Abstract
Many social animals live in stable groups. In contrast, African savannah elephants (Loxodonta africana) live in unusually fluid, fission–fusion societies. That is, ‘core’ social groups are composed of predictable sets of individuals; however, over the course of hours or days, these groups may temporarily divide and reunite, or they may fuse with other social groups to form much larger social units. Here, we test the hypothesis that genetic relatedness predicts patterns of group fission and fusion among wild, female African elephants. Our study of a single Kenyan population spans 236 individuals in 45 core social groups, genotyped at 11 microsatellite and one mitochondrial DNA (mtDNA) locus. We found that genetic relatedness predicted group fission; adult females remained with their first order maternal relatives when core groups fissioned temporarily. Relatedness also predicted temporary fusion between social groups; core groups were more likely to fuse with each other when the oldest females in each group were genetic relatives. Groups that shared mtDNA haplotypes were also significantly more likely to fuse than groups that did not share mtDNA. Our results suggest that associations between core social groups persist for decades after the original maternal kin have died. We discuss these results in the context of kin selection and its possible role in the evolution of elephant sociality.
Keywords: social behaviour, population genetics, African elephant, kin selection, relatedness
1. Introduction
The opportunity for kin selection to act on social behaviour is partially determined by how much time individuals spend with their genetic relatives (Hamilton 1964a,b; Maynard-Smith 1964; West-Eberhard 1975). In many social animals, kin are clustered into social groups that are stable from one generation to the next. However, some highly social species—notably humans, chimpanzees, dolphins and elephants—live in flexible, fission–fusion societies where group composition can change over the course of hours, days or weeks (Douglas-Hamilton 1972; Wursig 1978; Moss & Poole 1983; Goodall 1986; Whitehead & Christal 2001; Sukumar 2003). This flexibility may allow individuals to optimize the costs and benefits of group-living (Dunbar 1992; Kummer 1995; Van Schaik 1999). However, individuals in fission–fusion societies may not always be with their relatives; hence, opportunities for kin selection may be attenuated.
Here we address the question: to what extent does genetic relatedness predict the patterns of fission and fusion within and between social groups of female African elephants? Elephants are highly social, and their fission–fusion social structure has been well described from a behavioural perspective (Douglas-Hamilton 1972; Moss & Poole 1983; Moss 1988; Sukumar 2003; Wittemyer et al. 2005; Moss in press). Further, female elephants appear to have extensive knowledge about their relationships with many other animals in their population (Moss & Poole 1983; Moss 1988; McComb et al. 2000; McComb et al. 2001; Moss in press). Within elephant populations, ‘core’ social groups, often called ‘families’, are composed of predictable sets of individuals (Douglas-Hamilton 1972; Moss & Poole 1983; Moss 1988). Core groups may contain 1–20 adult females and their immature offspring. At maturity, males disperse and females generally remain with their natal core social group. Over the course of hours, days or weeks, these core groups may fission temporarily into smaller subgroups, or alternatively, whole core groups or subgroups may fuse to form a larger group with adult females from other core groups across the population. When two or more core social groups repeatedly and consistently fuse to form larger groups, the participating core groups are collectively known as a ‘bond group’ (Moss & Poole 1983). Individual elephants demonstrate long-term fidelity (over decades) for core social groups and bond groups (Moss in press), and it has been hypothesized that kinship may be one factor underlying these associations (Douglas-Hamilton 1972; Moss & Poole 1983).
Here we investigate the relationship between kinship and social structure in the wild population of elephants that lives in and around Amboseli National Park, Kenya. These elephants are individually known and have been studied by the Amboseli elephant research project (AERP) since 1972—longer than any other elephant population in the world. Hence their social structure is unusually well characterized, most recently in a detailed description of long-term, female social dynamics (Moss in press). The purpose of this study was to genetically confirm the observation that female elephants are matrilocal, and to test three long-held hypotheses concerning the nature of fission–fusion sociality in elephants (Douglas-Hamilton 1972; Moss & Poole 1983; Moss 1988; Moss in press): (i) that female elephants consistently remain with their closest kin when their core social groups fission temporarily into subgroups, (ii) that the core groups that make up a bond group were once part of a single core group that underwent a more permanent fission at some point in the past and (iii) that maternal kinship generally predicts the fission and fusion of all core social groups across the entire population. Positive results will indicate that female African elephants could accrue indirect fitness benefits from social relationships.
2. Material and methods
(a) Study area and population
Data were collected, between 1998 and 2003, from free-ranging, habituated, adult female elephants in and around Amboseli National Park, Kenya. The Amboseli ecosystem is semi-arid mixed savannah and woodlands. The park has permanent springs that provide a continuous source of water, but cyclical rainfall patterns cause annual wet and dry seasons. Mean annual rainfall is 346 mm in Amboseli, and most precipitation falls during biannual rainy seasons from March to May and November to December (Altmann et al. 2002).
The 390 km2 park and the surrounding dry lake basin support a population of around 1200 elephants (Moss 2001; Moss in press). All are individually recognizable from photographs of distinguishing characteristics, including tusk shape, body shape and holes or tears in the ears. The adult female elephants in Amboseli live in 55 core social groups (often called ‘families’) that range in size from 1 to 17 mother–calf units (Moss in press, mean±s.d. for this study=6.73±3.92). Because elephants respond flexibly to changes in their physical and social environment, and social groupings change as a consequence, core social groups in this study were not identified using a single threshold of association. Instead, core social groups were identified at the start of the AERP (between 1972 and 1978) based on repeated observations of (i) consistent spatial associations, (ii) coordinated activities, (iii) orientation around a single leader (i.e. matriarch) and (iv) high rates of affiliative behaviours that are exclusively exchanged among members of a core social group (such as alloparental care; Lee 1987; Moss in press). Since they were originally identified, most of the core social groups have grown in size and experienced phases of both tighter and looser cohesion. Seven have fissioned permanently to create new core social groups. However, the features listed above—especially orientation around the same leader and mutual offspring care—have remained consistent and exclusive features of core social groups since the inception of the long-term study. These features allow us to distinguish core social groups from other social units. The AERP maintains an ongoing list of all the permanent members of every core social group in the population (membership changes through birth, death, emigration—especially of maturing males—and very occasional immigration; see §3a).
For adult females born after observations began on the population in 1972, their familial relationships to one or more other core group members are known (their mother and often some of their sisters). However, elephants are very long-lived and even with 30 years of continuous observations, there are many pairs of females within core groups for whom we do not know pedigree relationships, including some mother–daughter pairs and sisters, and many more distant kin (aunts, nieces, cousins).
(b) Measuring association patterns
Elephants range widely and unpredictably; therefore, our sampling scheme was opportunistic. Each day we searched for elephants, and when we encountered them, we collected data on their association patterns. This data collection was restricted to daytime hours, but took place during all months of the year.
During data collection, we recorded spatial association at two levels of analysis: for individual females and for core social groups. For both individuals and core social groups, association indices (AI) were calculated using the ‘simple index’ where . In this equation, NA and NB are the total number of times either individual (or core social group) A or B was seen alone and NAB is the total number of times that A and B were seen together (Ginsberg & Young 1992). Differences in the way that individual and group level AI were calculated (explained in §2b(i),(ii)) mean that these values cannot be directly compared and were only used in separate analyses.
(i) Association patterns of individual females within core social groups
Patterns of association between adult female elephants were collected via scan sampling (Altmann 1974) of all adult females from ten focal core groups. As noted above, each core group consists of a set of permanent members, although these permanent members were not always found together. During any given observation session, we might find the members of a given core group all together in the same party or fissioned into two or more parties or fused with other core groups or subgroups into larger parties. When observers encountered any adult females from these focal core groups, they recorded the identity of all adult females present, as well as their spatial association patterns (i.e. how far they were from each other). For this analysis, individuals were considered to be in the same party and therefore ‘associated’, when no more than 100 m separated the most distant party member from her nearest neighbour. Scans were collected when the party was initially encountered, and at 10 min intervals thereafter, until observers left to search for a new party of elephants (10 min–2 h). Between July 2000 and July 2003 we collected 4868 scan samples of association among individuals.
(ii) Association patterns between core social groups
Patterns of association between core social groups were collected via scan sampling (Altmann 1974) as part of the AERP's long-term monitoring of the Amboseli elephant population. When observers encountered one or more adult female elephants in a party they recorded the identity of each female's core social group. The unit of analysis in the association index presented here is the core social group, not the individual, and any adult female in the core group could contribute to the final association index calculated between her core group and another core group. For this analysis, parties were defined as any aggregation of elephants where no single member or sub-group was at a distance from its nearest neighbour greater than the diameter of the main body of the group at its widest point. Between January 1998 and July 2003 we collected 5431 scan samples of association among core social groups.
(c) Quantifying group structure via cluster analysis
We visualized association patterns by applying cluster analysis to matrices of AI between individuals and core social groups. Previous studies of elephant social organization have suggested that elephant societies are hierarchical (e.g. Moss & Poole 1983), and cluster analysis is particularly useful for identifying such hierarchical clusters (Wittemyer et al. 2005). Before applying cluster analysis, we converted AI to distance measures; we did this by subtracting them from 1. Once the distance matrix is constructed, then hierarchical agglomerative clustering proceeds by joining the two individuals with the greatest similarity across the matrix first, and then successively joining individuals and groups of individuals in order of next closest similarity (thus, individuals who associate closely are at the tips of the tree). We used the un-weighted pair-group method using arithmetic averages (UPGMA) algorithm to join clusters, thus the distance between two clusters is the average distance among all members of each cluster. All cluster analyses were performed in S-Plus (v. 6.2 for Windows, Lucent Technologies Inc.).
(d) Genetic sample collection and constraints on the genetic data set
Genetic samples were either faeces or tissue collected from known individuals. Specific collection methods are outlined in Archie et al. (2003) and Buchan et al. (2005). DNA from faeces was isolated using the QIAamp DNA Stool Mini Kit (QIAGEN Inc, Valencia, CA) with some modifications (Archie et al. 2003). DNA from tissue was isolated using the Qiagen DNeasy Tissue Kit (QIAGEN Inc, Valencia, CA).
While it would have been ideal to perform genetic analyses on all adult females in all core social groups in Amboseli (approximately 400 individuals), this was impossible for several reasons: elephants range unpredictably, samples are difficult to collect in some habitats, and template DNA in faecal samples is often low quantity and quality. As a result, we were not able to collect samples from, or genotype all adult females in all core social groups. Thus, we had more complete genotype information in some core social groups than in others (see table 1 in electronic supplementary material), and this is reflected in our analyses.
(e) Mitochondrial and microsatellite DNA amplification
For 236 individuals we amplified a 672 bp sequence of mitochondrial control region using primers MDL 3 [5′-CCCACAATTAATGGGCCCGGAGCG-3′] and MDL 5 [5′-TTACATGAATTGGCAGCCAACCAG-3′] (Fernando & Lande 2000). PCR amplification was performed in 10 μl reactions containing 1 μl of DNA extract, 0.4 μl of each 5 μM primer, 2μl of 2 mM dNTP mix (Invitrogen, Carlsbad, CA), 1 μl of 100 mg ml−1 BSA, 1 μl 10× PCR buffer without MgCl2, 0.6 μl of 1.5 mM MgCl2 and 0.04 μl of Taq DNA polymerase (QIAGEN, Maryland, USA), and 3.56 μl of water. Reactions were amplified using a touchdown protocol in an MJ Research PTC-200 Thermocycler (MJ Research, Waltham, MA). Amplification was proceeded by a 4 min denaturation step at 95 °C, followed by 11 cycles of 1 min each at 68 °C annealing, 72 °C extension and 95 °C denaturation. For the next five cycles, annealing decreased 1 °C until it reached 63 °C. This 63 °C cycle was repeated 15 times and followed by 5 min at 72 °C. PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN, Valencia, CA) and eluted in 30 μl buffer EB. Sequencing was carried out using an ABI PRISM 3700 DNA analyzer using dye terminator cycle sequencing.
Complete microsatellite genotypes were determined for 236 individuals from 10 tetranucleotide (see table 2 in the electronic supplementary material; loci LaT05, LaT07, LaT08, LaT13, LaT16, LaT17, LaT18, LaT24, LaT25 and LaT26; Archie et al. 2003) and one dinucleotide locus (see table 2 in the electronic supplementary material; LaFMs02, Nyakaana & Arctander 1998). Amplification conditions and primer sequences are in Archie et al. (2003). PCR products were separated using an ABI PRISM 3700 DNA Analyzer. Allele sizes were determined using Genotyper software (v. 2.5, PE-Applied Biosystems, California, USA).
(f) Genotyping protocol and reliability
Mitochondrial DNA (mtDNA) haplotypes were sequenced for 236 individual adult female elephants. Since the concentration of mitochondrial template DNA is relatively high in faecal DNA extracts, amplification is generally successful and is less prone to the problems of non-invasive genetic analysis. However, in order to ensure accurate genotyping, all mtDNA genotypes were replicated with two independent PCR reactions, usually from two independently collected faecal samples (independent samples were collected from different defecations). Finally, although nuclear copies of the elephant mitochondrial genome have been reported (Greenwood & Paabo 1999), we have no reason to suspect this because each individual produced only one control region haplotype, and replicate PCRs always produced the same sequence.
Complete microsatellite genotypes were assigned for 236 individual adult female elephants. Of these genotypes, 227 were derived from faecal samples and 9 were derived from tissue samples. Each genotype derived from tissue was replicated twice. For faecal genotyping we used a modified version of the multi-tubes approach in order to ensure accurate genotypes (Navidi et al. 1992; Taberlet et al. 1996). Initially, two replicate positive PCRs, each from two independent extracts for the same individual (more than 1 extract was available for 217 of 227 individuals), were carried out for each individual at each locus. Results of the two initial replicate positive PCRs allowed individuals to be placed into one of three categories: true heterozygotes, possible heterozygotes and possible homozygotes. Animals were considered true heterozygotes if both PCRs produced identical heterozygote genotypes and we observed no Mendelian mismatches; in this case, no further replications were performed. Animals were considered possible heterozygotes when the first two PCRs produced both a heterozygote and homozygote genotype with a common allele between them, or two genotypes, each homozygous for a different allele. Possible heterozygotes were replicated until both alleles were observed at least twice. If one of the alleles observed in the initial replicates failed to appear again (after seven positive replicates) it was classified as an error. Animals were considered possible homozygotes if both initial replicates revealed a single, identical allele. Possible homozygotes were replicated until the same, single allele was observed in a total of seven PCRs. If an additional allele appeared two or more times in those replicates, the individual was considered a heterozygote. If an additional allele appeared only once, the individual was considered a homozygote and the unique allele was labelled an error.
All final genotypes were in Hardy–Weinberg equilibrium (Buchan et al. 2005). We also monitored the data for Mendelian errors (i.e. mother–offspring mismatches; maternity was known through observations shortly after parturition for all offspring born since 1972). We only observed one Mendelian mismatch between one mother–daughter pair at one locus. In this case, one of the daughter's mismatched alleles was one repeat unit larger than one of her mother's alleles, and we attributed this to a germ-line mutation. This low rate of inter-generational mismatch is within the published range of microsatellite mutation rates (e.g. Lai & Sun 2003).
(g) Statistical analyses of genetic relatedness and association
Average pairwise genetic relatedness within and between core social groups was estimated using the program Relatedness v. 5.0.8 (Queller & Goodnight 1989). Pairwise genetic relatedness is underestimated when the sample contains a large portion of relatives (e.g. Altmann et al. 1996), and consequently we used the bias-corrected value for relatedness, and all standard errors were determined by jack-knifing over loci (Queller & Goodnight 1989). Our estimates of relatedness fit the expectations of relatedness for various relationship categories (e.g. average pairwise genetic relatedness between mothers and offspring±s.e.=0.47±0.01, n=96 pairs; average pairwise genetic relatedness between maternal siblings±s.e.=0.28±0.02, n=58 pairs).
We used permutation tests to determine whether closely associating core groups and their matriarchs were significantly more related to each other as compared to random pairs of matriarchs and core groups drawn from the across the population. Permutation tests were carried out by writing a program in MatLab (v. 6.5, release 13, The Mathworks Inc.) that calculated the average relatedness among a specified number of randomly chosen pairs of core social groups. Average relatedness was re-calculated 1000 times to generate a distribution of means. In order to test significance, the observed mean was compared to this distribution.
In order to test whether relatedness generally predicted association across all core social groups, we used a Mantel test to correlate a matrix of pairwise AI with a matrix of pairwise relatedness among core social groups (Dietz 1983). Testing was carried out using the program PC-ORD (v. 4.0 for Windows, 1999, MjM software, Gleneden Beach, Oregon, USA) and significance was determined using Monte Carlo randomization.
3. Results
(a) Relatedness and association between individuals within the same core social group
(i) Most females are matrilocal
We sequenced mitochondrial control region haplotypes for 236 adult female elephants from 44 (of 55) core social groups (see electronic supplementary material). Among these sequences, 4 variable sites defined 4 haplotypes (NCBI GenBank accession numbers AY968043-6). The three most common haplotypes (frequencies; AMB1=0.34, occurring in n=79 individuals; AMB2=0.49, n=115 individuals; AMB3=0.14, n=33 individuals) each differed from one another by two mutations. The fourth and least common haplotype (AMB4, frequency=0.04, n=9 individuals) was found in only one core social group (QB) and differed by only one mutation from haplotype AMB3.
Consistent with the predictions of female matrilocality, we found that most members of the same core social group shared the same mtDNA haplotype. We genotyped at least two adult females in 39 core social groups (n=231 females, average percent of adult females genotyped per core social group=85%), and of these, 37 core social groups had complete uniformity of mtDNA haplotypes. However, two core social groups each contained one female that had mismatched mtDNA. These females were Jody from the JAs and Puff from the PAs. This result—that 5% of core social groups (2 of 39) contained an immigrant, or that 0.9% of females (2 of 231) immigrated to a new core social group—may slightly underestimate the actual rate of female migration between core social groups. Since there are only four mtDNA haplotypes among females in Amboseli, mtDNA haplotype diversity is a relatively coarse measure of migration. If we assume that emigrating females join core social groups at random, they would have around a 30% chance of joining a core social group that had their own haplotype (based on the haplotype frequencies that we observed), and so the actual rate of emigration may be closer to 1.8% of females (1.1%=0.9%+(0.3×0.9%)=3 of 232 females) and 8% of core social groups (3 of 39 core social groups). Migration rates may be higher if females do not join new social groups at random and prefer to join new groups that share their mtDNA haplotype.
Long-term observations in Amboseli indicate that females immigrate because they have lost all of their natal core social group members (Moss in press). When this happens, the decision to join a new core social group may be influenced by the number of relatives a female has—including paternal relatives—in their new group. It would be ideal to test this by comparing (i) the average pairwise genetic relatedness between the immigrant female and all her original core group members to (ii) the average pairwise relatedness between the immigrant and her new core social group. However, this was impossible because we did not know the original core social groups of the immigrant females. Instead, we compared (i) the average pairwise relatedness of the immigrant females to their new core social group members, to (ii) the average pairwise relatedness of all other non-immigrant females to their core social group members (n=221 females in 34 core groups, average percent of adult females genotyped per core social group=91%). We found little evidence that immigrant females are as closely related as natal females to members of their new core social groups. The average pairwise genetic relatedness of Puff and Jody to the other adult females in their new core social group was −0.03 (n=13) and 0.02 (n=5) respectively. These values were in the lowest 4 and 7% of all adult females. Furthermore, the maximum pairwise relatedness of Puff and Jody to another adult female in their new core group was 0.16 and 0.04, respectively. While Puff appears to be closely related to another core group member, she had higher pairwise relatedness values with adult females in 21 other core groups. Hence, while we cannot rule out the possibility that immigrant females join core groups where they have paternal kin, our data suggest that it is uncommon for females to do so.
(ii) Core social group members are close genetic relatives
Although a few females were not matrilocal, most females remained with their natal group, and as a result, average pairwise genetic relatedness within core social groups—as calculated from microsatellite genotypes—was high (average pairwise relatedness among adult female core social group members±s.e.=0.1502±0.0158, n=221 females in 34 core groups, average percent of adult females genotyped per core group=91%). However, stochastic demographic events (e.g. births and deaths) and occasional female migrants created considerable variability, and thus average relatedness within core groups varied widely (figure 1).
Figure 1.
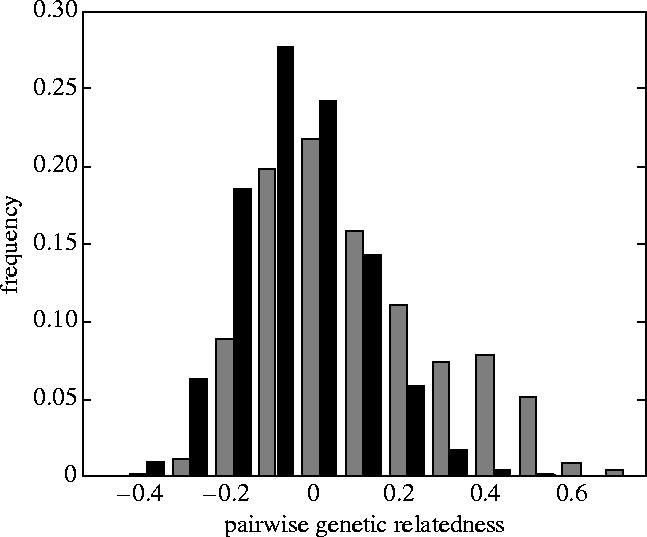
Histogram of the frequency of pairwise genetic relatedness values calculated for all pairs of adult females who were members of the same core social group (grey bars, n=865 pairs) and all pairs who were members of different core social groups (black bars, n=26 864 pairs). Genetic relatedness was calculated from the complete genotypes of 236 adult females at 11 microsatellite loci. Average pairwise genetic relatedness within the entire population is, by definition, zero.
(iii) The strength of association between core social group members is variable
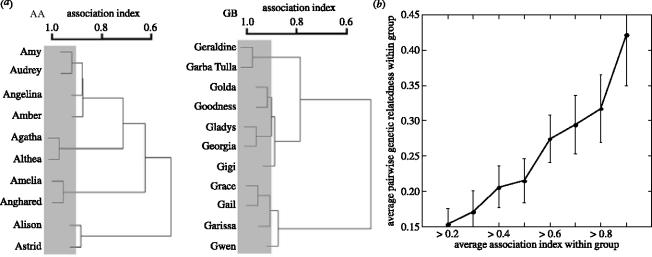
Because elephant social groups fission and fuse, members of our ten focal core groups varied in how often they were together in the same party. On average, a given pair of adult female core group members were in the same party about 2/3 of the time (average AI±s.d.=0.637±0.215, n=317 dyads in 10 focal core groups), but pairwise AI ranged widely within core social groups from 0.196 to 0.993 (see also Moss in press). In general, we found that individual adult females were usually in parties with a few other core group members, and associated less frequently with the rest of the adult females in their core social group. These patterns of association and sub-grouping within core social groups were also described using cluster analysis (see §3a(iv), and figure 2a for examples of two core social groups).
Figure 2.
(a) UPGMA trees of AI for the AA and the GB core social groups. The grey box encompasses the clusters that have average AI >0.9; average pairwise genetic relatedness within these clusters, across all ten focal core groups, was 0.42 (see b). (b) A plot created by ‘cutting’ the association distance trees at set association intervals, and then calculating average pairwise genetic relatedness within the resulting clusters. Values are calculated for 80 adult female elephants from 10 different focal core groups. Standard errors for genetic relatedness were calculated by jack-knifing across loci.
(iv) Relatedness predicts association between individual core social group members
In order to determine whether genetic relatedness predicted the observed variation in the strength of association, we ‘cut’ the association trees resulting from cluster analysis at 10% intervals and then calculated average pairwise genetic relatedness within the resulting clusters (figure 2b). We found that average pairwise genetic relatedness within clusters that spent at least 90% of their time in the same party was 0.42 (figure 2b). This high degree of relatedness indicates that these unusually close associates were almost always first order maternal relatives (mothers, daughters and maternal sisters). As associations expanded to include individuals that females associated with less often, average relatedness within clusters declined.
We also used Mantel matrix correlation to statistically demonstrate the correlation between a matrix of pairwise AI between all core social group members and a matrix of pairwise genetic relatedness values between all members in our 10 focal core groups. We found that variation in the strength of association was explained by genetic relatedness for almost all adult females in these core social groups (Monte Carlo randomization of Mantel tests, table 1). In two core social groups, relatedness was not a significant predictor of association. One was a very small core group (DB, n=4), in which pairs showed little variance in strength of association. In the other (FB), the correlation coefficient was positive but smaller in magnitude than coefficients in other core groups, reflecting some unusually close associations among individuals who were not each others' closest relatives.
Table 1.
Mantel test results for the correlation between a genetic relatedness and association (Z>observed out of 1000 Monte Carlo simulations, r is similar to the Pearson correlation statistic).
| core social group (number of adult females) | r | p | Z>observed |
|---|---|---|---|
| AA (10) | 0.46 | 0.004 | 2 |
| CB (6) | 0.73 | 0.01 | 3 |
| DB (4) | −0.89 | >0.1 | 914 |
| EA (9) | 0.67 | 0.001 | 0 |
| EB (10) | 0.68 | 0.001 | 0 |
| FB (6) | 0.34 | >0.5 | 68 |
| GB (11) | 0.62 | 0.001 | 0 |
| JA (6) | 0.65 | 0.014 | 11 |
| OA (9) | 0.81 | 0.001 | 0 |
| PC (9) | 0.7 | 0.001 | 0 |
(b) Relatedness and association between core social groups across the population
(i) Some core social groups have unusually close associations with other core social groups
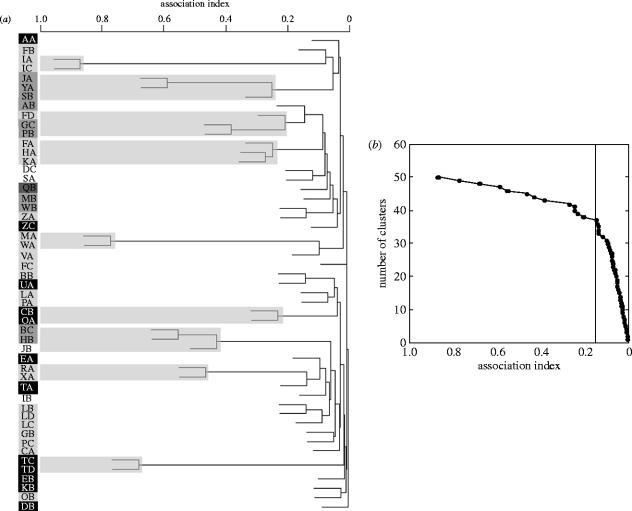
Core social groups of elephants sometimes merge to form large parties that contain many tens—and occasionally hundreds—of animals. Between 1998 and 2003, the parties we observed contained females from at least two different core social groups during approximately 40% of sightings. However, while most core social groups associate with each other relatively infrequently, some core social groups had unusually close associations. This pattern is depicted in a cluster diagram of AI between core social groups: most core social groups are clustered near the base of the tree, but a few long branches indicate pairs or trios of core social groups that had unusually high AI (figure 3a). A plot of the number of clusters as a function of association distance reveals a sharp change in slope between D=0.80–0.85 (figure 3b; see Wittemyer et al. (2005) for a statistically based approach to identifying the break point in a similar analysis). There are 9 clusters, containing 22 core social groups, with nodes beyond this breakpoint. Four of these clusters indicate three-way associations (BC/HB/JB, GC/PB/FD, FA/KA/HA and JA/YA/SB) and five indicate two-way associations (CB/OA, IA/IC, MA/WA, TC/TD and XA/RA). We consider these clusters to be bond groups (Moss & Poole 1983; see Moss (in press) for a description of bond group membership over a longer period of time, with similar relationships to those described here).
Figure 3.
(a) UPGMA tree of AI between 51 (out of 55) of the most frequently seen core social groups in Amboseli. Black and grey boxes on the left contain core group names and each greyscale combination represents a different mitochondrial DNA d-loop haplotype (black with white letters=AMB1, light grey with black letters=AMB2, dark grey with black letters=AMB3, dark grey with white letters=AMB4, and no box with grey letters=un-genotyped core group). Bond group clusters (defined in figure 2b) are encompassed by light grey boxes. (b) Plot of the accumulation of clusters (or nodes) on the tree in figure 2a as a function of association index. The vertical line at association index 0.17 indicates where the slope changes. The 13 points to the left of this are derived from all the clusters that comprise the nine bond groups in Amboseli.
(ii) Bond groups are maternal kin
Bond groups may form because their member core social groups were once part of the same core group that grew large and then underwent a relatively permanent fission (Douglas-Hamilton 1972; Moss & Poole 1983; Moss 1988; Moss in press). If true, bond group members may be distant maternal kin and should therefore share mtDNA haplotypes. We determined the mtDNA genotype of at least one adult female in all but two of the 22 core social groups that participated in bond groups (n=95 females; 80% of females genotyped per core social group). We observed complete uniformity of mtDNA haplotypes in eight of nine bond groups. In the FD/GC/PB bond group, the FD core social group had a different mtDNA haplotype than the other core social groups in that bond group (figure 3a); The AI of the FD core group with other core groups in its bond group were lower than any other AI within bond groups.
In further support of the hypothesis that bond group members were once part of the same core social group, we found that the matriarch (oldest female) of each core social group tended to be more closely related than expected to the matriarchs of the other core groups in her bond group. We calculated pairwise genetic relatedness between the matriarchs of 6 bond groups (i.e. the genetic relatedness between the 10 unique pairs of matriarchs from each core social group in these bond groups: CB/OA, JA/YA/SB, IA/IC, BC/HB, XA/RA and GC/PB/FD). Average pairwise relatedness between these matriarchs was 0.0848 (range=−0.1922 to 0.5156), which was significantly higher than pairwise relatedness among 10 randomly drawn pairs of matriarchs from the population (permutation test, average relatedness>observed in 31 of 1000 replicates, p=0.031).
Although matriarchs of core social groups in the same bond group are relatives, average pairwise relatedness between all adult females in the same bond group (but not the same core social group) was not significantly greater than expected by chance. To test this, we compiled complete microsatellite genotypes for all but two adult females in 11 core social groups (n=49 females, core groups and bond groups=JA/YA/SB, FD/PB, IA/IC, CB/OA and RA/XA). From these we were able to calculate the average pairwise genetic relatedness between core groups in five bond groups. Average relatedness among core groups in the same bond group was 0.0174 (s.e.±0.0137), which was not significantly greater than average relatedness between any five randomly chosen pairs of core groups in the population (permutation test, average pairwise relatedness>observed in 237 of 1000 replicates).
(iii) mtDNA predicts association among all core social groups in the population
If the daily, monthly and seasonal fission and fusion of all elephant social groups in the population reflects the long-term process of core social group growth and division, then genetic structure should predict association between all core social groups across the population. In support, we found that average pairwise AI between core groups with the same mtDNA haplotype were significantly higher than the average pairwise AI between core groups that did not share the same mtDNA haplotype. We used a Mantel test to correlate the matrix of association between 44 core social groups with a matrix of their mtDNA haplotypes, where within-haplotype comparisons were assigned a one and between-haplotype comparisons were assigned a zero. The resulting standardized Mantel statistic (r, which is similar to Pearson's correlation statistic) was small (r=0.14) but significantly positive (Monte Carlo randomization of Mantel test; Z>observed Z in 0 of 1000 runs, p<0.001; average association between core social groups with the same haplotype±s.e.=0.037±0.0052, average association between core social groups with different haplotypes±s.e.=0.019±0.0010). This trend persisted even if bond groups were excluded from the analysis (Mantel tests could not be performed on incomplete matrices; one-way ANOVA, average pairwise association index between core groups with the same haplotype=0.023±0.0018, n=323 pairs; average pairwise association index between core groups with different haplotypes=0.019±0.0010, n=613 pairs, F1935=4.6, p=0.032). These results may indicate that recently fissioned core groups have similar home ranges. However, they also suggest that either female elephants are able to recognize distant maternal kin, or alternatively, that these associations persist through associative learning—sometimes for several decades. Given the current number of core social groups in Amboseli (n=55) and a constant rate of permanent core group fission (approximately seven core groups have fissioned permanently in 30 years; Moss in press), the vast majority of core groups that share mtDNA in Amboseli probably fissioned permanently more than 100 years ago.
While mtDNA haplotype was significantly correlated with association between core social groups, neither the average pairwise genetic relatedness between core groups nor the pairwise genetic relatedness between the matriarch of each core group significantly predicted association between core groups (Monte Carlo randomization of Mantel test; matriarchs: Z>observed Z in 418 of 1000 runs, p>0.41; average pairwise genetic relatedness: Z>observed Z in 141 of 1000 runs, p>0.14).
4. Discussion
(a) Relationships within social groups: inclusive fitness and the costs and benefits of sociality
Until now, elephant social groups have primarily been defined using behavioural criteria (Douglas-Hamilton 1972; Moss & Poole 1983; Wittemyer et al. 2005). Our results show that elephant core social groups (often called ‘families’) are also genetic units. Most female elephants are matrilocal and remain with the group into which they were born; consequently, the average genetic relatedness within core social groups is relatively high—around the level of aunt–niece relationships—and the strongest social associations occur between mother–daughter pairs and maternal siblings.
This result, that close relatives are most often together in the same group, indicates that the most important social partners for female elephants are also usually their closest relatives. The benefits of such relationships are likely to include cooperative defence of calves against predators, parenting assistance from allomothers, resource defence and shared social and ecological knowledge from older and more experienced group members (Douglas-Hamilton 1972; Dublin 1983; Lee 1987; Moss 1988; McComb et al. 2001; Foley 2002; Sukumar 2003). Even if female elephants direct these behaviours randomly among group members, they will receive some indirect fitness benefits because relatedness within core social groups is relatively high. Further, females may maximize their indirect benefits by biasing cooperation towards their closest kin; this is the subject of ongoing research.
(b) Relationships between core social groups: kinship and long-term fidelity
Nearly half the core social groups in Amboseli (22 out of 51) were often together with one or two other specific core social groups. These social associations correspond to the bond groups first described by Douglas-Hamilton (1972), and have been hypothesized to form when existing core groups fission permanently (Douglas-Hamilton 1972; Moss & Poole 1983; Moss 1988; Moss in press). These permanent fissions are in contrast to the temporary fissions and fusions that occur normally in this society over the course of hours or days; permanent fissions occur rarely—approximately seven have been observed in Amboseli since 1972 (Moss in press).
Our data strongly support the hypothesis that bond group members were once part of the same core social group: closely associating core groups almost always had the same mtDNA haplotype, and the oldest members of each core group in the same bond group were more closely related to each other than expected by chance. Furthermore, maternal kinship explained some of the variance in association between core groups across the entire population. Core groups that shared the same mtDNA haplotype were slightly but significantly more likely to occur in the same party than core groups that did not share mtDNA. This occurred even when the members of one core group, including the matriarch, had no close genetic relatives (as measured by microsatellite markers) in the other core group.
The tendency for core social groups that share mtDNA to form groups probably persists because of shared range use or associative learning. In the latter case, when calves are young, they learn which other females and core groups in the population are familiar social partners, and in this way, the association patterns are maintained long after the females who were closely related have died. The low rate at which core groups permanently fission in Amboseli (Moss in press) means that these patterns of spatial association among core groups that share mtDNA may have persisted for several decades or perhaps hundreds of years. There is strong evidence that such associative learning is an important feature of elephant social organization (Moss 1988; Moss & Lee 1999; McComb et al. 2001). In particular, older females are thought to have accumulated the most knowledge about which social partners are familiar associates and where the best resources can be found when food and water are scarce (Moss 1988; McComb et al. 2001; Foley 2002). Core social groups with old matriarchs have higher female reproductive success than core groups with younger matriarchs (McComb et al. 2001).
However, while matrilineal relatedness predicted association between core social groups, female elephants are unlikely to accrue inclusive fitness benefits from these higher order associations. The only pattern of relatedness that predicted associations between core groups, other than shared mtDNA lineages, was the higher than expected relatedness between the oldest members of each core group in a bond group. In general, adult females were not closely related to females in other core groups. Thus, individuals are unlikely to accrue substantial inclusive fitness benefits from associations between core groups.
At least two processes contribute to the decay of the signal of genetic relatedness between the members of different core social groups. First, close maternal kin die after one or two generations. Second, males breed and produce offspring across multiple core groups (Hollister-Smith et al. in preparation), and this gene flow quickly lowers genetic differentiation between core groups and swamps the signal of maternal relatedness. Therefore, if individuals derive fitness benefits from associations with bond group members or other core groups, they must only accrue via direct fitness as opposed to indirect fitness (i.e. kin selection). One hypothesized direct benefit of associating in larger aggregations is lower rates of predation as a result of the dilution effect or of active cooperative defence (Douglas-Hamilton 1972; Dublin 1983; Moss 1988; Sukumar 2003). In particular, most elephant calves are born between November and May (Moss 2001), which coincides with the rainy season when elephants aggregate into groups that sometimes contain hundreds of animals.
(c) Conservation implications
These results—that the most important social partners for female elephants are probably close maternal kin, and that maternal kinship predicts the fusion of social groups—have conservation implications. In particular, if kin influence each other's reproductive success through cooperative relationships, as has been found in some species where this has been tested (Lambin & Yoccoz 1998; Pusenius et al. 1998; Armitage & Schwartz 2000; Dobson et al. 2000; Pope 2000; Rusu & Krackow 2004), then poaching may negatively impact elephant reproductive success by destroying an individual's relatives.
In Amboseli, our results show that when females lose all the members of their natal core social group, they often immigrate to a new core social group where they have no close kin. Data from other populations suggest that this is not unique to Amboseli (Nyakaana et al. 2001; Charif et al. 2005). In the 1970s, a combination of drought and poaching pressures reduced the size of the elephant population in Amboseli (Moss 2001). There is some indication that the immigrants in the PA and JA core social groups (Puff and Jody) were the sole surviving members of their natal core groups. They first associated peripherally with, and eventually joined the PA and JA core groups. Furthermore, in Sengwa, Zimbabwe, Charif et al. (2005) found, in contrast to our results, that closely associating core groups were not maternal kin. They report that three of four pairs of females that exhibited coordinated movements, but were from different core group groups (i.e. were bond group members), did not share the same mtDNA haplotype (Charif et al. 2005). This discrepancy is likely due to high rates of culling in Sengwa (Charif et al. 2005). Between 1978 and 1986, approximately 1400 elephants were culled from Sengwa, which reduced the total population by as much as 75% (Osborn 1998; Charif et al. 2005). In the process, many core social groups certainly lost their bond groups.
Hence, if kinship influences female elephant reproductive success, females with few kin may have lower reproductive rates. Whether or not this is true, it is clear that kinship is a primary determinant of elephant social relationships both within and across core social groups. We recommend that elephant conservation measures strive to keep patterns of maternal kinship intact.
Acknowledgments
We thank the Office of the President of Kenya for permission to work in Amboseli National Park under permit number MOES&T 13/001/30C 72/7. We thank the Kenya Wildlife Service for local sponsorship. The Amboseli Elephant Research Project provided invaluable scientific and logistical support, particularly the team of N. Njiraini, K. Sayialel and S. Sayialel who contributed greatly to the collection of genetic and behavioural data. This research was supported by the National Science Foundation (IBN0091612 to SCA), the African Trust for Elephants, the Amboseli Elephant Research Project and Duke University. All work was conducted with the approval of Duke University's Institutional Animal Care and Use Committee, registry #A436-00-09.
Supplementary Material
References
- Altmann J. Observational study of behavior: sampling methods. Behaviour. 1974;49:227–267. doi: 10.1163/156853974x00534. [DOI] [PubMed] [Google Scholar]
- Altmann J, et al. Behavior predicts genetic structure in a wild primate group. Proc. Natl Acad. Sci. USA. 1996;93:5797–5801. doi: 10.1073/pnas.93.12.5797. doi:10.1073/pnas.93.12.5797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann J, Alberts S.C, Altmann S.A, Roy S.B. Dramatic change in local climate patterns in the Amboseli basin, Kenya. Afr. J. Ecol. 2002;40:248–251. doi:10.1046/j.1365-2028.2002.00366.x [Google Scholar]
- Archie E.A, Moss C.J, Alberts S.C. Characterization of tetranucleotide microsatellite loci in the African savannah elephant (Loxodonta africana africana) Mol. Ecol. Notes. 2003;3:244–246. doi:10.1046/j.1471-8286.2003.00412.x [Google Scholar]
- Armitage K.B, Schwartz O.A. Social enhancement of fitness in yellow-bellied marmots. Proc. Natl Acad. Sci. USA. 2000;97:12 149–12 152. doi: 10.1073/pnas.200196097. doi:10.1073/pnas.200196097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.C, Archie E.A, Van Horn R.C, Alberts S.C. Locus effects and sources of error in non-invasive genotyping. Mol. Ecol. Notes. 2005;5:680–683. [Google Scholar]
- Charif R.A, Ramey R.R, Langbauer W.R, Payne K.B, Martin R.B, Brown L.M. Spatial relationships and matrilineal kinship in African savanna elephant (Loxodonta africana) clans. Behav. Ecol. Sociobiol. 2005;57:327–338. doi:10.1007/s00265-004-0867-5 [Google Scholar]
- Dietz J. Permutation tests for association between two distance matrices. Syst. Zool. 1983;32:21–26. [Google Scholar]
- Dobson F.S, Jacquot C, Baudoin C. An experimental test of kin association in the house mouse. Can. J. Zool. 2000;78:1806–1812. doi:10.1139/cjz-78-10-1806 [Google Scholar]
- Douglas-Hamilton I. Oxford University; Oxford, UK: 1972. On the ecology and behaviour of the African elephant: the elephants of Manyara. [Google Scholar]
- Dublin H.T. Cooperation and reproductive competition among female African elephants. In: Wasser S.K, editor. Social behavior of female vertebrates. Academic Press; New York: 1983. pp. 291–313. [Google Scholar]
- Dunbar R.I.M. Time: a hidden constraint on the behavioral ecology of baboons. Behav. Ecol. Sociobiol. 1992;31:35–49. doi:10.1007/BF00167814 [Google Scholar]
- Fernando P, Lande R. Molecular genetic and behavioral analysis of social organization in the Asian elephant (Elephas maximus) Behav. Ecol. Sociobiol. 2000;48:84–91. [Google Scholar]
- Foley C. Princeton University; Princeton: 2002. The effect of poaching on elephant social systems. [Google Scholar]
- Ginsberg J.R, Young T.P. Measuring association between individuals or groups in behavioural studies. Anim. Behav. 1992;44:377–329. doi:10.1016/0003-3472(92)90042-8 [Google Scholar]
- Goodall J. Harvard University Press; Cambridge: 1986. The chimpanzees of Gombe: patterns of behavior. [Google Scholar]
- Greenwood A.D, Paabo S. Nuclear insertion sequences of mitochondrial DNA predominate in hair but not in blood of elephants. Mol. Ecol. 1999;9:133–137. doi: 10.1046/j.1365-294x.1999.00507.x. doi:10.1046/j.1365-294X.1999.00507.x [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. The genetical evolution of social behaviour. I. J. Theor. Biol. 1964a;7:1–16. doi: 10.1016/0022-5193(64)90038-4. doi:10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. The genetical evolution of social behaviour. II. J. Theor. Biol. 1964b;7:17–52. doi: 10.1016/0022-5193(64)90039-6. doi:10.1016/0022-5193(64)90039-6 [DOI] [PubMed] [Google Scholar]
- Kummer H. Princeton University Press; Princeton: 1995. In quest of the sacred baboon. [Google Scholar]
- Lai Y, Sun F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 2003;20:2123–2131. doi: 10.1093/molbev/msg228. doi:10.1093/molbev/msg228 [DOI] [PubMed] [Google Scholar]
- Lambin X, Yoccoz N.G. The impact of population kin-structure on nestling survival in Townsend's voles, Microtus townsendii. J. Anim. Ecol. 1998;67:1–16. doi:10.1046/j.1365-2656.1998.00181.x [Google Scholar]
- Lee P.C. Allomothering among African elephants. Anim. Behav. 1987;35:278–291. [Google Scholar]
- Maynard-Smith J. Group selection and kin selection. Nature. 1964;201:1145–1147. [Google Scholar]
- McComb K, Moss C.J, Sayialel S, Baker L. Unusually extensive networks of vocal recognition in African elephants. Anim. Behav. 2000;59:1103–1109. doi: 10.1006/anbe.2000.1406. doi:10.1006/anbe.2000.1406 [DOI] [PubMed] [Google Scholar]
- McComb K, Moss C, Durant S.M, Baker L, Sayialel S. Matriarchs as repositories of social knowledge in African elephants. Science. 2001;292:491–494. doi: 10.1126/science.1057895. [DOI] [PubMed] [Google Scholar]
- Moss C.J. University of Chicago Press; Chicago: 1988. Elephant memories. [Google Scholar]
- Moss C.J. The demography of an African elephant (Loxodonta africana) population in Amboseli, Kenya. J. Zool. 2001;255:145–156. doi:10.1017/S0952836901001212 [Google Scholar]
- Moss, C. J. In press. Female social dynamics: fidelity and flexibility. In Amboseli elephants: a long-term perspective on a long-lived mammal (ed. C. J. Moss & H. Croze). Chicago: University of Chicago Press.
- Moss C.J, Lee P.C. The social context for learning and behavioural development among wild African elephants. In: Box H.O, Gibson K.R, editors. Mammalian social learning: comparative and ecological perspectives. vol. 72. Cambridge University Press; Cambridge: 1999. pp. 102–125. [Google Scholar]
- Moss C.J, Poole J.H. Relationships and social structure of African elephants. In: Hinde R.A, editor. Primate social relationships. Sinauer; Sunderland, MA: 1983. pp. 315–325. [Google Scholar]
- Navidi W, Arnheim N, Waterman M.S. A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am. J. Hum. Genet. 1992;50:347–359. [PMC free article] [PubMed] [Google Scholar]
- Nyakaana S, Arctander P. Isolation and characterization of microsatellite loci in the African elephant, Loxodonta africana. Mol. Ecol. 1998;7:1431–1439. doi:10.1111/j.1365-294X.1998.00445.x [PubMed] [Google Scholar]
- Nyakaana S, Abe E.L, Arctander P, Siegismund H.R. DNA evidence for social behaviour breakdown in Queen Elizabeth National Park, Uganda. Anim. Conserv. 2001;4:231–237. doi:10.1017/S1367943001001275 [Google Scholar]
- Osborn F.V. University of Cambridge; Cambridge, UK: 1998. The ecology of crop-raiding elephants in Zimbabwe. [Google Scholar]
- Pope T.R. Reproductive success increases with degree of kinship in cooperative coalitions of female red howler monkeys (Alouatta seniculus) Behav. Ecol. Sociobiol. 2000;48:253–267. doi:10.1007/s002650000236 [Google Scholar]
- Pusenius J, Viitala J, Marienberg T, Ritvanen S. Matrilineal kin clusters and their effect on reproductive success in the field vole Microtus agrestis. Behav. Ecol. 1998;9:85–92. [Google Scholar]
- Queller D.C, Goodnight K.F. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Rusu A.S, Krackow S. Kin-preferential cooperation, dominance-dependent reproductive skew, and competition for mates in communally nesting female house mice. Behav. Ecol. Sociobiol. 2004;56:298–305. doi:10.1007/s00265-004-0787-4 [Google Scholar]
- Sukumar R. Oxford University Press; Oxford, UK: 2003. The living elephants. [Google Scholar]
- Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits L.P, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. doi:10.1093/nar/24.16.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaik C.P. The socioecology of fission–fusion sociality in orangutans. Primates. 1999;40:69–86. doi: 10.1007/BF02557703. [DOI] [PubMed] [Google Scholar]
- West-Eberhard M.J. The evolution of social behavior by kin selection. Q. Rev. Biol. 1975;50:1–33. doi:10.1086/408298 [Google Scholar]
- Whitehead H, Christal J. Social affiliations within sperm whale (Physeter macrocephalus) groups. Ethology. 2001;107:323–340. doi:10.1046/j.1439-0310.2001.00666.x [Google Scholar]
- Wittemyer G, Douglas-Hamilton I, Getz W.M. The socioecology of elephants: analysis of the processes creating multitired social structures. Anim. Behav. 2005;69:1357–1371. doi:10.1016/j.anbehav.2004.08.018 [Google Scholar]
- Wursig B. Occurrence and group organization of Atlantic bottlenose porpoises (Tursiops truncatus) in Argentine Bay. Biol. Bull. 1978;154:348–359. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.