Abstract
We use a mathematical model to study the evolution of influenza A during the epidemic dynamics of a single season. Classifying strains by their distance from the epidemic-originating strain, we show that neutral mutation yields a constant rate of antigenic evolution, even in the presence of epidemic dynamics. We introduce host immunity and viral immune escape to construct a non-neutral model. Our population dynamics can then be framed naturally in the context of population genetics, and we show that departure from neutrality is governed by the covariance between a strain's fitness and its distance from the original epidemic strain. We quantify the amount of antigenic evolution that takes place in excess of what is expected under neutrality and find that this excess amount is largest under strong host immunity and long epidemics.
Keywords: influenza, antigenic drift, population dynamics, population genetics, herd immunity, immune escape
1. Introduction
Seasonal influenza A epidemics are a significant cause of morbidity and mortality in temperate zones of both hemispheres. In the Northern Hemisphere, annual epidemics occur between November and April and since the early 1900s have caused more cumulative mortality than the three major pandemic events of the twentieth century (Earn et al. 2002). The characteristics that make these annual influenza outbreaks unusual among non-childhood diseases is that they are periodic and sustained. Periodicity has often been attributed to seasonal changes in transmissibility or mixing patterns (Schulman & Kilbourne 1962; Davey & Reid 1972; Anderson & May 1991), and more recently to a possible dynamical resonance between low intrinsic seasonality and loss of host immune memory (Dushoff et al. 2004). Sustainability of annual epidemics is the result of viral immune escape through antigenic drift (Webster et al. 1992; Cox & Subbarao 2000; de Jong et al. 2000b; Hay et al. 2001; Hampson 2002).
Antigenic drift—the accumulation of point mutations in virus antigens—is easily detectable in sequence data covering almost four decades of influenza activity (Macken et al. 2001). The resulting immune escape, as measured by haemagglutinin inhibition (HI) tests, indicates that influenza can escape a significant amount of herd immunity after only 2–3 years (de Jong et al. 2000b; Coiras et al. 2001; Hay et al. 2001). Since hosts lose immunity gradually, the influenza virus population need not mutate to a completely new antigenic form. Rather, influenza benefits from each additional amino acid replacement in its surface proteins by becoming slightly less recognizable to the hosts on whom it previously conferred immunity. Mutations occur during replication in host epithelial cells, and the virus persists and replicates as long as host contacts sustain a chain of transmission in the host population. These chains of transmission enable influenza to accumulate mutations; the resulting mutated progeny viruses are often called antigenic drift variants or simply drift variants.
Here, we consider the forces that govern antigenic drift in influenza A. While it is well known that significant antigenic drift causes severe influenza outbreaks (Kilbourne 1973; Cox & Subbarao 2000; de Jong et al. 2000a; Hay et al. 2001), little is known about the effects of influenza outbreaks on antigenic drift. These two processes are of course concurrent and tightly coupled. Epidemic dynamics unroll a series of between-host transmission events, which increase viral population size and offer the influenza virus a means to reproduce, mutate, and escape host immunity. Viral immune escape then lowers the host population's effective immunity and adds momentum to the ongoing epidemic. The benefits of immune escape may be somewhat delayed as hosts maintain some short-term non-specific immunity (Ferguson et al. 2003; Xia et al. 2005).
To model antigenic drift during a single season of influenza, we use a standard susceptible–infected–recovered (SIR) framework (Kermack & McKendrick 1927; Anderson & May 1991). We classify the various influenza strains by their distance from a reference strain. Our model can be described as an -model, where the subscript denotes antigenic distance from the reference strain I0. This antigenic distance reflects a one-dimensional antigenic space as in previous models (Pease 1987; Sasaki 1994; Andreasen et al. 1996; Haraguchi & Sasaki 1997; Sasaki & Haraguchi 2000; Gog & Grenfell 2002; Andreasen 2003; Lin et al. 2003; Boni et al. 2004); however, it is important to remember that a realistic mapping of antigenic type onto immune escape would have higher dimensionality (Lapedes & Farber 2001).
We first introduce a neutral model with epidemic dynamics and mutation and show that the strain population has a Poisson distribution whose mean moves forward in time according to a molecular clock (Zuckerkandl & Pauling 1965; Kimura 1968, 1969). A second model includes host immunity, where strains that escape host immunity through antigenic drift have higher transmissibility. This non-neutral model has high dimensionality and persistent nonlinearities; we solve it numerically. Fortunately, the model's population dynamics can be naturally expressed in a population-genetic framework, which allows us to extract key viral fitness components and analyse their effects on antigenic drift. Using the neutral model as a baseline, we are able to study the forces that drive influenza antigenic drift in human populations.
2. Neutral model
We first consider a many-strain, single-season influenza epidemic model, where all strains are equally fit. Once recovered, hosts cannot become reinfected, and the epidemic ends when the susceptible pool is depleted. The epidemic begins with a particular strain which we call the epidemic strain or the zero-strain; individuals infected with the zero-strain are said to be in the population class I0. The zero-strain can mutate, and when it acquires one amino acid change the harbouring individual becomes a member of the population class I1. The I1 class represents those hosts that are infected with any strain which is exactly one amino acid different from the original epidemic strain; if a host's infecting virus undergoes another mutation event, that host would move into the I2 class. In general, hosts in the class Ik are infected with a strain that is k amino acids different from the original epidemic strain.
Real individuals are most likely to be infected with a virus population of great diversity, but we can approximate the genetic distance between a host's infecting influenza virions and the original epidemic-causing strain by considering the mean distance of a host's virus population from the original strain. In this model, we neglect within-host evolution and place individuals in population classes according to the strain they are most likely to transmit at a given moment.
We assume a homogeneous mutation rate across the HA1 segment (987 nt) of influenza's haemagglutinin surface protein. We focus on the HA1 because of its rapid evolution and importance to immune escape (Fitch et al. 1997; Bush et al. 1999; Plotkin & Dushoff 2003). Since back mutation is highly unlikely and recurrent mutations (double hits) are somewhat unlikely, individuals in the Ik class are assumed to move only to the class Ik+1 when an amino acid replacement occurs.
Using S to denote susceptible hosts, we write the neutral dynamical model as
| 2.1 |
where β is the compound parameter describing the transmission rate and the host contact rate, and ν is the hosts' recovery rate from infection. The class In denotes individuals infected with a strain at least n amino acids away from the zero-strain. The parameter μ is the non-synonymous mutation rate in the HA1. For the purposes of our model, μ is the RNA polymerase's error rate, times the proportion of possible mutations in the HA1 that cause amino acid changes, times the proportion of new mutations that are not lost due to stochastic fluctuations. We will refer to μ simply as the mutation rate. For low population sizes and slow transmission, stochastic extinction of new mutants would need to be modelled explicitly.
To investigate relative strain frequencies, we write and . Using (2.1), the dynamic equations for the strain frequencies are
| 2.2 |
which is a linear and autonomous system, whose dynamics are governed by a subdiagonal matrix and which is independent of the population dynamic variables S and I. If all the strains at time zero are of type zero, and for k>0, the solution to (2.2) is
| 2.3 |
for all k<n and t>0; the trajectory of in is determined by noting that . The strain frequencies are Poisson-distributed with mean μt. Thus, even in the presence of host population dynamics, we obtain the standard result that neutral mutation produces a molecular clock (we use the term loosely since our clock follows the mean number of changes in a heterogeneous population, rather than the number of fixation events of new variants). If μ=0.1 per day and the epidemic lasts 120 days, the strain population at the end of the epidemic will be a mean distance 12 amino acids away from the original epidemic strain. Our neutral system has the same assumptions and behaviour as a standard Poisson process.
3. Non-neutral model
In this section, we remove the neutrality assumption by allowing for immune structure in the host population and viral fitness differences based on host immunity. In our non-neutral model, influenza strains cause weaker infections in immune hosts and are thus less transmissible by these hosts; a strain's fitness (transmissibility) in a particular host depends on the host's immunity to that particular strain. As the virus population mutates, variants that are distant from this season's epidemic strain will be able to cause increasingly transmissible infections, even in hosts whose immunity to earlier variants may have been quite strong. As in the neutral model, hosts cannot become reinfected and the epidemic ends when it runs out of susceptibles.
We extend the p–q equations from our previous model (Boni et al. 2004, p. 180) to include multiple strains; susceptibles are denoted by the variables qi, where
qi=frequency of hosts who are susceptible and whose last infection was i amino acids away from this season's zero-strain; .
Susceptible hosts have decreasing immunity as the subscript increases. Infected individuals require two subscripts: the current infecting strain and the previous immunizing strain. We define
pjk=frequency of hosts whose last infection was j amino acids away from this season's zero-strain and who are currently infected with strain k; , .
This season's strain k differs by k amino acids from this year's zero-strain, and in a one-dimensional amino acid space, in accordance with our assumptions, individuals in class pjk have a distance of j+k amino acids between their immunizing strain and their current infecting strain. As the distance between challenging strain and immunizing strain increases, immunity decreases (Gill & Murphy 1976; Smith et al. 2004). We assume that immunity wanes exponentially with antigenic distance. Individuals in the class pjk have their transmissibility reduced to , where ; the scaling parameter a describes the amount of immune escape conferred by each additional amino acid change. We use the number of amino acid changes as a proxy for immune escape, although their location also plays an important role. An example of this is the 18 strongly selected codons identified by Bush et al. (1999), which are known to be associated with antibody-combining sites.
Our dynamical equations for susceptible individuals are:
| 3.1 |
where the parenthetical term represents the total force of infection in the population. The dynamic equations for the infected individuals are constructed similarly, but we need to take into account the boundary situations k=0 and k=n. The equations are:
| 3.2 |
| 3.3 |
| 3.4 |
where
| 3.5 |
is a number between 0 and 1. This definition is a fairly close approximation to cjk=1, which would be the natural way to write down the model from first principles. We define cjk as in (3.5) for mathematical convenience; the result of this approximation is a slightly slower rate of antigenic drift. The parameters β, ν and μ are defined as before.
Equations (3.1)–(3.4) now define a complete, infinite-dimensional dynamical system. As in our neutral model, we collapse some of our variables by defining:
The variable S denotes the total amount of susceptibility in the population (or the total amount of potential infectivity) and is a number between 0 and 1. Q denotes the total fraction of hosts that are susceptible, irrespective of their immune histories. Ik is the force of infection of strain k, while I is influenza's total force of infection. Q and S obey the dynamical equations:
which means that the ratio S/Q does not change with time. We call the immunity in the host population; θ measures the herd immunity to the zero-strain of the susceptible individuals in the host population. Alternatively, θ can be viewed as the expected immunity of any susceptible individual in the population.
As before, ik=Ik/I is the frequency of strain k, and equations (3.1)–(3.4) reduce to:
| 3.6 |
| 3.7 |
| 3.8 |
With the dynamical equations for Q and I,
| 3.9 |
| 3.10 |
Equations (3.6)–(3.10) now describe an (n+2)-dimensional dynamical system, which keeps track of the strain frequencies, the total force of infection and the number of susceptibles.
(a) Population genetics
The quantity has the natural population-genetic interpretation as the fitness of strain k, and
| 3.11 |
is thus the mean fitness of the entire virus population. The dynamical equations (3.6)–(3.10) then resemble standard population-genetic equations where the key determinant of a variant's increase or decrease in frequency is its fitness (wk) relative to the population's mean fitness (W).
To see how mean fitness behaves as a function of time, we differentiate equation (3.11) and approximating ,
which, if we set μ=0, is the continuous analogue to Fisher's fundamental theorem of natural selection (Fisher 1930).
To investigate the dynamic properties of antigenic drift, we define:
| 3.12 |
which is the mean antigenic distance from the strain population at time t to the zero-strain. The quantity D follows the dynamics of system (3.6)–(3.10). Approximating , we have:
| 3.13 |
which is a form of the Price equation (Price 1970, 1972). Distance refers to the number of amino acid replacements a strain is away from the original invading strain, and fitness refers to . Since strain fitness always increases with added distance from the zero-strain, the covariance term—which in the above equation is calculated across the strain frequencies at time t—will always be non-negative. At t=0, the covariance is zero, and its derivative with respect to time is . At the beginning of the epidemic, when there are strains of high and low fitness, the covariance term will be positive and increasing; then, as Q decreases and as antigenic drift causes most of the strains in the virus population to have a fitness close to one, the covariance term will tend back towards zero. When there are no fitness differences among strains (neutral mutation), the covariance term is always zero and
| 3.14 |
which can also be derived from (2.3).
Once all the population-genetic structure is extracted from our population-dynamic influenza model, the intensity of selection for antigenically distant strains can be measured by the size of the covariance term in (3.13) relative to the mutation rate μ. The mutation rate μ is responsible for neutral mutation accumulation and sets the baseline pace of the molecular clock, while the covariance term changes throughout the epidemic and accelerates the clock to varying degrees (see figure 1).
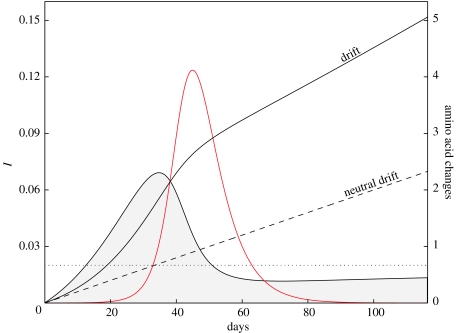
Figure 1.
Results from integrating the modified model (3.6)–(3.10). β=1.0, θ=0.6, a=0.1, N=105; μ=0.02 is marked by the dashed line. The red line represents the force of infection I (left-hand scale). The curve bounding the filled area is βQ Cov (left-hand scale) from equation (3.13) and shows here that selection for new variants is more intense in the beginning phases of the epidemic. Comparing the covariance term with the dotted line (μ) shows the relative non-neutral and neutral contributions to total antigenic drift. The two drift lines correspond to values on the right-hand vertical scale. After about 116 days, the epidemic is over and the mean number of amino acid changes is around five; under neutral drift we would expect about two.
(b) Excess antigenic drift
The amount of antigenic drift that occurs during one season is highly dependent on μ−1—the mean number of days it takes for a neutrally mutating flu population to acquire, on the average, one additional amino acid change. Models of within-host flu evolution (Sasaki 1994; Haraguchi & Sasaki 1997) have calculated a drift speed that scales with μ, while some between-host models (Andreasen et al. 1996; Gog & Grenfell 2002; Lin et al. 2003) have found a drift speed that scales approximately with . In this investigation, we focus on the excess antigenic drift, δ, which we define as the difference between the amount of drift occurring under neutral conditions and the amount of drift occurring under non-neutral conditions. We show that δ is relatively insensitive to the mutation rate μ; this means that we can study the factors that affect δ without knowing the true mutation rate.
Let DS be the amount of antigenic drift that occurs when there is selection for immune escape, and let DN be the amount of neutral antigenic drift that would be expected to occur. We calculate DS by numerically integrating equations (3.6)–(3.10) with some initial condition, or inoculum, I(0) that gives the force of infection at time t=0. We set and use N as a proxy for population size. Equations (3.6)–(3.10) are numerically integrated from time t=0 until a time tf such that ; we say that the epidemic ends at time tf. Using definition (3.12), we let ; this is the mean amount of antigenic drift that occurs when selective pressure causes the virus to mutate away from the epidemic strain. To calculate for an epidemic of the same length, we use equation (3.14) and get .
It is then natural to define δ as:
| 3.15 |
The covariance term under the integral is the term from the Price equation (3.13), and is the time at which the epidemic ends. Since the integrand is always non-negative, we see that a longer epidemic results in a larger δ, since it allows selection more time to operate; this phenomenon has been analysed in the context of pathogen emergence by Antia et al. (2003). At first glance, it seems that increasing transmissibility and host contact rates (via β) should yield more excess drift; however, higher β-values correspond to shorter epidemics which can in turn yield less excess drift. We will characterize the behaviour of δ as we change the model parameters described in table 1.
Table 1.
Parameter descriptions and probable ranges for the non-neutral model.
| parameter | description | range |
|---|---|---|
| a | immune-escape parameter; when a is large, immune escape is rapid (cross-immunity is weak) | 0.01≤a≤0.50 |
| β | transmissibility and host contact rate; assumes ν=0.2, hence 1.2≤0≤6.0 | 0.24≤β≤1.20 |
| μ | non-synonymous mutation rate in the HA1: error rate in the RNA polymerase, times proportion of possible non-silent changes, times proportion of mutants that survive stochastic loss | 0.001≤μ≤0.05 |
| ν | recovery rate from infection; this is set to ν=0.2, so that an infection lasts 5 days; fixing this parameter has no qualitative effect on model results; realistic range shown at right | 0.1≤ν≤0.3 |
| θ | population-wide immunity or herd immunity; θ=1 means that the population has full immunity (i.e. everyone is completely immune); θ=0 means that every host is completely naive | 0≤θ≤1 |
| N−1 | inoculum; the initial force of infection that begins the epidemic; the parameter N can be thought of as the host population size | 10−9≤N−1≤10−3 |
(c) Parameter ranges
The high dimensionality of our system forces us to study it numerically. The dynamical system (3.6)–(3.10) has five parameters (a, β, μ, ν and θ), although ν can be scaled out if we wish; we simply set , fixing the mean infection length at 5 days. The initial condition must be set, and we consider it a model parameter. The number of strains n in all simulations is 60. The parameters β, θ and N can be reasonably varied to simulate strains with basic reproduction ratios in the range (Mills et al. 2004), and populations of various sizes with varying levels of herd immunity. The tested ranges for these and other parameters are summarized in table 1.
The parameters a and μ are more difficult to measure and can vary over a wide range of values. The mutation rate in influenza's haemagglutinin has been measured by Fitch et al. (1997) and Bush et al. (1999) who estimated the observed, rather than neutral, rate of evolution. Moreover, depending on whether one calculates distance from a root strain or mean distance between pairs of strains isolated in consecutive years, estimates of mutation rates can vary by an order of magnitude. Worldwide (Macken et al. 2001) and local (Coiras et al. 2001; Pyhälä et al. 2004) HA1 datasets suggest that the observed mutation rate corresponds to between 1 and 13 amino acid changes per year; the neutral rate can of course be lower. In our numerical simulations, we test the range of μ-values , which corresponds to between 0.4 and 18 non-synonymous mutations per year. Since δ is not highly sensitive to μ, the choice of range for the neutral mutation rate has little effect on our results.
Finally, a measures immune escape per amino acid change. The range entails that it takes between 5 and 20 amino acid replacements to evade 50% host immunity. This seems reasonable based on published HI tables (de Jong et al. 2000b; Coiras et al. 2001; Hay et al. 2001) and the antigenic map in Smith et al. (2004). The tested range for a will be slightly wider: . Note that parameter estimates of a and μ are a function of the length of the HA1 molecule (987 nt).
4. Results
According to our model, the keys to generating a large amount of excess antigenic drift are strong herd immunity and long epidemics. Host immunity forces the virus population to mutate to a distant variant so that it can begin spreading efficiently. Figure 2 shows immunity driving antigenic drift within the context of epidemic dynamics, while figure 3 shows excess antigenic drift (δ) increasing as a function of immunity (θ). A slow (and thus long) epidemic allows selection pressure to operate for a longer period of time and allows the virus population to drift further than under a short epidemic. The two key characteristics of a host–parasite system that can lengthen an epidemic are large host population size and low . In our model, if N is large or if our effective is close to 1, the epidemic will be long and excess drift will be large.
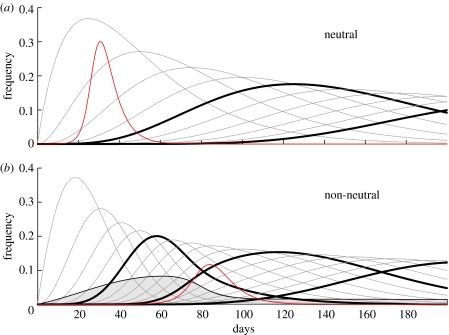
Figure 2.
The solid black lines represent the frequencies i1, i2, i3 and so on (i0 is not shown). Every fifth variant has a bold line so that (a) and (b) can be compared. In (a), i5 and i10 are bold; in (b), i5, i10 and i15 are bold. In (a) and (b), β=0.6, ν=0.2, μ=0.04, a=0.1 and I(0)=10−5. In (a), θ=0.0, . In (b), θ=0.6, and the effective . The red line represents the force of infection I. In (b), the solid curve bounding the filled area corresponds to the term βQ Cov from the Price equation (3.13).
Figure 3.
Excess drift as a function of herd immunity. In (a) and (b), the two curves bounding the filled area show the value of δ for μ=0.05 (upper curve) and μ=0.005 (lower curve). In (a) and (b), β=1.0, ν=0.2 and a=0.05. The negatively sloped straight line in both panels shows as a function of θ. When θ>0.8, the effective basic reproduction ratio is less than 1 and there is no epidemic.
Thus, the parameters β, θ and N have intuitive effects on δ. Decreasing β or increasing θ lowers , lengthens the epidemic, and increases the amount of excess antigenic drift. In addition, increasing θ drives antigenic drift by augmenting the strength of selection for escape mutants. An increase in the population size N decreases the relative size of the inoculum, lengthens the epidemic, and leads to more excess antigenic drift (higher δ).
For the range of a-values that we test, lowering a decreases the amount of excess drift during the course of an epidemic. This happens because when a is small enough, the initial populations of strains that are 1, 2 or 3 amino acids away from the zero-strain are not much more fit than the zero-strain, and natural selection has little fitness variation on which to act. In the case of very small a, antigenic drift is close to neutral. On the other hand, if a is very large, δ will also be small since fit variants are achieved with few mutations and the epidemics are generally short. Figure 4 shows this behaviour of our model as a function of a; in general, intermediate values of a maximize δ.
Figure 4.
In (a)–(c), β=0.6, ν=0.2, θ=0.3 (effective for the zero-strain), μ=0.02 and I(0)=10−5; a=0.1, 0.5, 3.0 in (a), (b) and (c), respectively. The red line represents the force of infection I and corresponds to the red axis on the right-hand side of the graph. The solid curve bounding the filled area corresponds to the term βQ Cov from the Price equation (3.13). The shaded area under this curve is the value of the integral of βQ Cov from 0 to tf; this is the excess antigenic drift. Graph (b) maximizes the area under the covariance curve. The value a=3.0 is used for illustrative effect only.
Similarly, intermediate values of μ appear to maximize δ. Again, low μ yields little variation, and thus slow natural selection. High μ yields lots of variation and much antigenic drift, but most of this antigenic drift can be explained by the fast mutation rate rather than selective pressure on the virus population—total antigenic drift is high, but excess drift is low.
In general, the mutation rate μ and immune-escape parameter a have relatively little effect on the excess drift δ. Over a set of 83 994 runs using distinct parameter combinations, δ exhibited partial correlations of 0.12 with μ and −0.05 with a; this suggests low sensitivity of δ to μ and a. There is no way to test for statistical significance since the correlated quantities are the results of deterministic simulations (see table 2 and electronic supplementary material, Appendix A). Also, δ–μ sensitivity is dependent on the choice of cross-immunity function τ. Alternate functional forms of τ can produce a noticeable sensitivity of δ to μ (electronic supplementary material, Appendix B).
Table 2.
Partial correlations between a parameter and a dynamic quantity, when the other four model parameters are held constant. The last column is the partial correlation between δ and a parameter when the other four parameters as well as the length of the epidemic are held constant. For each parameter, the first row uses all simulations (83 994 in all) where the epidemic length was less than 1000 days. The second row, removes some outliers and uses those simulations (71 227) whose epidemics were shorter than 250 days. Note that accounting for the epidemic length does not explain away a correlation between θ and δ. Abbreviations: es: epidemic size, 1−Q(tf); wes: weighted epidemic size, 1−θ−S(tf); len: epidemic length, tf ; D: total antigenic drift.
| es | wes | len | D | δ | δ(len) | |
|---|---|---|---|---|---|---|
| β | 0.76 | 0.76 | −0.54 | −0.44 | −0.32 | −0.06 |
| 0.70 | 0.76 | −0.79 | −0.48 | −0.34 | −0.02 | |
| θ | −0.39 | −0.94 | 0.25 | 0.49 | 0.59 | 0.57 |
| −0.45 | −0.98 | 0.65 | 0.73 | 0.78 | 0.63 | |
| a | 0.37 | 0.25 | −0.20 | −0.12 | −0.05 | 0.09 |
| 0.37 | 0.31 | −0.33 | 0.04 | 0.18 | 0.26 | |
| μ | 0.15 | 0.11 | −0.09 | 0.56 | 0.12 | 0.21 |
| 0.15 | 0.13 | −0.13 | 0.84 | 0.33 | 0.36 | |
| log N | 0.17 | 0.11 | 0.42 | 0.35 | 0.25 | −0.02 |
| 0.33 | 0.29 | 0.85 | 0.55 | 0.39 | 0.00 |
From these explorations of the effects of the model parameters on excess drift, we note two curious behaviours of our single-season model of influenza evolution.
First, the epidemic usually peaks when much of the drift or excess drift has already happened. Therefore, sampling isolates during what we believe to be the beginning of the epidemic may lead us to overestimate the amount of drift that happened between seasons, when in fact, the observed drift may have happened early in the current season. In figure 2a, the neutral epidemic peaks after 30 days, having undergone about 1.2 replacements. In the non-neutral dynamics in figure 2b, the epidemic peaks after 84 days having undergone 7.9 replacements, 4.6 of which are in excess of what can be explained by neutral mutation during that time period. Sampling during the beginning of the non-neutral epidemic (e.g. between days 60 and 70) may lead to an incorrect conclusion about that year's epidemic strain.
This result may help explain a phenomenon described by Schweiger et al. (2002), namely, that ‘comparable major antigenic differences may result in a severe outbreak—not necessarily during the first epidemic season [of] their appearance, but during the second.’ A slow and mild epidemic can be accompanied by a lot of excess drift in its early phases; in such an epidemic, distant variants may be observed in collected flu isolates. If a distant variant at the end of a mild season starts the epidemic at the beginning of next season, it will benefit by having escaped much of the host population's immunity and may be able to cause a large epidemic. This pattern was observed in Germany during the mild 1997/1998 season and the more severe 1998/1999 season. In general, amino acid changes accumulate within an epidemic season, but short-term non-specific host immunity may prevent their effects from being felt until the following season.
Second, we note that the total size of the epidemic, , measured as the total fraction of hosts infected, as well as the weighted size of the epidemic, , do not always correlate positively with the excess drift δ (partial correlations are −0.15 and −0.06, respectively). Large epidemics do not always result in a lot of antigenic drift in part because larger epidemic sizes correlate negatively (−0.61 and −0.62, respectively) with epidemic length, and epidemic length correlates positively (+0.60) with δ. This suggests that a scenario of annual epidemics with runaway antigenic drift (Boni et al. 2004) would have to be revisited under the assumption that long epidemics, rather than large epidemics, yield a lot of drift. In such a scenario, the strain distribution at the end of one epidemic and the ‘choice’ of a particular strain to start next season's epidemic may be critical.
5. Discussion
We analysed a neutral and a non-neutral model of influenza spread and evolution in a single epidemic season in order to investigate the forces that drive antigenic drift in influenza. We solved the neutral model analytically, which provided a basis for comparison of the numerical results of the non-neutral model. In the non-neutral model, we examined the conditions that cause the most excess antigenic drift, which we defined as the drift that occurs beyond that expected under neutral mutation. We found that strong host immunity and long epidemics result in greater excess antigenic drift, that significant amounts of antigenic drift can occur in the early phases of the epidemic when there are still relatively few infected hosts, and that large epidemics tend to be short, generating little excess drift.
We used a standard deterministic SIR formulation with multiple strains; our model had no host births and no immigration so that the epidemic ended when the virus ran out of susceptibles. This restricts our results to closed panmictic populations. Antigenic drift on a global scale would require a meta-population model, which describes human populations exposed to influenza. In particular, the stochastic nature of (i) migration between sub-populations, (ii) summertime transmission dynamics of influenza in temperate zones (Gog et al. 2003) and (iii) local extinction of epidemics and new mutants when infected numbers are small (Girvan et al. 2002; Park et al. 2002) would all need to be better understood.
Our non-neutral model (3.6)–(3.10) fits elegantly into Price's covariance formulation of natural selection. Using Price's population-genetic framework, we can track changes in the mean antigenic drift in the virus population via the covariance between mutations accumulated (k) and immunity escaped (wk). This covariance term increases initially and then wanes, reflecting the differential selection pressure on the influenza virus population during the course of an epidemic. Price's formulation describes the forces that govern the progression of the virus population's mean antigenic distance from the original epidemic-causing strain, but it tells us nothing about other properties of the strain distribution. An important and open problem is the characterization of the differences between the observed (non-neutral) strain distribution at the end of an epidemic and the neutral Poisson distribution.
We identified the parameters in our system that caused the greatest departures from neutrality. It appears that immunity (θ) has the largest effect on excess antigenic drift (δ). A high level of host immunity puts significant selection pressure on the virus population, and in addition, it slows the epidemic, giving natural selection more time to select for distant antigenic variants. The relationship between θ and δ may have important public health consequences as it indicates that vaccinated populations, as long as they can still sustain epidemics, can cause significant antigenic drift (as suggested by Pease 1987, p. 445). Public health officials may wish to investigate whether the benefits of vaccination during one season conflict with the feasibility of vaccination for the following season. If antigenic drift is indeed greater in more immune populations, preparedness for influenza pandemics (Webby & Webster 2003) may need to include vaccination strategies for the second year after a pandemic with consideration to the effect this will have on the third year after a pandemic.
The δ–θ relationship has further importance due to the discontinuity that appears at in figure 3. The stochastic nature of mutation, transmission, vaccination efficacy and population interactions may cause our system to fall on either side of this discontinuity, either yielding an unexpected amount of antigenic drift (to the left of the vertical dashed line) or preventing an epidemic entirely (to the right of the vertical dashed line). The consequences of this particular threshold property will need to be explored with a stochastic model.
With a wealth of sequence data and a high mutation rate, influenza virus ecology and evolution have a broad and important intersection with the growing field of measurably evolving populations (Drummond et al. 2003). Techniques for the accurate estimation of mutation rates could be applied to detailed, localized influenza datasets such as the one described by Schweiger et al. (2002). A precise estimate of influenza's mutation rate would be a significant step towards accurate predictions of near-term antigenic drift. Similarly, the effects of local population structure during influenza epidemics could be measured with a technique based on allelic mismatch distributions as developed by Fraser et al. (2005); this type of study may help determine whether the observed strain distributions result more from host population immunity or host population structure. These methods, along with the techniques presented in this paper, will help quantify the driving forces behind antigenic drift in influenza A.
Acknowledgements
Thanks to F. B. Christiansen, J. M. Macpherson and two anonymous reviewers for their valuable comments. Authors are supported by NIH grant GM28016 (M.F.B, M.W.F), The Royal Society (J.R.G.) and NIH grant GM607929 (V.A.).
Supplementary Material
Numerical simulations and reverse-sigmoidal cross-immunity function
References
- Anderson R.M, May R.M. Oxford Science Publications; Oxford, UK: 1991. Infectious diseases of humans: dynamics and control. [Google Scholar]
- Andreasen V. Dynamics of annual influenza A epidemics with immuno-selection. J. Math. Biol. 2003;46:504–536. doi: 10.1007/s00285-002-0186-2. 10.1007/s00285-002-0186-2 [DOI] [PubMed] [Google Scholar]
- Andreasen V, Levin S, Lin J. A model of influenza A drift evolution. Z. Angew. Math. Mech. 1996;76:421–424. [Google Scholar]
- Antia R, Regoes R.R, Koella J.C, Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. 10.1038/nature02104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F, Gog J.R, Andreasen V, Christiansen F.B. Influenza drift and epidemic size: the race between generating and escaping immunity. Theor. Popul. Biol. 2004;65:179–191. doi: 10.1016/j.tpb.2003.10.002. 10.1016/j.tpb.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Bush R.M, Fitch W.M, Bender C.A, Cox N.J. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 1999;16:1457–1465. doi: 10.1093/oxfordjournals.molbev.a026057. [DOI] [PubMed] [Google Scholar]
- Coiras M, Aguilar J, Galiano M, Carlos S, Gregory V, Lin Y, Hay A, Pérez-Breña P. Rapid molecular analysis of the haemagglutinin gene of human influenza A H3N2 viruses isolated in Spain from 1996 to 2000. Arch. Virol. 2001;146:2133–2147. doi: 10.1007/s007050170025. 10.1007/s007050170025 [DOI] [PubMed] [Google Scholar]
- Cox N.J, Subbarao K. Global epidemiology of influenza: past and present. Annu. Rev. Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. 10.1146/annurev.med.51.1.407 [DOI] [PubMed] [Google Scholar]
- Davey M.L, Reid D. Relationship of air temperature to outbreaks of influenza. Br. J. Prev. Soc. Med. 1972;26:28–32. doi: 10.1136/jech.26.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J.C, Beyer W.E.P, Palache A.M, Rimmelzwaan G.F, Osterhaus A.D.M.E. Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J. Med. Virol. 2000a;61:94–99. 10.1002/(SICI)1096-9071(200005)61:1%3C94::AID-JMV15%3E3.0.CO;2-C [PubMed] [Google Scholar]
- de Jong J.C, Rimmelzwaan G.F, Fouchier R.A.M, Osterhaus A.D.M.E. Influenza virus: a master of metamorphosis. J. Infect. 2000b;40:218–228. doi: 10.1053/jinf.2000.0652. 10.1053/jinf.2000.0652 [DOI] [PubMed] [Google Scholar]
- Drummond A.J, Pybus O.G, Rambaut A, Forsberg R, Rodrigo A.G. Measurably evolving populations. Trends Ecol. Evol. 2003;18:481–488. 10.1016/S0169-5347(03)00216-7 [Google Scholar]
- Dushoff J, Plotkin J.B, Levin S.A, Earn D.J.D. Dynamical resonance can account for seasonality of influenza epidemics. Proc. Natl Acad. Sci. USA. 2004;101:16 915–16 916. doi: 10.1073/pnas.0407293101. 10.1073/pnas.0407293101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earn D.J.D, Dushoff J, Levin S.A. Ecology and evolution of the flu. Trends Ecol. Evol. 2002;17:334–340. 10.1016/S0169-5347(02)02502-8 [Google Scholar]
- Ferguson N.M, Galvani A.P, Bush R.M. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. doi: 10.1038/nature01509. 10.1038/nature01509 [DOI] [PubMed] [Google Scholar]
- Fisher R.A. Clarendon Press; Oxford, UK: 1930. The genetical theory of natural selection. [Google Scholar]
- Fitch W.M, Bush R.M, Bender C.A, Cox N.J. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl Acad. Sci. USA. 1997;94:7712–7718. doi: 10.1073/pnas.94.15.7712. 10.1073/pnas.94.15.7712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Hanage W.P, Spratt B.G. Neutral microepidemic evolution of bacterial pathogens. Proc. Natl Acad. Sci. USA. 2005;102:1968–1973. doi: 10.1073/pnas.0406993102. 10.1073/pnas.0406993102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P.W, Murphy A.M. Naturally acquired immunity to influenza type A: a clinical and laboratory study. Med. J. Aust. 1976;2:329–333. doi: 10.5694/j.1326-5377.1976.tb130219.x. [DOI] [PubMed] [Google Scholar]
- Girvan M, Callaway D.S, Newman M.E.J, Strogatz S.H. A simple model of epidemics with pathogen mutation. Phys. Rev. E. 2002;65:031 915. doi: 10.1103/PhysRevE.65.031915. 10.1103/PhysRevE.65.031915 [DOI] [PubMed] [Google Scholar]
- Gog J.R, Grenfell B.T. Dynamics and selection of many-strain pathogens. Proc. Natl Acad. Sci. USA. 2002;99:17 209–17 214. doi: 10.1073/pnas.252512799. 10.1073/pnas.252512799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gog J.R, Rimmelzwaan G.F, Osterhaus A.D.M.E, Grenfell B.T. Population dynamics of rapid fixation in cytotoxic T lymphocye escape mutants of influenza A. Proc. Natl Acad. Sci. USA. 2003;100:11 143–11 147. doi: 10.1073/pnas.1830296100. 10.1073/pnas.1830296100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson A.W. Influenza virus antigens and ‘antigenic drift’. In: Potter C.W, editor. Influenza. Elsevier Science B.V.; Amsterdam, The Netherlands: 2002. pp. 49–86. [Google Scholar]
- Haraguchi Y, Sasaki A. Evolutionary pattern of intra-host pathogen antigenic drift: effect of cross-reactivity in immune response. Phil. Trans. R. Soc. B. 1997;352:11–20. doi: 10.1098/rstb.1997.0002. 10.1098/rstb.1997.0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A.J, Gregory V, Douglas A.R, Lin Y.P. The evolution of human influenza viruses. Phil. Trans. R. Soc. B. 2001;356:1861–1870. doi: 10.1098/rstb.2001.0999. 10.1098/rstb.2001.0999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermack W.O, McKendrick A.G. Contributions to the mathematical theory of epidemics 1. Proc. R. Soc. A. 1927;115:700–721. doi: 10.1007/BF02464423. [DOI] [PubMed] [Google Scholar]
- Kilbourne E.D. The molecular epidemiology of influenza. J. Infect. Dis. 1973;127:478–487. doi: 10.1093/infdis/127.4.478. [DOI] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The rate of molecular evolution considered from the standpoint of population genetics. Proc. Natl Acad. Sci. USA. 1969;63:1181–1188. doi: 10.1073/pnas.63.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapedes A, Farber R. The geometry of shape space: application to influenza. J. Theor. Biol. 2001;212:57–69. doi: 10.1006/jtbi.2001.2347. 10.1006/jtbi.2001.2347 [DOI] [PubMed] [Google Scholar]
- Lin J, Andreasen V, Casagrandi R, Levin S.A. Traveling waves in a model of influenza A drift. J. Theor. Biol. 2003;222:437–445. doi: 10.1016/s0022-5193(03)00056-0. [DOI] [PubMed] [Google Scholar]
- Macken C, Lu H, Goodman J, Boykin L. The value of a database in surveillance and vaccine selection. In: Osterhaus A.D.M.E, Cox N, Hampson A.W, editors. Options for the control of influenza IV. Elsevier Science; Amsterdam: 2001. pp. 103–106. [Google Scholar]
- Mills C.E, Robins J.M, Lipsitch M. Transmissibility of the 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. 10.1038/nature03063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A.W, Gubbins S, Gilligan C.A. Extinction times for closed epidemics: the effects of host spatial structure. Ecol. Lett. 2002;5:747–755. 10.1046/j.1461-0248.2002.00378.x [Google Scholar]
- Pease C.M. An evolutionary epidemiological mechanism, with applications to type A influenza. Theor. Popul. Biol. 1987;31:422–452. doi: 10.1016/0040-5809(87)90014-1. 10.1016/0040-5809(87)90014-1 [DOI] [PubMed] [Google Scholar]
- Plotkin J.B, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of the influenza A virus. Proc. Natl Acad. Sci. USA. 2003;100:7152–7157. doi: 10.1073/pnas.1132114100. 10.1073/pnas.1132114100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price G.R. Selection and covariance. Nature. 1970;227:520–521. doi: 10.1038/227520a0. 10.1038/227520a0 [DOI] [PubMed] [Google Scholar]
- Price G.R. Extension of covariance selection mathematics. Ann. Hum. Genet. Lond. 1972;35:485–490. doi: 10.1111/j.1469-1809.1957.tb01874.x. [DOI] [PubMed] [Google Scholar]
- Pyhälä R, Visakorpi R, Ikonen N, Kleemola M. Influence of antigenic drift on the intensity of influenza outbreaks: upper respiratory tract infections of military conscripts in Finland. J. Med. Virol. 2004;72:275–280. doi: 10.1002/jmv.10552. 10.1002/jmv.10552 [DOI] [PubMed] [Google Scholar]
- Sasaki A. Evolution of antigenic drift/switching: continuously evading pathogens. J. Theor. Biol. 1994;168:291–308. doi: 10.1006/jtbi.1994.1110. 10.1006/jtbi.1994.1110 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Haraguchi Y. Antigenic drift of viruses within a host: a finite site model with demographic stochasticity. J. Mol. Evol. 2000;51:245–255. doi: 10.1007/s002390010086. [DOI] [PubMed] [Google Scholar]
- Schulman J.L, Kilbourne E.D. Airborne transmission of influenza virus infection in mice. Nature. 1962;195:1129–1130. doi: 10.1038/1951129a0. [DOI] [PubMed] [Google Scholar]
- Schweiger B, Zadow I, Heckler R. Antigenic drift and variability of influenza viruses. Med. Microbiol. Immunol. 2002;191:133–138. doi: 10.1007/s00430-002-0132-3. 10.1007/s00430-002-0132-3 [DOI] [PubMed] [Google Scholar]
- Smith D.J, Lapedes A.S, de Jong J.C, Bestebroer T.M, Rimmelzwaan G.F, Osterhaus A.D.M.E, Fouchier R.A.M. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. 10.1126/science.1097211 [DOI] [PubMed] [Google Scholar]
- Webby R.J, Webster R.G. Are we ready for pandemic influenza? Science. 2003;302:1519–1522. doi: 10.1126/science.1090350. 10.1126/science.1090350 [DOI] [PubMed] [Google Scholar]
- Webster R.G, Bean W.J, Gorman O.T, Chambers T.M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Gog J.R, Grenfell B.T. Semiparametric estimation of the duration of immunity from infectious disease time series: influenza as a case-study. J. R. Stat. Soc. C. 2005;54:659–672. 10.1111/j.1467-9876.2005.05383.x [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel H.J, editors. Evolving genes and proteins. Academic Press; New York: 1965. pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Numerical simulations and reverse-sigmoidal cross-immunity function