Abstract
Recently, it has been demonstrated that several complex systems may have simple graph-theoretic characterizations as so-called ‘small-world’ and ‘scale-free’ networks. These networks have also been applied to the gross neural connectivity between primate cortical areas and the nervous system of Caenorhabditis elegans. Here, we extend this work to a specific neural circuit of the vertebrate brain—the medial reticular formation (RF) of the brainstem—and, in doing so, we have made three key contributions. First, this work constitutes the first model (and quantitative review) of this important brain structure for over three decades. Second, we have developed the first graph-theoretic analysis of vertebrate brain connectivity at the neural network level. Third, we propose simple metrics to quantitatively assess the extent to which the networks studied are small-world or scale-free. We conclude that the medial RF is configured to create small-world (implying coherent rapid-processing capabilities), but not scale-free, type networks under assumptions which are amenable to quantitative measurement.
Keywords: reticular formation, small world, scale-free, networks, computational neuroanatomy
1. Introduction
Many real-world systems can be represented as networks (a set of nodes joined by links indicating an interaction). Recently, graph-theorists have demonstrated that even the most complex of these systems may have simple characterizations. So-called ‘small-world’ (Watts & Strogatz 1998) and ‘scale-free’ (Barabasi & Albert 1999) networks have been found within such diverse structures as food webs, the internet, and power grids (Albert & Barabasi 2002). These two network types are of interest because of the special properties that are known to ensue if the underlying network satisfies the criteria for either or both. Recently, several authors have studied these network types in the context of gross neural connectivity between primate cortical areas (Hilgetag et al. 2000; Sporns et al. 2002) and between C. elegans nervous system components (Watts & Strogatz 1998).
Our aim here is to extend this work to a specific network of the vertebrate brain, making two key contributions. First, this work constitutes the first graph-theoretic analysis of vertebrate brain connectivity at the neural network level: we analyse the structure of the medial reticular formation (RF) of the brainstem due to its extraordinary configuration of sensory and motor connections (see below) and for its relevance to our work on action selection (see §5). Second, this work constitutes the first model—and quantitative review—of this important brain structure for over three decades. In addition, by applying graph-theoretic analysis to an exploration of plausible neural network structural models, this work contributes new methods to the nascent field of computational neuroanatomy (Ascoli 1999). We believe it is useful to analyse neural networks for their small-world and scale-free properties because each network type conveys a set of functional advantages compared to a true random network, and yet the determination of network type can be made primarily using anatomical data.
A small-world network is characterized by the following two features. (i) Dense interconnectivity within small groups of nodes: two common neighbours of one node are more likely to be neighbours of each other than two nodes selected at random. Note that if the nodes exist in physical space, for example people or neurons, then the nodes of a highly inter-connected group will tend to be physically close in space. (ii) The average shortest path length is small: to connect any two nodes only a small number of intermediate nodes are typically traversed, due to long-range links between the small groups of nodes (Watts & Strogatz 1998).
In a real network, nodes are not uniformly connected. A given node has β links—the node's degree. Over all nodes in the network, the degree distribution P(β) defines the probability that a randomly selected node has β links in the network. The corresponding cumulative degree distribution F(β) defines the probability that a randomly selected node has at most β links. For many real networks, this distribution is best fitted by a power-law (F(β)∼β−τ, τ>0; Barabasi & Albert 1999), which is a straight line on a log–log plot. The power-law fit implies that: (i) the network has no ‘typical’ node, in the sense that a Gaussian distribution would have a mean node; (ii) the distribution is scale-invariant. Thus, networks with a power-law distribution have been dubbed ‘scale-free’.
The identification of either small-world or scale-free topologies implies particular dynamic properties of the network, e.g. stability (Li & Chen 2003), which may be beneficial to biological neural networks (see §5). The plan of the paper is as follows. First, we review the available data on medial RF anatomy, and propose a new structural organization. We define two models which generate the set of plausible structures, then attempt to discern why this structure exists. The graph-theoretic analysis is applied to determine whether the structure could be explained by the need to give the neural network the functional advantages associated with each network type.
2. The vertebrate brainstem reticular formation
The vertebrate RF lies within the centre of the brainstem and midbrain. It extends rostrally from the spinal cord's junction with the medulla, through the pons, to terminate at the border with caudal-most intralaminar thalamus, beneath the superior colliculus (figure 1a). The RF is sub-divided into a bewildering array of fields and nuclei (Jones 1995). Many of these have been assigned distinct functional roles (see electronic supplementary material A), except for a major component of the RF: the fields comprising the medial core running the length of the medulla and pons.
Figure 1.
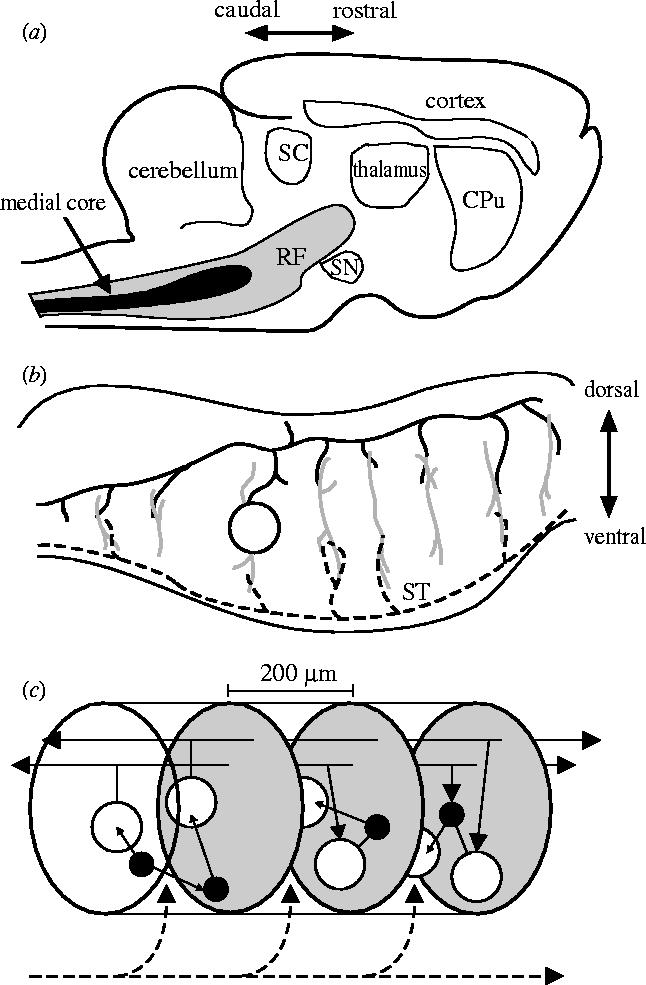
Schematic summary of the vertebrate reticular formation's anatomical organization. Directional arrows apply to all panels. (a) Sagittal section of cat brain, showing relative size and location of reticular formation (RF) and medial core. Abbreviations: CPu, caudate-putamen; SC, superior colliculus; SN, substantia nigra. (b) Sagittal section of the brainstem; the dendritic trees (grey lines) of the projection neurons (one cell body shown, open circle) extend throughout the medial RF along the dorso-ventral axis but extend little along the rostro-caudal axis. These dendritic trees contact axon collaterals of both ascending sensory systems (black dashed line) and far-reaching axons of the projection neurons (the axon of the depicted cell body is shown by the solid black line); ST is the spinothalamic tract. (c) The cluster model of RF organization. The medial RF comprises stacked clusters (three shown) containing medium-to-large projection neurons (open circles) and small-to-medium inter-neurons (filled circles); cluster limits (grey ovals) are defined by the initial collaterals from the projection neuron axons. Their radial dendritic fields allow sampling of ascending and descending input from both other clusters (solid black lines) and sensory systems (dashed black line). The interneurons project predominantly within their parent cluster.
In a landmark paper (Kilmer et al. 1969), Warren McCulloch and colleagues proposed that this region was the substrate for the selection of an organism's global behavioural state. They also described a computational model which demonstrated that the known neuroanatomy of the medial RF supported this function (Kilmer et al. 1969). Recently, we have demonstrated that this model supports action selection when implemented as a robot control architecture, but concluded that it warrants substantial revision as a model of the medial RF (Humphries et al. 2005b). Despite numerous unique and intriguing features (detailed below), and its comparatively large areal extent, this region has received no modern functional evaluation. However, before we can evaluate its potential functional roles in simulation, we must establish its structure. What follows describes our collation of modern studies of the medial RF into a coherent quantitative model of its anatomy.
(a) Physical boundaries and neuron types
We include only the major neuron types of, and sensory inputs to, the medial RF between, rostrally, the border between the caudal and oral pons and, caudally, the spinal cord. The rostral limit is imposed because medial fields within the oral pons have been ascribed specific rather than general functions and the characteristic giant-bodied cells rarely appear rostral to the caudal/oral pons border (Jones 1995).
(i) Projection neurons
Numerous staining studies (for example, Scheibel & Scheibel 1967; Newman 1985) have demonstrated that the medial RF contains a medium-to-giant-bodied neuron characterized by far-reaching bifurcating axons running rostro-caudally, for which the long branch reaches either the forebrain or spinal cord (the latter being dominant); extensive collateralization along the axon, the first occurring at least 100 μm from the cell body; a radial dendritic tree, flattened along the rostro-caudal axis so that the dendritic zone is disc shaped; and with dendritic branches specifically directed at sources of passing fibres (for example, the descending sensory trigeminal or ascending spinothalamic tract axons). A recent review suggests that these cells are glutamatergic (Jones 1995). As all of these cells appear to project outside the medial RF, we refer to them as projection neurons. Electrophysiological studies have demonstrated that synaptic connections between these neurons, formed by terminals of the axon collaterals, are sufficient to induce excitatory post-synaptic potentials (Ito & McCarley 1987). Therefore, these projection neurons appear both anatomically and functionally connected.
The number of projection neurons is suggested by an analysis of data presented in a recent study (Holmes et al. 1994). Holmes et al. (1994) estimated that GABAergic cells form approximately 10–20% of the cells in this area.
Given that spinally projecting medial RF cells are rarely GABAergic (Jones et al. 1991), and thus none of the GABAergic cells are likely to be a projection neuron, then the projection cells constitute at least 70% of the cell population of the medial RF.
(ii) Interneurons
The existence of an interneuron cell-type in the medial RF is more controversial, but there is substantial evidence for its existence. Stained examples of small-to-medium sized neurons with sparse dendritic trees and oval bodies have been reported within this region (Bowsher & Westman 1970; Mason & Fields 1989; Jones et al. 1991), and these neurons are morphologically distinct from the projection neurons described above. From the limited data available, it appears that their axons preferentially project medio-laterally, rather than rostro-caudally, with collateral terminals concentrated within the parent cell's dendritic field (Mason & Fields 1989). Jones et al. (1991) stained morphologically similar cells for GAD, suggesting that these interneurons may be GABAergic. Holmes et al. (1994) demonstrated that selective destruction of GABAergic cells in medial RF resulted in a corresponding proportional decrease in GABAergic terminals; the decrease was confined to within a 1 mm radius from the individual damaged cell bodies, which concurs with the limited axon terminal extent of the neurons described by Mason & Fields (1989). Moreover, Jones et al. (1991) report that roughly 45% of synapses on the dendrites of a typical large/giant cell are GABAergic. As sensory input to the medial RF comes from regions known to use excitatory neurotransmitters, and retrogradely labelled spinally projecting GABAergic projection neurons are rare within the medulla RF (Jones et al. 1991), these synapses may originate from an interneuron. We conclude from this data that there is a small-to-medium sized inhibitory interneuron within the medial RF, which constitutes the 10–20% of GABAergic cells (detailed above) in this region. We also note that researchers have demonstrated the actions of presumed locally projecting GABAergic interneurons in other regions of the RF (e.g. Hayar et al. 1996).
(b) Sensory input
Although we intend to study here only the structural properties of our model, a review of the patterning of sensory input is necessary both to emphasize why the medial RF is of importance and to explain the model's basic structural elements. Numerous studies have demonstrated that medial RF cells respond to a wide variety of sensory stimuli, and that many are multi-modal (Siegel 1979). The pattern of afferent inputs suggests that the medial RF receives information on every sense available to an animal, including balance (vestibular), pain, proprioception (via collaterals from the gracile and cuneate nuclei), auditory, and whisker systems (Salibi et al. 1980; Yates & Stocker 1998; Kleinfeld et al. 1999; Cant & Benson 2003).
The combination of multiple sources of sensory input to, and the dendritic organization of the projection neuron (§2) is strong evidence that they all receive some form of direct sensory input from peripheral sources. Moreover, the collaterals of ascending and descending tracts from which the input originates are arranged in a striking manner (figure 1b), running perpendicular to the main axon, and seemingly in juxtaposition with the axon collaterals from the giant cells themselves (Scheibel 1984).
3. The model architecture
Based on the anatomy described above, we propose that there exists within the medial RF a quasi-independent grouping of cells which we term a cluster. The neural architecture resulting from this hypothesis is depicted in figure 1c and has some similarities to the ‘poker-chip’ anatomy proposed by Scheibel & Scheibel (1967).
Each cluster contains a set of neighbouring medium-to-giant bodied projection neurons and small-to-medium bodied local interneurons. We define the extent of a cluster as the region in which there are no axon collaterals from any of its projection neurons (and, therefore, projection neurons make no connections within their own clusters). The limited rostro-caudal extent of collateral input from the passing sensory tracts, and corresponding flattening of the projection neurons dendritic fields, suggest that their cell bodies are roughly adjacent in the medio-lateral, dorsal-medial plane (that is, in coronal section). Thus, given the anatomical data reviewed above, a single cluster would be approximately 200 μm long in the medial RF, corresponding to the initial axon collateral of the projection neurons. The interneurons are assumed to project only within their parent cluster, due to their limited projection radius (1 mm), to the concentration of the majority of their axon terminals within their own dendritic fields, and to the predominantly medio-lateral (rather than rostro-caudal) projections of their axons. They make connections with both other interneurons and projection neurons within their cluster.
We define two models which generate the proposed cluster structure of the medial RF. The first is a stochastic model, which creates a structure equivalent to the adult configuration of the neural tissue. The second is a generative model which approximates the post-natal development of the structure, and uses the first model to generate its initial state.
(a) The stochastic model
Every one of the Nc clusters in the model has n neurons (or nodes); the total number of nodes T within the model is thus T=Nc×n. Within each cluster a certain proportion ρ of nodes are deemed to be the projection neurons, the remainder are deemed to be interneurons. From the data reviewed above, we set bounds 0.7≤ρ<0.9.
Given an estimated size for the clusters, we may determine a rough estimate for the number of clusters within the medial RF. From the UCLA Laboratory Of Neural Imaging (LONI) rat brain atlases (www.loni.ucla.edu), we have estimated the rostro-caudal extent of the medial RF to be at least ∼7 mm, If we assume that the clusters are stacked end-to-end rostro-caudally and each is 200 μm long, then there are at least 35 of them. Thus, we set bounds 35≤Nc≤75.
Three parameters define the stochastic connectivity between nodes. For each projection neuron node, the probability of forming a connection c between itself and another cluster is P(c). If a connection is made, then P(p) is the probability that the projection neuron forms a connection p with a given neuron in that cluster. Finally, P(l) denotes the probability of connection l between an interneuron node and any other node in its cluster. The probabilities P(l), P(p) are constant, independent of particular clusters and nodes within clusters. However, two model variants were defined by two choices of distribution for P(c). Data from Grantyn et al. (1987) suggest a spatially uniform model for which we assigned P(c)=0.25 for all clusters. In contrast, McCulloch and colleague's RF model (Kilmer et al. 1969) used a distance-dependent distribution (William Kilmer, personal communication), typical of models of neural connectivity (Hellwig 2000). Thus, if there are d intervening clusters between the projection neuron node and the target cluster (so for adjacency d=1), then P(c)=d−a; we use a=1 throughout.
As we are interested here only in the network structure of the model, we need not assign weights to these connections (as is common practice in neural network models) but simply use the above probabilities to define (directed) edges in the connectivity graph.
(b) Pruning model
An alternative model was constructed in which the connections were defined by a procedure analogous to the development process, rather than a stochastic model of the final, adult configuration of the neural structure. Unlike many other neural structures, most, if not all, medial RF cells exist at birth (Hammer et al. 1981). Thus, developmental changes wrought by experience-dependent plasticity must be mainly restricted to cell connectivity.
One dominant form of post-natal connectivity change is that of synaptic pruning (Bourgeois & Rakic 1993). This process is characterized by synaptic overgrowth and subsequent pruning to adult levels during the neo-natal period. Within the developing medial RF, synaptic pre-cursors in the form of proto-spines significantly increase in number immediately following birth, then decline to immediate pre-birth levels by post-natal day 20 (Hammer et al. 1981). This suggests that synaptic overgrowth occurs immediately after birth, followed by pruning.
Simple models of the pruning process propose that it maximizes the potential for Hebbian learning given known metabolic constraints (Chechik et al. 1998). Assuming that a network could support only a limited number of synapses, Chechik et al. (1998) report that optimal synaptic pruning can be achieved using a minimal-value deletion scheme—removing all connections below a given absolute weighted value. We adopt that scheme here to generate an alternate ‘pruning’ anatomical model of the medial RF, using the following algorithm.
Generate an over-growth anatomical model: using either collateral probability variant, create a stochastic anatomical model at the upper limits of its connection parameter intervals (here using P(p)=P(l)=0.9).
Assign each resulting graph edge a weight value wij (Gamma distribution, a=10, b=0.02; a Gamma distribution with these parameters approximates a normal distribution, and is used so that all weight values are initially positive—sign is then assigned according to connection type, inter-neuron or projection neuron).
- Loop until the expected synapse total E(Ns) is reached (see electronic supplementary material B), computed from target values of P(l) and P(p), labelled tl and tp, respectively.
- Learn: adjust proportion ϕ units' input synapses with random values from normal distribution (μ=0, σ=0.025)—this variance is chosen to be smaller than the approximate variance of the Gamma distribution used to generate the initial weight values. Units are chosen with probability proportional to total weighted input. (This is a statistical approximation of Hebbian-style learning. Given sufficient time, for a given series of sensory input patterns, neurons with the greatest synaptic efficacy on their inputs are more likely to respond (or not respond, depending on the sign of total) and, therefore, their inputs are more likely to be strengthened or weakened according to the correctness of that response. Given the central limit theorem, for a sufficiently large set of input patterns and a sufficiently large network, the total synaptic change will tend towards a normal distribution).
- Prune: remove all |wij|<t.
Thus, we begin with a structure representing the overgrown synaptic density with all cells in position, and then repeat a process of synaptic weight change—representing plastic changes due to learning and sensory experience—and synaptic pruning. Again, the final resulting model is a graph of connectivity—final weights are converted to a binary connection matrix.
4. Analyses of the architecture
For the graph-theoretic analyses, the stochastic model parameters were chosen from the following sets: Nc={35, 45, 55, 65, 75}, n={30, 40, 50}, ρ={0.7, 0.8, 0.9}, P(p)={0.1, 0.5, 0.9} and P(l)={0.1, 0.5, 0.9}. Each possible combination {Nc, n, ρ, P(p), P(l)} formed by selecting from these parameter sets defined a class of models, giving 5×34=405 classes in total. The entire set of classes was tested with a single instantiation of each of the spatially uniform and distance-dependent collateral variants (totalling 810 instantiations).
For the pruning model, we set ϕ=0.3 and t=0.2, and chose the target values from the following sets: tp={0.1, 0.3, 0.5} and tl={0.1, 0.3, 0.5}. Values for Nc, n, and ρ were taken from the sets above. Again, each possible combination of these parameters defined 405 classes in total; a single model instantiation of each of the collateral variants was analysed per class. Thus, a total of 810 instantiations were also assessed for the pruning model—giving a total of 1620 instantiations tested across both anatomical models for small-world and scale-free topologies.
(a) Small-world analysis
A cluster model forms a graph with T nodes and a mean of k links per node. To be quantitatively defined as a small-world network, values for two network properties must be compared to their values for the equivalent random graph. The characteristic path length L is the average length of the shortest path between any two nodes in the graph; the clustering coefficient C is a measure of how much neighbours of each node are also neighbours of each other (for quantitative definitions see, e.g. Watts & Strogatz 1998). When comparing a small-world network s to a random network r of the same T, k value, it is found that Ls≈Lr and that Cs≫Cr. For networks with small numbers of nodes (∼200–3000), it is known that Cs>Cr is sufficient to demonstrate small-world properties (Montoya & Sole 2002). Typically, these relationships are calculated for datasets forming a single network, and so the presence of a small-world topology is decided by inspection of the generated L, C values. As we are generating large numbers of networks, we propose a simple quantitative definition of these relationships. This will allow us to explore the extent to which the small-world topology changes with parameter variation: in other words, it is a measurement of ‘small-world-ness’.
For each instantiated cluster RF model ci, we created a random graph ri with the same T, k values as ci. Let the characteristic path lengths for ci and ri be , , respectively, and the corresponding clustering coefficients be , . These values were computed for the two graphs and used to form the following ratios: , and . Thus, to meet the small-world criteria given above, the model ci must fulfil the conditions: γi>1 and Si>1. The latter condition indicates that the magnitude ratio between C for model and random graph is greater than the magnitude ratio between L for model and random graph. It is this value we use as an indicator of comparative ‘small-world-ness’. For comparison, we computed S for small-world neural networks already evaluated in the literature: from Watts & Strogatz (1998), for C. elegans's entire neural net, S=4.75; from Hilgetag et al. (2000) S values for cortical area connectivity are: macaque visual, S=1.81; macaque somatosensory, S=1.77; macaque whole cortex, S=2.78; cat whole cortex, S=1.86.
Models were grouped by {ρ, P(l), P(p)} or {ρ, tl, tp} combination for analysis, yielding 27 groups with 15 models per group for each anatomical model and collateral variant combination, ordered by T.
(i) Results: stochastic model
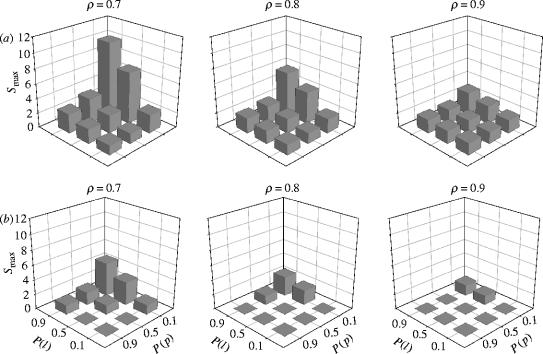
For the spatially uniform collateral variant, 11 (out of 27) of the groups fulfilled all the criteria for a small-world topology for all 15 of the member models. The maximum value of was obtained for parameter combination {ρ=0.7, P(l)=0.9, P(p)=0.1}. The distance-dependent collateral variant had Si>1 for all 27 of its parameter combinations. The maximum was for the same parameter combination as the spatially uniform models. In fact, for both collateral probability variants, ordering the parameter combinations by mean, median, or maximum S results in the same first six parameter combinations (see electronic supplementary material C). The variation in small-world topology (figure 2) was consistent across both collateral variants, with higher S for ascending P(l) (resp. P(p)) for a given value of P(p) (resp. P(l)). Taken together, this is evidence that the small-world topology is a robust property of the network with respect to the distribution of collaterals.
Figure 2.
Variations in small-world topology for the stochastic anatomical model. (a) Distance-dependent collaterals. (b) Spatially uniform collaterals. A value of zero indicates that the model did not meet the minimum criteria for the topology.
(ii) Results: pruning model
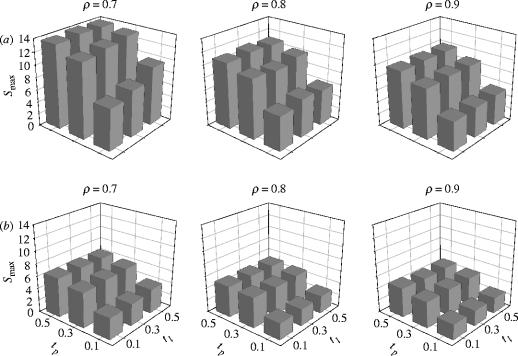
For both the spatially uniform and distance-dependent collateral variants, every tested model fulfilled the small-world topology criteria above (figure 3). Again, the same parameter combination {ρ=0.7, tl=0.3, tp=0.5} for both collateral variants resulted in the maximum S value, of and for the distance-dependent and spatially uniform collaterals, respectively. Ordering the parameter combinations by Smax results in the same nine top parameter combinations for both collateral variants (except positions 2 and 3, which are inverted). And, again, the variation in small-world topology (figure 3) was consistent across both collateral variants: for a given value of tl, S increased with increasing tp; however, S was roughly constant with increasing tl.
Figure 3.
Variations in small-world topology for the pruning anatomical model. (a) Distance-dependent collaterals. (b) Spatially uniform collaterals. Every model met the criteria for the topology.
(iii) Robustness of the results
Each of the Si values computed for the analysis above was based, directly or indirectly, on a single instantiation of the stochastic anatomical model per {Nc, n, ρ, P(l), P(p)} class. Given that three of the stochastic model parameters are probabilities, there could be considerable variation of topology across instantiations of a particular group. We therefore tested the robustness of the small-world topology for the parameter group and collateral variant which resulted in maximum Si for each anatomical model (values reported above). For both anatomical models' Smax parameter groups, two sets of 50 instantiations were created. One set had the minimum quantity of cells (Nc=35, n=30) and one set had the maximum quantity of cells (Nc=75, n=50) to assess the variation at the extremes of the models. A corresponding set of equivalent random networks (same k, T) was created for each set of 50 models.
We computed the mean and standard error of each model set's S values to determine the robustness of the reported values. We also tested the corresponding distributions of model and random network clustering coefficients to verify that they were drawn from significantly different populations. Each distribution was tested for normality using the Lillefors test at the p=0.05 confidence limit. If either distribution of C significantly deviated from normality, then the distributions were compared using the Mann–Whitney U-test, otherwise a standard Student's t-test was used.
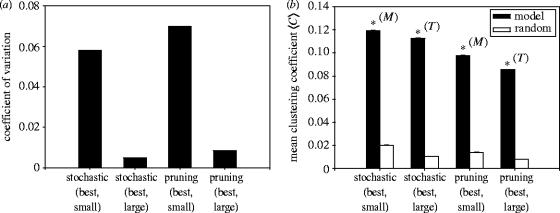
The S values were remarkably invariant for all tested models. The coefficients of variation for the model sets (figure 4a) show that the variability in S was at least one order of magnitude smaller than the mean value. All tested models also had significantly greater mean clustering coefficients 〈C〉 than the corresponding random network populations (p<0.001). Figure 4b illustrates the difference in population 〈C〉, and the small variation in C for the tested networks. Thus, we conclude that the topology is present in every instantiation of the models tested, and that it is robust across the instantiations of a particular parameter class.
Figure 4.
Robustness of the small-world topology. (a) Coefficient of variation of S for each model set tested. (b) Mean clustering coefficients for model-types and equivalent random networks. Small and large refer to the minimum and maximum quantities of cells, respectively. Asterisks indicate significance at the p<0.001 level; (M)=Mann–Whitney U-test; (T)=Student's t-test; Error bars are ±3 s.e.
(b) Scale-free analysis
Previous analyses of the scale-free properties of real-world networks have assumed them to be undirected graphs (Amaral et al. 2000). However, as a neural network is by definition a directed graph, we look separately at F(β) (the cumulative degree distribution) for input and output links, and for the undirected links (counting any connection made on a node). We compute F(β) (input, output, and undirected) for each model instantiation from a class, invert it (see electronic supplementary material D), and fit with exponential, power-law, truncated power law and Gaussian distributions as these are all typical fits to patterns found in real-world connection distribution data (Amaral et al. 2000). The goodness-of-fit for each of the model curves was quantified using (corrected) Akaike's information criterion (AICc; Motulsky & Christopoulos 2004), an information-theoretic score which accounts for the different number of coefficients in the fitted curves. By definition, the curve with the lowest AICc value is the closest fit of those tested.
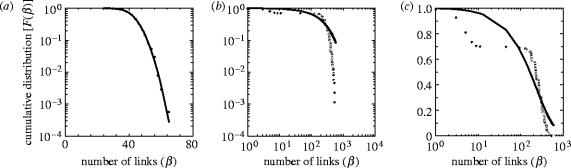
The input and undirected degree distributions were all best-fit by a Gaussian for the distance-dependent and spatially uniform collateral versions of both the stochastic and pruning models (example in figure 5a); of the output degree distributions, all were best-fit by a Gaussian except two instantiations of the distance-dependent collateral version of the stochastic model which were both best-fit by an exponential. An exponential fit is shown in figure 5b—a poor fit to the extreme end of the distribution tail is evident. Re-plotting on a log-linear scale shows that the exponential fit to the output distribution was the least-worst rather than an accurate fit (figure 5c). By fitting curves to the corresponding degree distributions P(β), we have determined that these poorly fitted F(β) follow a double Gaussian distribution (see electronic supplementary material D). The lack of genuine exponential fits to F(β) means that none of the models were classic random graphs (Albert & Barabasi 2002). We conclude that, to the extent that the anatomical models reflect its organization, the medial RF is unlikely to have a scale-free topology.
Figure 5.
Best-fit curves to example cumulative degree distributions F(β), all from the stochastic model. (a) Linear-log plot showing an example of Gaussian fit to input distribution from the distance-dependent collateral variant—the dominant best-fit of the tested model curves. (b) Log–log plot showing an example of an exponential best-fit to an output distribution from the distance-dependent model: tail of the data was not fitted by any tested curve. The absence of the characteristic power-law tail is clear. (c) The same data and fit shown as a log-linear plot make clear that the exponential fit was the least-worst of those tested, rather than an accurate fit.
5. Discussion
We have provided the first quantitative review and structural model of the medial RF based on modern data. Considered as a graph, the proposed cluster model is likely to have small-world but not scale-free properties. Therefore, if the cluster structure is an accurate model of medial RF anatomy, this work is the first identification of small-world topologies at the vertebrate neuronal network level.
No power-law or truncated power-law fits to the cumulative degree distributions of either anatomical model were found. The absence of these fits over the 1620 separate model instantiations across the full range of the connection probability parameter space suggests that this result is robust. The anatomical models, therefore, predict that the medial RF does not have the properties of a scale-free network. The dominance of Gaussian fits to the cumulative degree distribution was unsurprising, given that the combination of statistical distributions used to generate the models would tend to a normal distribution according to the central limit theorem. Amaral et al. (2000) found that entire neural net of C. elegans followed an exponential cumulative degree distribution, but there is no other evidence to suggest that a normal cumulative distribution of neural connectivity is unexpected for vertebrate brain regions (we briefly address the question of the likelihood of ever finding a scale-free network in neural tissue in electronic supplementary material E).
The small-world topology was most robustly identified for the pruning rather than the stochastic anatomical model, for the distance-dependent collateral version of both, and for the lower likely proportion of projection neurons. The results for the anatomical models were also remarkably consistent across both collateral probability variants (see electronic supplementary material F for consideration of the P(p) and P(l) dependencies).
Confirmation that the actual medial RF conforms to a small-world topology is thus dependent only on showing that the combination of a spatially uniform collateral distribution and stochastic anatomical model is invalid. Analysis of connectivity between primate visual cortex pyramidal cells has shown the distribution of contacts to be distance-dependent, falling as a Gaussian with increasing separation of the cell bodies (Hellwig 2000). A recent theoretical result has shown that by assuming distance-dependent long-range connections within cortex, there is a trade-off between neuron separation and total axonal wiring length that can be optimized according to brain size (Karbowski 2001), which cortex at least seems to achieve (Cherniak 1994; Laughlin & Sejnowski 2003). Thus, the data based on individual stained cells suggesting a spatially uniform collateral arrangement in the medial RF (Grantyn et al. 1987) are at odds with the body of work demonstrating the advantage of distance-dependent projections. A detailed quantitative study of collateralization within the medial RF, specifically the density of axonal branching points as a function of distance from the cell body, is required to resolve this incongruency.
How then should we decide between the anatomical models? Though grossly simplified, the developmental model has some appealing features—it accounts for the medial RF's structure as a result of immediate post-birth experience, and thus more closely follows the network's development. A complete model would incorporate initial cell placement and axon growth, but that is both beyond the scope of this paper, and lacking the necessary supporting studies in the experimental literature. Nevertheless, there is an enticing congruency in that synaptic pruning by minimal deletion—as used here—results in optimal Hebbian learning (Chechik et al. 1998), that small-world networks are more effective at learned pattern recall than regular or random networks (Morelli et al. 2004), and that, as we have demonstrated, a small-world network can be generated using a statistical approach to synaptic pruning. The pruning model could be invalidated by demonstrating that synaptic pruning does not occur in the developing medial RF, which could be shown by changes in density of pre-synaptic markers immediately following birth.
(a) Functional implications
The demonstration of probable small-world but not scale-free topologies allows us to make some general hypotheses about functional properties of the medial RF, which may in turn explain the existence (and evolution) of the cluster structure. Scale-free networks are typified by a few highly connected nodes—‘hubs’—and are thus resilient to randomly placed damage or failure, but susceptible to targeted attacks on the hubs, which remove a disproportionate amount of the links in the network (Albert & Barabasi 2002). Putative neural networks with this topology may thus be resilient to diseases, which cause cell death, as they would typically require a high percentage of cell loss before overt functional effects are seen. The lack of a scale-free topology, therefore, implies that such hubs do not exist, and that random cell death would cause a proportional loss of network connectivity, with its associated functional effects.
Neural networks based on the canonical small-world network model have been studied for their dynamic properties using a variety of artificial neurons and connection types. Rapid cross-network synchronization (Lago-Fernandez et al. 2000; Masuda & Aihara 2004), consistent stabilization (Li & Chen 2003) and increased persistence of activity (Roxin et al. 2004) have all been reported for small-world networks, when compared to equivalent regular and random networks. Each of these properties has been found to be desirable in specific neural systems—for example, the rapid synchronization is a property of processing in the locust olfactory system. Each of these properties may also be desirable in a system, such as the medial RF, which directly associates sensory input to motor output.
To elaborate: the afferent and efferent connectivity of the projection neurons creates a single synaptic relay between the ascending sensory systems and the spinal motor circuits. Yet there is considerable behavioural and neurophysiological evidence that the medial RF is the neural substrate of both action selection (Humphries et al. 2005a) and instrumental conditioning (Buchwald 1975) in the isolated brainstem. Thus, the internal processing of the medial RF appears to support the representation and learning of an action repertoire. The dynamic properties just listed may each contribute to this: rapid cross-network communication could facilitate both competition between and simultaneous recruitment of action representations competing for selection; consistent stabilization could ensure that some representation is always recalled; and persistent activity could in turn ensure that the recalled representation remains active to drive the appropriate motor response. The accuracy of such speculation remains to be established in dynamic exploration of the structural models proposed here, and in the extension of current small-world neural network research to cases with more realistic neural constraints (such as distance-dependent transmission delays).
In conclusion, this study has proposed that the medial RF has a cell-cluster based structure, which is likely to have a small-world but not scale-free topology, and that this topology is robust across considerable parameter variation and invariant across instantiations of a parameter set.
Acknowledgments
This work was funded by EPSRC grant GR/R95722/01. We thank T. Stafford, B. Mitchinson, and two anonymous referees for comments on earlier versions of this manuscript.
Supplementary Material
References
- Albert R, Barabasi A.-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97. doi:10.1103/RevModPhys.74.47 [Google Scholar]
- Amaral L.A, Scala A, Barthelemy M, Stanley H.E. Classes of small-world networks. Proc. Natl Acad. Sci. USA. 2000;97:11 149–11 152. doi: 10.1073/pnas.200327197. doi:10.1073/pnas.200327197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli G. Progress and perspectives in computational neuroanatomy. Anat. Rec. 1999;257:195–207. doi: 10.1002/(SICI)1097-0185(19991215)257:6<195::AID-AR5>3.0.CO;2-H. doi:10.1002/(SICI)1097-0185(19991215)257:6<195::AID-AR5>3.0.CO;2-H [DOI] [PubMed] [Google Scholar]
- Barabasi A.-L, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. doi:10.1126/science.286.5439.509 [DOI] [PubMed] [Google Scholar]
- Bourgeois J.P, Rakic P. Changes of synaptic density in the primary visual cortex of the macaque monkey from fetal to adult stage. J. Neurosci. 1993;13:2801–2820. doi: 10.1523/JNEUROSCI.13-07-02801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowsher D, Westman J. The gigantocellular reticular region and its spinal afferents: a light and electron microscope study in the cat. J. Anat. 1970;106:23–36. [PMC free article] [PubMed] [Google Scholar]
- Buchwald J.S. Brainstem substrates of sensory information processing and adaptive behavior. UCLA Forum Med. Sci. 1975;18:315–333. [PubMed] [Google Scholar]
- Cant N.B, Benson C.G. Parallel auditory pathways: projection patterns of the different neuronal populations in the dorsal and ventral cochlear nuclei. Brain Res. Bull. 2003;60:457–474. doi: 10.1016/s0361-9230(03)00050-9. doi:10.1016/S0361-9230(03)00050-9 [DOI] [PubMed] [Google Scholar]
- Chechik G, Meilijson I, Ruppin E. Synaptic pruning in development: a computational account. Neural Comput. 1998;10:1759–1777. doi: 10.1162/089976698300017124. doi:10.1162/089976698300017124 [DOI] [PubMed] [Google Scholar]
- Cherniak C. Component placement optimization in the brain. J. Neurosci. 1994;14:2418–2427. doi: 10.1523/JNEUROSCI.14-04-02418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantyn A, Ong-Meang Jacques V, Berthoz A. Reticulo-spinal neurons participating in the control of synergic eye and head movements during orienting in the cat. II. Morphological properties as revealed by intra-axonal injections of horseradish peroxidase. Exp. Brain Res. 1987;66:355–377. doi: 10.1007/BF00243310. [DOI] [PubMed] [Google Scholar]
- Hammer R.P.J, Lindsay R.D, Scheibel A.B. Development of the brain stem reticular core: an assessment of dendritic state and configuration in the perinatal rat. Dev. Brain Res. 1981;1:179–190. doi: 10.1016/0165-3806(81)90106-1. doi:10.1016/0165-3806(81)90106-1 [DOI] [PubMed] [Google Scholar]
- Hayar A, Piguet P, Feltz P. GABA-induced responses in electrophysiologically characterized neurons within the rat rostro-ventrolateral medulla in vitro. Brain Res. 1996;709:173–183. doi: 10.1016/0006-8993(95)01238-9. doi:10.1016/0006-8993(95)01238-9 [DOI] [PubMed] [Google Scholar]
- Hellwig B. A quantitative analysis of the local connectivity between pyramidal neurons in layers 2/3 of the rat visual cortex. Biol. Cybern. 2000;82:111–121. doi: 10.1007/PL00007964. [DOI] [PubMed] [Google Scholar]
- Hilgetag C.C, Burns G.A, O'Neill M.A, Scannell J.W, Young M.P. Anatomical connectivity defines the organization of clusters of cortical areas in the macaque monkey and the cat. Phil. Trans. R. Soc. B. 2000;355:91–110. doi: 10.1098/rstb.2000.0551. doi:10.1098/rstb.2000.0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C.J, Mainville L.S, Jones B.E. Distribution of cholinergic, GABAergic and serotonergic neurons in the medial medullary reticular formation and their projections studied by cytotoxic lesions in the cat. Neuroscience. 1994;62:1155–1178. doi: 10.1016/0306-4522(94)90351-4. doi:10.1016/0306-4522(94)90351-4 [DOI] [PubMed] [Google Scholar]
- Humphries M.D, Gurney K, Prescott T.J. Action selection in a macroscopic model of the brainstem reticular formation. In: Bryson J.J, Prescott T.J, Seth A.K, editors. Modelling natural action selection. AISB Press; Brighton, UK: 2005a. pp. 61–68. [Google Scholar]
- Humphries M.D, Gurney K, Prescott T.J. Is there an integrative center in the vertebrate brainstem? A robotic evaluation of a model of the reticular formation viewed as an action selection device. Adapt. Behav. 2005b;13:97–113. doi:10.1177/105971230501300203 [Google Scholar]
- Ito K, McCarley R.W. Physiological studies of brainstem reticular connectivity. I. Responses of mPRF neurons to stimulation of bulbar reticular formation. Brain Res. 1987;409:97–110. doi: 10.1016/0006-8993(87)90745-1. doi:10.1016/0006-8993(87)90745-1 [DOI] [PubMed] [Google Scholar]
- Jones B.E. Reticular formation: cytoarchitecture, transmitters, and projections. In: Paxinos G, editor. The rat nervous system. 2nd edn. Academic Press; New York: 1995. pp. 155–171. [Google Scholar]
- Jones B.E, Holmes C.J, Rodriguez-Veiga E, Mainville L. GABA-synthesizing neurons in the medulla: their relationship to serotonin-containing and spinally projecting neurons in the rat. J. Comp. Neurol. 1991;313:349–367. doi: 10.1002/cne.903130210. doi:10.1002/cne.903130210 [DOI] [PubMed] [Google Scholar]
- Karbowski J. Optimal wiring principle and plateaus in the degree of separation for cortical neurons. Phys. Rev. Lett. 2001;86:3674–3677. doi: 10.1103/PhysRevLett.86.3674. doi:10.1103/PhysRevLett.86.3674 [DOI] [PubMed] [Google Scholar]
- Kilmer W, McCulloch W.S, Blum J. A model of vertebrate central command system. Int. J. Man Mach. Stud. 1969;1:279–309. [Google Scholar]
- Kleinfeld D, Berg R.W, O'Connor S.M. Anatomical loops and their electrical dynamics in relation to whisking by rat. Somatosens. Mot. Res. 1999;16:69–88. doi: 10.1080/08990229970528. doi:10.1080/08990229970528 [DOI] [PubMed] [Google Scholar]
- Lago-Fernandez L.F, Huerta R, Corbacho F, Siguenza J.A. Fast response and temporal coherent oscillations in small-world networks. Phys. Rev. Lett. 2000;84:2758–2761. doi: 10.1103/PhysRevLett.84.2758. doi:10.1103/PhysRevLett.84.2758 [DOI] [PubMed] [Google Scholar]
- Laughlin S.B, Sejnowski T.J. Communication in neuronal networks. Science. 2003;301:1870–1874. doi: 10.1126/science.1089662. doi:10.1126/science.1089662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen G. Stability of a neural network model with small-world connections. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;68:052901. doi: 10.1103/PhysRevE.68.052901. [DOI] [PubMed] [Google Scholar]
- Mason P, Fields H.L. Axonal trajectories and terminations of on- and off-cells in the cat lower brainstem. J. Comp. Neurol. 1989;288:185–207. doi: 10.1002/cne.902880202. doi:10.1002/cne.902880202 [DOI] [PubMed] [Google Scholar]
- Masuda N, Aihara K. Global and local synchrony of coupled neurons in small-world networks. Biol. Cybern. 2004;90:302–309. doi: 10.1007/s00422-004-0471-9. doi:10.1007/s00422-004-0471-9 [DOI] [PubMed] [Google Scholar]
- Montoya J.M, Sole R.V. Small world patterns in food webs. J. Theor. Biol. 2002;214:405–412. doi: 10.1006/jtbi.2001.2460. doi:10.1006/jtbi.2001.2460 [DOI] [PubMed] [Google Scholar]
- Morelli L.G, Abramson G, Kuperman M.N. Associative memory on a small-world neural network. Eur. Phys. J. B. 2004;38:495–500. doi:10.1140/epjb/e2004-00144-7 [Google Scholar]
- Motulsky H, Christopoulos A. Oxford University Press; Oxford, UK: 2004. Fitting models to biological data using linear and non-linear regression. A practical guide to curve fitting. [Google Scholar]
- Newman D.B. Distinguishing rat brainstem reticulospinal nuclei by their neuronal morphology. II. Pontine and mesencephalic nuclei. J. Hirnforsch. 1985;26:385–418. [PubMed] [Google Scholar]
- Roxin A, Riecke H, Solla S.A. Self-sustained activity in a small-world network of excitable neurons. Phys. Rev. Lett. 2004;92:19 801. doi: 10.1103/PhysRevLett.92.198101. doi:10.1103/PhysRevLett.92.198101 [DOI] [PubMed] [Google Scholar]
- Salibi N.A, Saade N.E, Banna N.R, Jabbur S.J. Dorsal column input into the reticular formation. Nature. 1980;288:481–483. doi: 10.1038/288481a0. doi:10.1038/288481a0 [DOI] [PubMed] [Google Scholar]
- Scheibel A.B. The brainstem reticular core and sensory function. In: Brookhart J.M, Mountcastle V.B, editors. Handbook of physiology. Section 1: the nervous system. American Physiological Society; Bethesda, MD: 1984. [Google Scholar]
- Scheibel M.E, Scheibel A.B. Anatomical basis of attention mechanisms in vertebrate brains. In: Quarton G.C, Melnechuk T, Schmitt F.O, editors. The neurosciences, a study program. The Rockefeller University Press; New York: 1967. pp. 577–602. [Google Scholar]
- Siegel J.M. Behavioral functions of the reticular formation. Brain Res. Rev. 1979;1:69–105. doi: 10.1016/0165-0173(79)90017-1. doi:10.1016/0165-0173(79)90017-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edelman G.M. Theoretical neuroanatomy and the connectivity of the cerebral cortex. Behav. Brain Res. 2002;135:69–74. doi: 10.1016/s0166-4328(02)00157-2. doi:10.1016/S0166-4328(02)00157-2 [DOI] [PubMed] [Google Scholar]
- Watts D.J, Strogatz S.H. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. doi:10.1038/30918 [DOI] [PubMed] [Google Scholar]
- Yates B.J, Stocker S.D. Integration of somatic and visceral inputs by the brainstem: functional considerations. Exp. Brain Res. 1998;119:269–275. doi: 10.1007/s002210050342. doi:10.1007/s002210050342 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.