive selection or purifying selection. The latter scenarios would have generated significantly negative values of DT, D* and F*. Positive selection would also be expected to yield a significantly positive Fay and Wu's H statistic. Evidence for balancing selection at PRNP in humans (Mead et al. 2003) has recently been disputed on the grounds that an ascertainment bias may have led the investigators to preferentially genotype polymorphisms where derived alleles were at intermediate frequencies (Kreitman & Di Rienzo 2004). If polymorphisms with alleles of intermediate frequency are over-represented then statistics such as DT can be inflated, giving the apparent signature of balancing selection. A bias of this nature does not apply to this analysis because all polymorphisms were detected in the same sequences that were used in the analysis. In other words, there was no initial pre-selection of intermediate frequency alleles.
The most compelling evidence of balancing selection comes from the allele frequency tests. However, these tests are sensitive not only to selection but also to demographic processes such as undetected population structure or bottlenecks. The 16 sheep (32 haplotypes) for which PRNP sequences were obtained came from 9 Norwegian breeds. Population structure among Norwegian breeds has not been estimated, although data from other Northern European breeds suggest that 15–20% of genetic variation is partitioned between breeds (Stahlberger-Saitbekova et al. 2001; Ibeagha-Awemu & Erhardt 2004). However, if population structure were present in this sample, then different breeds would be expected to reside within distinct lineages of the PRNP genealogy. This is clearly not the case (see electronic supplementary material, part C). Thus, the data are more consistent with balancing selection than population structure, although the definitive test to distinguish these two possibilities could not be performed.
Several different forms of balancing selection have been proposed to maintain ovine PRNP variation (Woolhouse et al. 2001). First, susceptible genotypes could confer greater fitness on their host in the absence of scrapie, i.e. the fitness of alternative genotypes could vary temporally, depending on the prevalence of scrapie in the population. However, associations between PRNP genotype and other traits have not been reported, to my knowledge. Thus, there is no empirical support for this hypothesis at present. Second, PRNP variation could be maintained by heterozygote advantage, either because heterozygotes show greater scrapie resistance than homozygotes, or due to LD between PRNP and a linked gene influencing fitness. Recent epidemiological and experimental evidence supports the idea that PRNP heterozygotes are more TSE-resistant than homozygotes in both humans (Mead et al. 2003) and sheep (Houston et al. 2003). A third possibility is that the relative fitness of different PRNP genotypes depends on the strain of infective scrapie that the host is most likely to encounter. There is empirical evidence that the conversion of the normal form of PrP (PrPc) to the infectious form (PrPSc) is most efficient when the host and infective prion are of the same genotype (Bossers et al. 1997, 2000). Therefore the relative susceptibility of PRNP genotypes may vary temporally and/or spatially depending on which scrapie strains are common, and PRNP variation may be maintained by negative frequency-dependent selection. Notably, ARR homozygous (i.e. ‘resistant’) sheep have tested positive for scrapie in Japan (Ikeda et al. 1995) and France (World Organisation for Animal Health, http://www.oie.int/), although in each case the genotype of the infective prion was unknown. Of the three previously proposed forms of balancing selection, heterozygote advantage and frequency-dependence appear most likely, although all three mechanisms are not necessarily mutually exclusive.
An additional, previously undiscussed, hypothesis is worthy of consideration. LD is greatest at sites approximately 2 kb downstream of the PRNP coding region (figure 2b). This observation is consistent with balancing selection acting at sites in the 3′ untranslated region of exon 3, rather than at the coding sequence. In support of this hypothesis, there is evidence that variation in the 3′ untranslated region of PRNP is associated with levels of gene expression (Goldmann et al. 1999), which in turn might be associated with scrapie susceptibility or even some other trait. Thus balancing selection at the 3′ UTR of PRNP could help to maintain variation in the coding region though LD.
Regardless of what form(s) of balancing selection are acting at PRNP, these findings may have far-reaching consequences for scrapie eradication programs. Existing strategies stipulate that all breeding rams must carry the ARR allele. Therefore positive selection is being imposed on PRNP, causing the depletion of existing genetic PRNP variation. If balancing selection is attributable to negative-frequency dependence, then a genetically uniform population of ARR homozygous sheep could be susceptible to rare scrapie strains. Until the cause(s) of balancing selection are known it would be prudent to preserve existing PRNP variation, both as frozen semen and in managed populations. This study highlights the need for further investigations of the role of PRNP variation in resistance to rare scrapie strains, as well as in fitness and production traits.
Concerns about scrapie eradication programs aside, detecting the signature of selection in livestock genomes is of more general interest. Modern animal breeding programs utilise semen from elite sires with the highest breeding values at traits of economic importance. Consequently a limited number of males sire a large proportion of progeny each generation, ensuring that effective population sizes are small and genetic variation can rapidly be lost by drift. A classic example is Holstein dairy cattle in the USA, which have an actual population size of around 4 million but an effective population size as small as 40. The Food and Agriculture Organisation of the United Nations has recently recognised the need to preserve diversity at genes relevant to production traits (see http://www.fao.org/ag/cgrfa/AnGR.htm). An important initial step is to identify genes that have been under historical selection in our domestic livestock (Bruford et al. 2003). This study provides a rare example of the footprint of natural selection being detected in the genomes of livestock.
Acknowledgements
I thank Brian Charlesworth, Dave Coltman, Wilfred Goldmann, Jake Gratten, Laurence Hurst, Loeske Kruuk, Gil McVean, Josephine Pemberton, Francis Ratnieks and Rhonda Snook for useful discussion and/or comments on the manuscript.
Supplementary Material
References
- Abecasis G.R, Cookson W.O.C. GOLD–graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- Anisimova M, Nielsen R, Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics. 2003;164:1229–1236. doi: 10.1093/genetics/164.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.J, Forster P, Sykes B.C, Richards M.B. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billinis C, Panagiotidis C.H, Psychas V, Argyroudis S, Nicolaou A, Leontides S, Papadopoulos O, Sklaviadis T. Prion protein gene polymorphisms in natural goat scrapie. J. Gen. Virol. 2002;83:713–721. doi: 10.1099/0022-1317-83-3-713. [DOI] [PubMed] [Google Scholar]
- Bossers A, Belt P, Raymond G.J, Caughey B, deVries R, Smits M.A. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl Acad. Sci. USA. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. 10.1073/pnas.94.10.4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossers A, de Vries R, Smits M.A. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J. Virol. 2000;74:1407–1414. doi: 10.1128/jvi.74.3.1407-1414.2000. 10.1128/JVI.74.3.1407-1414.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford M.W, Bradley D.G, Luikart G. DNA markers reveal the complexity of livestock domestication. Nat. Rev. Genet. 2003;4:900–910. doi: 10.1038/nrg1203. 10.1038/nrg1203 [DOI] [PubMed] [Google Scholar]
- Collinge J, Beck J, Campbell T, Estibeiro K, Will R.G. Prion protein gene analysis in new variant cases of Creutzfeldt-Jakob disease. Lancet. 1996;348:56–56. doi: 10.1016/s0140-6736(05)64378-4. 10.1016/S0140-6736(05)64378-4 [DOI] [PubMed] [Google Scholar]
- DeSilva U, Guo X, Kupfer D.M, Fernando S.C, Pillai A.T.V, Najar F.Z, So S, Fitch G.Q, Roe B.A. Allelic variants of ovine prion protein gene (PRNP) in Oklahoma sheep. Cytogenet. Genome Res. 2003;102:89–94. doi: 10.1159/000075731. 10.1159/000075731 [DOI] [PubMed] [Google Scholar]
- Fay J.C, Wu C.I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford M.J. Applications of selective neutrality tests to molecular ecology. Mol. Ecol. 2002;11:1245–1262. doi: 10.1046/j.1365-294X.2002.01536.x. 10.1046/j.1365-294X.2002.01536.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann W, Hunter N, Foster J.D, Salbaum J.M, Beyreuther K, Hope J. Two alleles of a neural protein gene linked to scrapie in sheep. Proc. Natl Acad. Sci. USA. 1990;87:2476–2480. doi: 10.1073/pnas.87.7.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann W, O'Neill G, Cheung F, Charleson F, Ford P, Hunter N. PrP (prion) gene expression in sheep may be modulated by alternative polyadenylation of its messenger RNA. J. Gen. Virol. 1999;80:2275–2283. doi: 10.1099/0022-1317-80-8-2275. [DOI] [PubMed] [Google Scholar]
- Heaton M.P, Leymaster K.A, Freking B.A, Hawk D.A, Smith T.P.L, Keele J.W, Snelling W.M, Fox J.M, Chitko-McKown C.G, Laegreid W.W. Prion gene sequence variation within diverse groups of US sheep, beef cattle, and deer. Mamm. Genome. 2003;14:765–777. doi: 10.1007/s00335-003-2283-y. 10.1007/s00335-003-2283-y [DOI] [PubMed] [Google Scholar]
- Hill W.G, Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968;38:226–231. doi: 10.1007/BF01245622. 10.1007/BF01245622 [DOI] [PubMed] [Google Scholar]
- Hills D, Schlaepfer J, Comincini S, MacLean I, Dolf G, Ferretti L, Olsaker I, Williams J.L. Sequence variation in the bovine and ovine PRNP genes. Anim. Genet. 2003;34:183–190. doi: 10.1046/j.1365-2052.2003.00977.x. 10.1046/j.1365-2052.2003.00977.x [DOI] [PubMed] [Google Scholar]
- Houston F, Goldmann W, Chong A, Jeffrey M, Gonzalez L, Foster J, Parnham D, Hunter N. Prion diseases: BSE in sheep bred for resistance to infection. Nature. 2003;423:498–498. doi: 10.1038/423498a. 10.1038/423498a [DOI] [PubMed] [Google Scholar]
- Hunter N. PrP genetics in sheep and the implications for scrapie and BSE. Trends Microbiol. 1997;5:331–334. doi: 10.1016/s0966-842x(97)01081-0. 10.1016/S0966-842X(97)01081-0 [DOI] [PubMed] [Google Scholar]
- Ibeagha-Awemu E.M, Erhardt G. Genetic variations between African and German sheep breeds, and description of a new variant of vitamin D-binding protein. Small Ruminant Res. 2004;55:33–43. 10.1016/j.smallrumres.2004.01.002 [Google Scholar]
- Ikeda T, Horiuchi M, Ishiguro N, Muramatsu Y, Kai-Uwe G, Shinagawa M. Amino acid polymorphisms of PrP with reference to onset of scrapie in Suffolk and Corriedale sheep in Japan. J. Gen. Virol. 1995;76:2577–2581. doi: 10.1099/0022-1317-76-10-2577. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge, UK: 1983. [Google Scholar]
- Kreitman M, Di Rienzo A. Balancing claims for balancing selection. Trends Genet. 2004;7:300–304. doi: 10.1016/j.tig.2004.05.002. 10.1016/j.tig.2004.05.002 [DOI] [PubMed] [Google Scholar]
- McDonald J.H, Kreitman M. Adaptive protein evolution at the adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S, et al. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science. 2003;300:640–643. doi: 10.1126/science.1083320. 10.1126/science.1083320 [DOI] [PubMed] [Google Scholar]
- Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. 10.1046/j.1365-2540.2001.00895.x [DOI] [PubMed] [Google Scholar]
- Niu T.H, Qin Z.H.S, Xu X.P, Liu J.S. Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am. J. Hum. Genet. 2002;70:157–169. doi: 10.1086/338446. 10.1086/338446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke K.I, Besser T.E, Miller M.W, Cline T.F, Spraker T.R, Jenny A.L, Wild M.A, Zebarth G.L, Williams E.S. PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J. Gen. Virol. 1999;80:2765–2769. doi: 10.1099/0022-1317-80-10-2765. [DOI] [PubMed] [Google Scholar]
- O'Rourke K.I, Spraker T.R, Hamburg L.K, Besser T.E, Brayton K.A, Knowles D.P. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J. Gen. Virol. 2004;85:1339–1346. doi: 10.1099/vir.0.79785-0. 10.1099/vir.0.79785-0 [DOI] [PubMed] [Google Scholar]
- Otto S.P. Detecting the form of selection from DNA sequence data. Trends Genet. 2000;16:526–529. doi: 10.1016/s0168-9525(00)02141-7. 10.1016/S0168-9525(00)02141-7 [DOI] [PubMed] [Google Scholar]
- Parry H. Scrapie disease in sheep. Academic Press; London: 1983. [Google Scholar]
- Peden A.H, Head M.W, Ritchie D.L, Bell J.E, Ironside J.W. Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet. 2004;364:527–529. doi: 10.1016/S0140-6736(04)16811-6. 10.1016/S0140-6736(04)16811-6 [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio J.C, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. 10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Seabury C.M, Honeycutt R.L, Rooney A.P, Halbert N.D, Derr J.N. Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3. PNAS. 2004;101:15142–15147. doi: 10.1073/pnas.0406403101. 10.1073/pnas.0406403101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D, Nickle D, Jensen M.A, Mullins J.I. Potential impact of recombination on sitewise approaches for detecting positive natural selection. Genet. Res. 2003;81:115–121. doi: 10.1017/s0016672303006128. 10.1017/S0016672303006128 [DOI] [PubMed] [Google Scholar]
- Stahlberger-Saitbekova N, et al. Genetic relationships in Swiss sheep breeds based on microsatellite analysis. J. Anim. Breed. Genet. 2001;118:379–387. 10.1046/j.1439-0388.2001.00312.x [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheede T, Smolenaars M.M.W, Madsen O, de Jong W.W. Molecular evolution of the mammalian prion protein. Mol. Biol. Evol. 2003;20:111–121. doi: 10.1093/molbev/msg014. 10.1093/molbev/msg014 [DOI] [PubMed] [Google Scholar]
- Watterson G. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. 10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Woolhouse M.E.J, Coen P, Matthews L, Foster J.D, Elsen J.M, Lewis R.M, Haydon D.T, Hunter N. A centuries-long epidemic of scrapie in British sheep? Trends Microbiol. 2001;9:67–70. doi: 10.1016/s0966-842x(00)01912-0. 10.1016/S0966-842X(00)01912-0 [DOI] [PubMed] [Google Scholar]
- Wopfner F, Weidenhofer G, Schneider R, von Brunn A, Gilch S, Schwarz T.F, Werner T, Schatzl M. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J. Mol. Biol. 1999;289:1163–1178. doi: 10.1006/jmbi.1999.2831. 10.1006/jmbi.1999.2831 [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.