Abstract
Cotton fibers are differentiated epidermal cells originating from the outer integuments of the ovule. To identify genes involved in cotton fiber elongation, we performed subtractive PCR using cDNA prepared from 10 days post anthesis (d.p.a.) wild-type cotton fiber as tester and cDNA from a fuzzless-lintless (fl) mutant as driver. We recovered 280 independent cDNA fragments including most of the previously published cotton fiber-related genes. cDNA macroarrays showed that 172 genes were significantly up-regulated in elongating cotton fibers as confirmed by in situ hybridization in representative cases. Twenty-nine cDNAs, including a putative vacuolar (H+)-ATPase catalytic subunit, a kinesin-like calmodulin binding protein, several arabinogalactan proteins and key enzymes involved in long chain fatty acid biosynthesis, accumulated to greater than 50-fold in 10 d.p.a. fiber cells when compared to that in 0 d.p.a. ovules. Various upstream pathways, such as auxin signal transduction, the MAPK pathway and profilin- and expansin-induced cell wall loosening, were also activated during the fast fiber elongation period. This report constitutes the first systematic analysis of genes involved in cotton fiber development. Our results suggest that a concerted mechanism involving multiple cellular pathways is responsible for cotton fiber elongation.
INTRODUCTION
Cotton is one of the most important industrial crops in the world and is the most prevalent natural fiber used in textile production. Cotton fibers, or seed hairs, consist of single cells of 30–40 mm in length and 15 µm in thickness. Fiber development consists of four overlapping stages: fiber initiation, fiber elongation, secondary cell wall deposition and maturation/dehydration (1). The fiber initiation stage occurs around the time of anthesis [from –3 to 1 day post anthesis (d.p.a.)] and is characterized by the enlargement and protrusion of epidermal cells from the ovular surface. Though all epidermal cells have the potential to become fibers, only about 30% will eventually differentiate into mature seed hairs (2). During the fiber elongation period (5–25 d.p.a.) cells demonstrate vigorous polarized expansion with peak growth rates of >2 mm/day until the fiber reaches its final dimensions (3,4). During secondary cell wall deposition (20–45 d.p.a.) cellulose biosynthesis predominates until the fiber contains ∼90% cellulose (3). The final stage of maturation/dehydration (45–50 d.p.a.) is associated with the accumulation of minerals and a simultaneous decrease in water potential, resulting in a mature cotton fiber (4). Taken together, these stages include the biomechanical processes of rapid cell division, differentiation and expansion, suggesting that a large number of genes are involved in the regulation of fiber development (5,6).
Cell expansion is crucial for plant growth and development in general, and for cotton production in particular, since the length and the fineness of the fiber determine its quality. A large amount of experimental evidence shows that flow of water into the central vacuole of an expanding cell and consequent cell enlargement are initiated by relaxation of the cell wall (7). This important process is thought to be controlled by two different mechanisms, namely tip growth and diffuse growth. In the former type of growth, post-Golgi vesicles are targeted and fused to a defined region of the plasma membrane, resulting in a unidirectional extension of both the cell membrane and cell wall. Pollen tubes and root hairs are known to rely on tip growth for their morphogenesis (8). Using maize root hairs as a model system, scientists found that local assembly of expansin-dependent F-actin meshworks was required for initiation of tip growth (9).
Most cell types in plants may expand via the diffuse growth mechanism, for which cell expansion is driven by turgor pressure and occurs diffusely throughout the entire cell surface. The primary wall of a plant cell possesses a remarkable combination of strength and pliancy to sustain the large mechanical forces that arise from cell turgor pressure, while at the same time permitting a controlled polymer extension to create space for enlargement of the protoplast (7). The pattern of wall polymer deposition and the site of polymer loosening control polarity (or direction) of diffuse growth (7,10,11). Etiolated Arabidopsis seedlings under-expressing the profilin gene displayed an overall dwarf phenotype with their hypocotyls significantly shorter than those of wild-type plants (12), indicating the importance of profilin in cell elongation. Plant cells may also secrete expansins to unlock the cross-linking of wall polysaccharides to allow the turgor-driven cell expansion during the active growth period (13).
Apart from two recent reports that implicated the involvement of β-tubulins during early fiber development (6,14), no systematic studies on the mechanism(s) of cotton fiber elongation are available in the literature. However, a large number of genes are necessary for the regulation of fiber development since many biochemical processes, such as cell division, differentiation and expansion, are involved (5,6). Here we report cloning and analysis of a set of cotton genes that are related to fiber elongation by using a fuzzless-lintless mutant as the driver for suppressive subtraction. We believe that elucidation of expression patterns for all genes involved in early fiber development is a prerequisite to our understanding of the molecular mechanisms controlling this important process and will ultimately provide novel target genes for improvement of fiber length and strength as well as other qualities of industrial applications.
MATERIALS AND METHODS
Accession numbers
All EST sequences have been submitted to the EST division of GenBank with accession numbers CB350396–CB350561 and can be accessed at the NCBI EST database (http://www.ncbi.nlm.nih.gov/dbEST/). The sequences of the following putative full-length or putative ORF-containing cDNAs were submitted to GenBank with accession numbers: cotton arabinogalactan protein (AGP), AY218846; cotton auxin-binding protein (ABP), AY189968; cotton mitogen-activated protein kinase (MAPK), AY207316; cotton profilin, AY189970; cotton expansin, AY189969; cotton xyloglucan endotransglycosylase (XET), AY189971; cotton ubiquitin, AY189972.
Plant materials
Upland cotton (Gossypium hirsutum L. cv. Xuzhou 142) and fuzzless-lintless mutant (fl) plants were field grown during the summer of 2001. The mutant was originally discovered in an upland cotton (cv. Xuzhou 142) field in China. Immediately after harvest, developing ovules were excised from each boll and fiber cells were carefully scraped from the epidermis of the ovules. All harvested plant materials were frozen in liquid nitrogen and stored at –80°C before use.
Isolation of total RNA and mRNA
For PCR-select cDNA subtraction, large amounts of total RNA were prepared from cotton fibers and fl mutant ovules by the modified hot borate method (15), using 1 g of fresh fibers and mutant ovules as starting materials. mRNA was purified from total RNA using the PolyATtract mRNA isolation system (Promega, Madison, WI) according to the manufacturer’s instructions.
PCR-select cDNA subtraction
PCR-select cDNA subtraction was employed using a kit (Clontech, Palo Alto, CA) according to the manufacturer’s protocol. The tester (samples prepared from 10 d.p.a. wild-type cotton fibers) and driver (samples from the same growth stage fl mutant ovules) cDNA were reverse transcribed from 2 µg mRNA of cotton fiber and fl mutant ovule, respectively, digested with RsaI and then ligated to different adaptors. Two rounds of hybridization and PCR amplification were performed to enrich the differentially expressed sequences. The subtracted cDNAs were inserted directly into the T/A cloning vector pGEM®-T Easy Vector (Promega) and then transformed into Escherichia coli DH5α cells, producing the resultant subtractive cDNA library.
Sequence analyses
Colonies were randomly picked from the plated subtractive cDNA library for sequence analysis (TaKaRa, Dalian, China). Fourteen groups of 50 independent clones were successively sequenced. All sequences were compared to the GenBank database using BLASTN, BLASTX and BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/). Cotton cDNAs were named according to homologous sequences in the database, and cDNAs with BLAST scores lower than 45 bits (no homologous stretch longer than 50 bp) were designated ‘unknown’. Various metabolic pathways were identified using the website of KEGG Metabolic Pathways (http://fire2.scl.genome.ad.jp/kegg/metabolism.html).
Amplification of cDNA inserts
The recovered clones were cultured in 200 µl of LB-Amp medium in 96-well plates at 37°C. The cDNA inserts were amplified by PCR in a 384-well PTC200 Peltier Thermal Cycler (MJ Research, Waltham, MA) with nested PCR primers 1 and 2R included in the PCR-select cDNA subtraction kit, which are complementary to sequences flanking the cDNA insert. PCR reactions (30 µl) contained 22.8 µl distilled water, 0.3 µl each of nested PCR primers 1 and 2R (10 µM each), 3 µl 10× ExTaq buffer (TaKaRa), 2.4 µl dNTP mix (2.5 mM each), 0.2 µl ExTaq polymerase (TaKaRa) and 1 µl bacterial culture (all reagents were added sequentially as listed). Samples were first denatured at 94°C for 5 min, followed by 30 cycles of 95°C for 30 s, 68°C for 30 s and 72°C for 40 s, with final extension at 72°C for 5 min. All PCR products were analyzed by agarose gel electrophoresis and quantified using a spectrophotometer.
cDNA macroarray preparations
DNA (0.3 µg) from each of the 280 recovered clones was printed from 384-well PCR plates onto nylon membranes (Roche, Basel, Switzerland) using the Biomek2000 Laboratory Automation Workstation (Beckman Coulter, Fullerton, CA). Each clone was printed in quadruplicate with all spots 1.125 mm in diameter and 1.25 mm apart. After air drying, membranes (with nucleic acid spots facing up) were denatured in 0.6 M NaOH for 3 min, neutralized in 0.5 M Tris–HCl (pH 7.5) for 3 min and rinsed in distilled water for 30 s. Samples were cross-linked to membranes using a low energy UV source (Stratagene, La Jolla, CA), and the membranes were stored at –20°C. Cotton ubiquitin and α-tubulin cDNAs were also printed on each membrane as internal controls. Distilled water, nested PCR primers and vector DNA were used as negative controls.
Hybridization and washing of cDNA macroarrays
RNA prepared from 0, 5, 10 and 20 d.p.a. cotton fibers were used as probes for expression pattern analysis. 33P-labeled probes were synthesized and purified using the GeneFilters® probe labeling and purification kit (ResGen, Huntsville, AL). Membranes were first prehybridized in ExpressHyb™ hybridization solution supplemented with blocking solution (Clontech) for 40–60 min at 68°C. After addition of probes to the solution, hybridization was carried out overnight at 68°C. The washing procedure was as follows: wash 1 (2× SSC, 1% SDS), 68°C, 4 × 30 min; wash 2 (0.1× SSC, 0.5% SDS), 68°C, 30 min; final wash (2× SSC), room temperature, 5 min. Membranes were then exposed to PhosphorImager screens (Amersham Biosciences, Piscataway, NJ) for 3 days.
Image acquisition and analysis
Images were acquired by scanning the membranes with a Typhoon 9210 scanner (Amersham Biosciences). Data analysis was performed using ArrayVision 6.0 software (Imaging Research, Ontario, Canada). The radioactive intensity of each spot was quantified as volume values and the levels of the local background were subtracted, resulting in the subtracted volume values designated as sVOL. Ubiquitin cDNA was used as the internal control whose subtracted volume value was termed sRef. Normalization among all images was performed by dividing sVOL of each spot by the sRef value within the same image, resulting in a normalized volume value (nVOL) for each spot. nVOL values were comparable between all images. Differential screening and expression pattern analysis data were averages of three and two independent experiments, respectively. Therefore, the expression levels for each EST in either 10 d.p.a. fiber cells or fl mutant ovules represent the average of 12 nVOL values and the levels for each EST in 0, 5, 10 and 20 d.p.a. fiber cells represent the average of 8 nVOL values.
Semi-quantitative RT–PCR analysis
First-strand cDNA was synthesized from 5 µg total RNA using the SUPERSCRIPT™ first-strand synthesis system for RT–PCR (Invitrogen, Carlsbad, CA). One-tenth of the first-strand cDNA was used as a template in 50 µl PCR reactions. Gene-specific RT–PCR primers were designed according to the cDNA sequences and synthesized commercially (TaKaRa). Parallel reactions using cotton ubiquitin primers served to normalize the amount of template added.
In situ hybridization
Paraffin-embedded sections were used for in situ hybridization following the procedure described previously (14,16).
RESULTS
Identification of cotton fiber-specific cDNA fragments by PCR-select cDNA subtraction
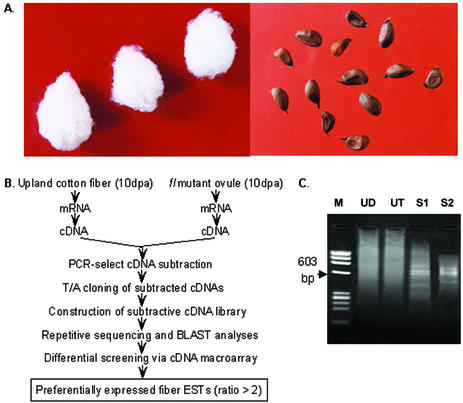
Using cDNA prepared from wild-type cotton (Fig. 1A, left) fibers as testers and that of the fl mutant (Fig. 1A, right) ovules as drivers, we performed PCR-select cDNA subtraction as outlined in Figure 1B. A pool of putative fiber-specific cDNA fragments was obtained after two rounds of subtraction. These cDNAs ranged from ∼300 bp to ∼1 kb, with most of the fragments distributed between 400 and 600 bp (Fig. 1C). Fragments from the second round of subtraction were inserted into the T/A cloning vector and the resultant subtractive library was used for sequence analysis.
Figure 1.
Phenotypes, experimental procedures for and results of PCR-select cDNA subtraction. (A) Seeds of wild-type upland cotton Xuzhou 142 (left) and fl (fuzzless-lintless) mutant (right). (B) Strategy for isolating and identifying cotton fiber development-related genes via PCR-select cDNA subtraction and differential screening. (C) Products of PCR-select cDNA subtraction. M, Φ174/HaeIII digested molecular marker; UD, unsubtracted driver cDNA; UT, unsubtracted tester cDNA; S1, PCR products obtained after one round of subtraction; S2, PCR products obtained after two rounds of subtraction.
Sequence analysis of genes in the subtractive library
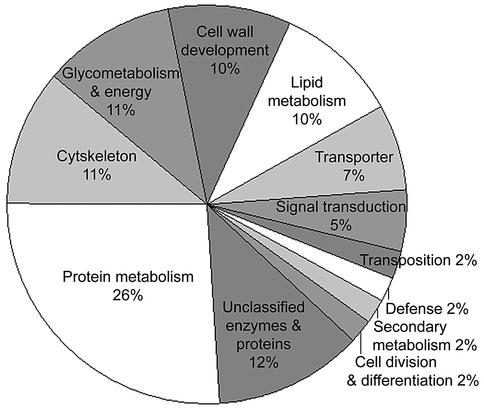
Groups of 50 independent colonies were randomly picked and sequenced. BLAST analysis showed that 189 sequences representing 133 non-redundant cDNA fragments were obtained from the first 200 clones tested (pool of groups 1–4; Table 1). As more clones were sequenced the percentage of new and independent cDNAs discovered in each group decreased sharply from 60% in groups 5 and 6 to only 9% in group 14 (Table 1). This suggests that the 280 independent fiber cDNAs obtained by repeated sequencing may constitute over 80–90% of the genes present in our subtracted library that are specifically expressed during the fiber elongation period. Database and literature searches revealed that we recovered most of the previously reported cotton fiber-related genes (data not shown), confirming the comprehensiveness of our subtractive library. The fiber-associated cDNAs were classified into 12 functional categories according to partial sequence identities to known proteins and enzymes (Fig. 2). A complete list of these cotton fiber development-related cDNA clones is available at http://cottonfiber.cbi.pku.edu.cn.
Table 1. Complete sequence analysis of the subtractive cDNA library.
| Groups | 1–4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Successful sequencesa | 189 | 40 | 42 | 43 | 43 | 42 | 45 | 44 | 43 | 41 | 43 | 615 |
| Independent new ESTs | 133 | 24 | 25 | 21 | 19 | 16 | 12 | 11 | 8 | 7 | 4 | 280 |
| Percentb | 70 | 60 | 60 | 49 | 44 | 38 | 26 | 25 | 18 | 17 | 9 |
aGroups 1–4 indicate pooled sequence information from the first four groups and groups 5–14 report data from each group separately.
bThe percentage of independent new cDNAs recovered from each column relative to the total successful sequences of the same group.
Figure 2.
Functional classification of genes included in the subtractive cDNA library. Cotton cDNAs were classified by referring to the SRB embryonic EST project (http://www.mcdb.ucla.edu/Research/Goldberg), in which ∼20 000 ESTs representing messages active 6–7 days after fertilization in embryos of scarlet runner bean were identified and were assigned to 15 different functional categories.
Macroarray analysis of fiber-specific cDNAs
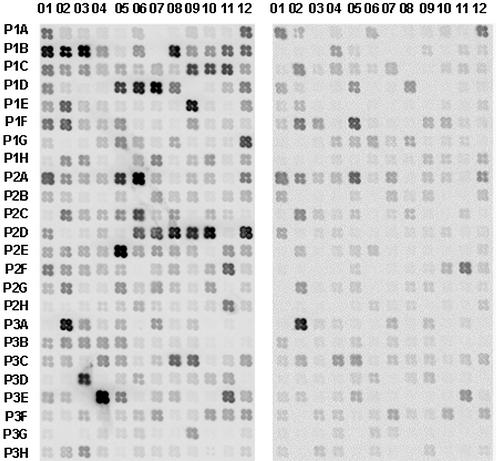
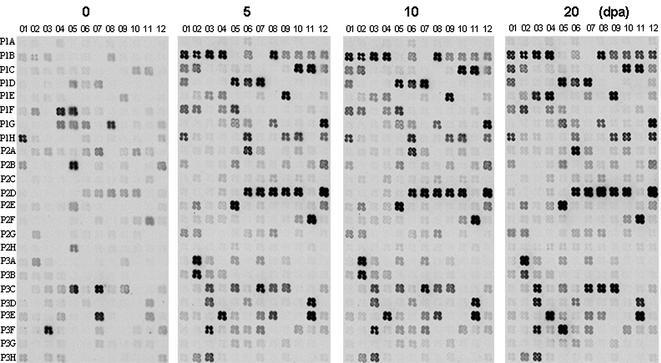
Although PCR-select cDNA subtraction is a powerful tool for identifying differentially expressed genes, subtractive products may contain some cDNAs that are common to or have similar levels in both tissue types. To avoid analysis of false-positive clones and to provide further data on relative expression level of the cloned cDNAs, we performed a further screen using a cDNA macroarray. Out of 280 cDNA fragments tested, 172 were either expressed only in cotton fibers or displayed significantly higher levels (>2-fold) in fibers compared to mutant ovules (Fig. 3).
Figure 3.
Differential screening of the 280 cDNAs by macroarray, using fiber cDNA (left) and fl ovule cDNA (right) as probes. A cotton ubiquitin (P2D01) gene was used as the positive control. Negative controls include PCR primers (P1D09), ddH2O (P1B07) and vector DNA (P1D10).
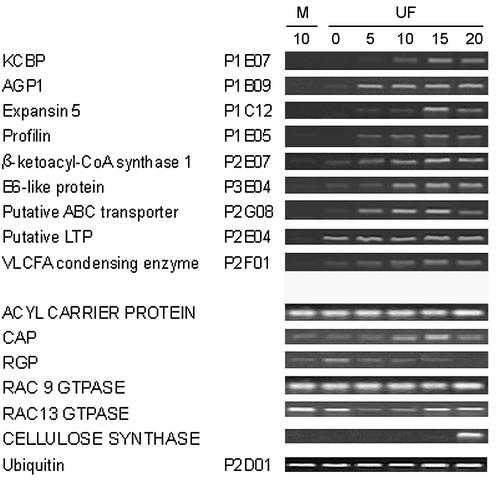
We next carried out RT–PCR to analyze the cDNAs that were found to be specifically or preferentially expressed in elongating cotton fibers (representative data shown in Fig. 4). We also tested several previously reported fiber genes that were not recovered by our subtractive library, such as the acyl carrier protein, adenyl cyclase associated protein, reversibly glycosylated polypeptide and two Rac GTPases. RT–PCR detected no significant differences between levels in the fl mutant ovule and wild-type cotton fiber (Fig. 4). Despite the fact that cellulose synthesis is central to cotton fiber development, no cellulose synthase cDNAs were recovered from our library. Therefore, RT–PCR was performed using primers specific for several known cellulose synthase genes. In samples prepared from fiber cells younger than 20 d.p.a., cellulose synthase mRNA levels were low and showed no significant difference between wild-type fiber and the mutant ovule (Fig. 4).
Figure 4.
RT–PCR analysis of cotton fiber cDNAs. Total RNA (5 µg) isolated from fl mutant ovules and from fiber cells at different developmental stages was used to synthesize first-strand cDNA for RT–PCR using gene-specific primers. A cotton ubiquitin cDNA was used to normalize the amount of templates added in PCR reactions. M, fl mutant ovule; UF, upland cotton fiber. Days post anthesis (d.p.a., days 0–20) are shown at the top. Capitalized gene names indicate previously reported fiber genes that were not recovered from our subtractive library. These genes did not show significant differences in expression between 10 d.p.a. fiber and fl mutant ovules. KCBP, kinesin-like calmodulin binding protein; VLCFA, very-long chain fatty acid; CAP, adenyl cyclase associated protein; RGP, reversibly glycosylated polypeptide.
Analysis of cDNAs preferentially expressed during the fiber elongation phase
The printed cDNA arrays were next hybridized to 33P-labeled probes prepared from poly(A)+ RNA that had been isolated from 0, 5, 10 and 20 d.p.a. wild-type cotton fibers (Fig. 5). Expression profiles (see Supplementary Material) showed that 121 (type II) of 172 fiber-preferential cDNAs reached their highest levels around 10 d.p.a. and decreased significantly thereafter. The remaining 51 cDNAs were categorized into three groups. The levels of type I genes peaked around 5 d.p.a. with a gradual decrease observed as early as 10 d.p.a. Type III genes generally reached their highest expression around 10 d.p.a., but maintained similar RNA levels from 10 to 20 d.p.a. Type IV genes, such as the cotton profilin homolog (P1E05), displayed continuously increasing expression levels from 0 to 20 d.p.a. (Fig. 5). In addition to type II genes being involved primarily in elongation, we suggest that type I genes may be important in early events, particularly in fiber initiation, while type III and IV genes may participate in primary cell wall synthesis and deposition stages.
Figure 5.
Analysis of cotton fiber preferentially expressed genes during early fiber development. Macroarrays were prepared as for Figure 3. 33P-labeled cDNA probes were prepared from RNA isolated from 0, 5, 10 and 20 d.p.a. cotton fibers.
Twenty-nine cDNAs accumulated more than 50-fold during the rapid fiber elongation period (Table 2). For example, mRNA encoding a putative vacuolar ATPase catalytic subunit (P2G01) was 996-fold higher and that of several cotton AGPs (P1B09, P1B10 and P1B12) accumulated 163- to 423-fold in 10 d.p.a. fiber cells than in 0 d.p.a. ovules. A kinesin-like calmodulin binding protein (KCBP) homolog was 330-fold and the cytochrome P450-like protein was 280-fold more abundant in 10 d.p.a. fiber cells. mRNAs encoding one putative importin, one homolog of periodontal ligament fibroblast gene, one Arabidopsis trichome-specific FIDDLEHEAD homolog, one putative RING zinc finger protein and several proline-rich proteins and heat shock proteins as well as expansins were all increased to very high levels (Table 2), indicating their potential involvement during this growth phase.
Table 2. Most significantly accumulated cDNAs during rapid fiber elongation.
| Clone No. | Annotation | Ratio (10 d.p.a./0 d.p.a.) |
|---|---|---|
| P2G01 | V-ATPase catalytic subunit | 996 |
| P1B12 | AGP 4 | 423 |
| P1B10 | AGP 2 | 400 |
| P1E07 | KCBP | 330 |
| P3H02 | Unknown | 288 |
| P3A05 | Cytochrome P450-like protein 1 | 280 |
| P3G05 | Unknown | 260 |
| P1B06 | Proline-rich protein 5 | 256 |
| P1H10 | Unknown | 253 |
| P3F05 | FbLate-2 gene | 195 |
| P2D04 | Putative importin | 186 |
| P2F03 | LCFA elongation enzyme 1 | 175 |
| P2C08 | Unknown | 174 |
| P3F06 | Homolog of periodontal ligament fibroblast gene | 171 |
| P1B09 | AGP1 | 163 |
| P2C04 | 70 kDa heat shock protein 3 | 160 |
| P3E08 | Unknown | 123 |
| P1C12 | Expansin 5 | 120 |
| P2C10 | 26S proteasome ATPase subunit | 106 |
| P1D01 | XET | 99 |
| P3B02 | Unknown | 93 |
| P2E11 | β-ketoacyl CoA synthase 4 | 89 |
| P2E07 | β-ketoacyl CoA synthase 1 | 75 |
| P1F01 | ABP | 59 |
| P1D12 | β-tubulin 8 | 59 |
| P2F02 | Arabidopsis FIDDLEHEAD homolog | 57 |
| P1H03 | Putative RING zinc finger protein | 53 |
| P2G08 | Putative ABC transporter | 52 |
| P3D03 | MAPK | 51 |
Very long chain fatty acid (VLCFA) biosynthetic enzymes are involved in elongation
In fiber cells, we found that many genes encoding enzymes involved in lipid metabolism and wax biosynthesis, such as LTP (P2D06–P2E06; Fig. 5 and Supplementary Material), aldehyde decarbonylase (P3B06), acyl transferase (P2F07), VLCFA condensing enzyme (P2F01), fatty acid elongase (P2F05 and P2F06), β-ketoacyl CoA synthase (P2E07, P2E08, P2E09 and P2E11) and long chain fatty acid (LCFA) elongation enzyme (P2F03, P2F04), were significantly activated during the elongation period. We suggest that increased activity of fatty acid condensing enzymes may facilitate membrane biosynthesis, which is likely a limiting factor during the elongation period.
Two major types of wall-loosening enzymes are up-regulated in elongating fiber cells
XET and expansins are considered major wall-loosening enzymes in different plant cells (17). Interestingly, we found that mRNAs encoding both types of enzymes accumulated to significantly higher levels in wild-type preparations (3.2- to 94-fold) than in samples taken from the fl mutant. A cotton XET homolog [P1D01; 48–80% sequence identity with known plant XETs (data not shown)] and several expansins (P1C08, P1C10, P1C11 and P1C12 with sequence identities >90% at the nucleic acid level) recovered from our library were all expressed at high levels at around 10 d.p.a. and showed decreases in 20 d.p.a. samples (Fig. 5). The mRNA levels of XET and one expansin gene (P1D01 and P1C12) increased about two orders of magnitude in 10 d.p.a. fiber cells when compared to that in 0 d.p.a. ovules (Table 2).
Some fiber-preferential cDNAs are similar to known cell expansion proteins
At the deduced amino acid level, clone P1F01 shares 68–69% sequence identity with Prunus persica ABP19 and ABP20 and clone P3D03 shares 55% sequence identity with an Arabidopsis mitogen-activated protein kinase (MAPK) gene (data not shown). Macroarray analysis showed that the cotton ABP was 59-fold and the MAPK homolog was 51-fold more abundant in 10 d.p.a. fiber cells than in 0 d.p.a. ovules, respectively (Table 2). One cotton profilin cDNA that accumulated continuously from 0 to 20 d.p.a. (P1E05; Fig. 5) shared 81–84% identity with other plant profilins accumulated 26-fold in 10 d.p.a. and 64-fold in 20 d.p.a. fiber cells (Fig. 5 and Supplementary Material).
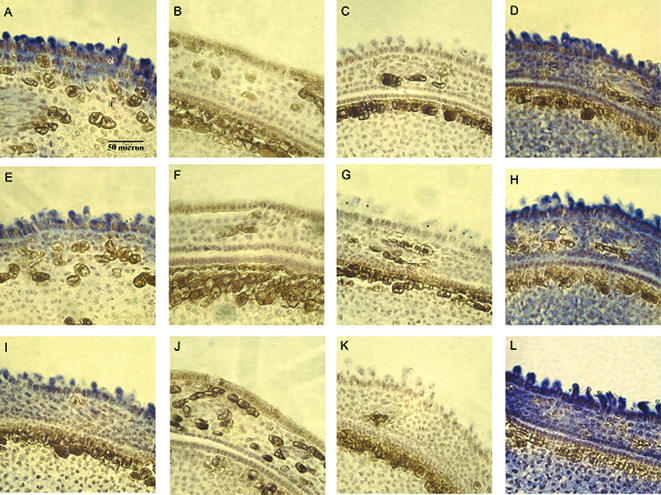
In situ determination of early fiber-specific expression of several cell expansion genes
To determine the expression of several potentially important genes at an early stage of development, we carried out in situ hybridization using thin sections prepared from 2 d.p.a. ovules of either wild-type cotton or the fl mutant. When the antisense strand of the cotton MAPK was used as a probe, strong hybridization was localized mainly in elongating fiber cells but not in non-differentiated epidermal, outer integument or inner integument cells (Fig. 6A). No hybridization was observed with either age-matched fl mutant ovules (Fig. 6B) or the sense strand of the same cDNA (negative control, Fig. 6C). Similar results were obtained when sections were probed with either the ABP19/20 homolog (Fig. 6E–G) or cotton profilin (Fig. 6I–K) cDNAs. In contrast to the fiber-specific hybridization of the three genes, the cotton ubiquitin gene (as a positive control) resulted in strong and universal hybridization in all cell types (Fig. 6D, H and L).
Figure 6.
In situ determination of early fiber-specific expression of three important genes. In situ hybridization: (A–C) a putative MAPK (P3D03); (E–G) a putative ABP (P1F01); (I–K) a profilin (P1E05). (A), (E) and (I) show antisense hybridization to cross-sections of 2 d.p.a. wild-type upland cotton. (B), (F) and (J) show antisense hybridization to 2 d.p.a. fl mutant ovules. (C), (G) and (K) show negative controls of 2 d.p.a. wild-type cotton with the sense strand of each cDNA. (D, H and L) Positive controls of 2 d.p.a. wild-type cotton with the antisense strand of a cotton ubiquitin. Blue signals represent mRNA of the tested genes. f, fiber cells; oi, outer integument; ii, inner integument.
DISCUSSION
Identification of novel fiber-specific or fiber-preferential genes
The complexity of cotton fiber development suggests that large numbers of plant genes are involved, especially during initiation, elongation and maturation (5,16). However, only about 40 such genes have been reported to date (data not shown). Therefore, we performed an in-depth search for these genes using PCR-select cDNA subtraction with cDNA from a recessive fuzzless-lintless cotton mutant (18) as the driver and wild-type cotton fibers as the tester. We selected cotton fibers and age-matched fl mutant ovules at 10 d.p.a. Fiber cells elongate most rapidly at ∼10 d.p.a. (5,14,19), suggesting that the majority of early fiber development genes, especially those involved in expansion and elongation, should be expressed at high levels at this time. A total of 280 independent cotton fiber cDNAs were identified when the subtractive library was sequenced to near completion (Table 1). When these cDNAs were printed on a macroarray membrane and screened with probes prepared from either 10 d.p.a. wild-type cotton fiber or mutant ovule, 172 (excluding possible overlapping or redundant sequences) were found to be preferentially expressed in cotton fibers with a ratio greater than 2.0 (Fig. 3). Although this value was chosen arbitrarily, genes with a differential expression ratio less than 2 often behave similarly in RT–PCR and in situ analyses (data not shown). Macroarray analysis of expression patterns indicated that most of the cDNAs recovered by our suppressive PCR peaked at ∼10 d.p.a. (Fig. 5 and Supplementary Material, type II) and a small number leveled off between 10 and 20 d.p.a. (Fig. 5 and Supplementary Material, type III). We suggest that these genes are most directly involved in fiber elongation since their accumulation precedes or correlates with increased fiber growth. Other cDNAs displayed ever-increasing expression (Fig. 5 and Supplementary Material, type IV), suggesting possible involvement in late fiber development. Several genes, such as P1A01, P1A12 and P1B04, were not fully reproduced in compatible panels of Figures 3 and 5. We think that this apparent inconsistency was probably due to the fact that the RNA used for probing genes expressed in elongating fibers (Fig. 5) came from cotton samples harvested from fully automated growth chambers. RNA used for probing hairless mutant and fibered wild-type cotton (Fig. 3) came directly from field grown cotton during the summer. Experimental evidence showed that some of these spots might be stress-induced genes that bore no relationship to fiber development (data not shown).
ABP, MAPK, KCBP and profilin may relay developmental signals for cotton fiber cell elongation
Several upstream pathways were activated during early fiber development. The mRNA encoding a KCBP homolog (P1E07) increased 330-fold while a putative auxin receptor-like protein (P1F01) homologous to Prunus persica ABP19/20 increased 59-fold, and that of a fiber-specific MAPK (P3D03) was 51-fold higher in 10 d.p.a. fiber cells than in 0 d.p.a. ovules (Table 2). Likewise, a 41-fold increase was found for a fiber-specific profilin cDNA (P1E05; Supplementary Material). In plant cells, cortical microtubule arrays are known to play a vital role in cell expansion by controlling the orientation of newly synthesized cellulose microfibrils (20). The Arabidopsis KCBP/ZWI encodes a microtubular motor protein that was shown to be involved in directed transport of secretory vesicles to regions of localized growth (21,22). MAPK was required for appressoria turgor generation and plant cuticle breaching during Magnaporthe grisea invasion of the host plant, and for root hair elongation in alfalfa (23,24). Other experiments suggested that MAPK might interact with F-actin meshworks to promote polar vesicular trafficking of materials needed for wall assembly (25). Since transgenic tobacco plants overexpressing ABP1 showed auxin-dependent cell expansion (26) and since the MAPK cascade(s) may mediate cellular responses to auxin directly by protein phosphorylation (27,28), we therefore anticipate that fiber-specific production of ABPs will facilitate cell expansion processes once bound to the plant hormone auxin. A fiber-specific ABP-like protein in concert with the up-regulation of profilin, KCBP and the MAPK pathway may provide a plausible mechanism(s) to account for cotton fiber elongation. Further experiments are necessary to verify the biochemical and physiological functions of these important signaling molecules during this process.
Vacuolar ATPase, AGPs and two major types of wall-loosening enzymes are important for fiber development
Vacuolar (H+)-ATPases may constitute nature’s most versatile proton pumps that are involved in regulation of the pH in various intracellular compartments (29). In Arabidopsis, the det3 phenotype was shown to be the result of a weak mutation in the gene encoding for V-ATPase subunit C, indicating its importance in plant cell expansion (30). AGPs are proteoglycans that have important functions in various cellular processes such as embryogenesis, cell proliferation and expansion (31–33). A fucosylated AGP was required for root hair elongation and SOS5, the Arabidopsis AGP homolog, was able to promote leaf cell expansion (32,33). The finding in our current work that mRNAs encoding for the V-ATPase catalytic subunit and for several AGP homologs accumulated to two to three orders of magnitude in fast elongating fiber cells (Table 2) seems also to indicate their involvement in regulating cell expansion. The mRNA levels for both types of the most extensively studied wall-loosening enzymes, XET and expansins, were about 100-fold higher in 10 d.p.a. fiber cells when compared to those in 0 d.p.a. ovules (Table 2), corresponding to their proposed role in cell enlargement (34–36). Expansins are required for disruption of the non-covalent bonding between cellulose and hemicellulose, thereby allowing cell wall polymers to yield to the turgor-generated growth force (17). They may accelerate wall hydrolysis by enhancing the accessibility of wall polymers to hydrolytic enzymes (37). A tomato XET was expressed abundantly in rapidly expanding regions of the etiolated hypocotyls and the Arabidopsis homolog was localized to the root elongation zone or at the root hair initiation site (34,35). Using etiolated Arabidopsis seedlings, Ma and colleagues found that XET accumulation coincided with the high rate of hypocotyl elongation (38). Fiber-specific and high level accumulation of messengers encoding different wall-loosening enzymes, V-ATPase and AGPs may provide host cells with greater extensibility.
Conclusions
PCR-select cDNA subtraction with wild-type cotton as the tester and a fiberless-lintless mutant as the driver was used to create a subtractive cDNA library containing genes associated with fiber development. A total of 280 independent cDNAs were obtained by sequencing the library to near completion and 172 fiber-specific or fiber-preferential cDNAs were identified using cDNA macroarrays. Some of the cDNA encoded upstream components related directly to cell expansion as well as several downstream pathways important for lipid biosynthesis and cell wall loosening. Isolation and analysis of genes related to cotton fiber development may help our understanding of the mechanisms responsible for cell elongation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Prof. Mei-hua Liu for her assistance with the cDNA macroarray and PhosphorImager analyses. This work was supported by a grant from the Chinese Ministry of Science and Technology (J99-A-03).
DDBJ/EMBL/GenBank accession nos+ CB350396–CB350561, AY189968–AY189972, AY207316, AY218846
REFERENCES
- 1.Basra A.S. and Malik,C.P. (1984) Development of the cotton fiber. Int. Rev. Cytol., 89, 65–113. [Google Scholar]
- 2.Tiwari S.C. and Wilkins,T.A. (1995) Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can. J. Bot., 73, 746–757. [Google Scholar]
- 3.John M.E. and Crow,L.J. (1992) Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc. Natl Acad. Sci. USA, 89, 5769–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.John M.E. and Keller,G. (1996) Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc. Natl Acad. Sci. USA, 93, 12768–12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smart L.B., Vojdani,F., Maeshima,M. and Wilkins,T.A. (1998) Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol., 116, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X.-B., Cai,L., Cheng,N.-H. and Liu,J.-W. (2002) Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol., 130, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove D.J. (2001) Wall structure and wall loosening. A look backwards and forwards. Plant Physiol., 125, 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones M.A., Shen,J.-J., Fu,Y., Li,H., Yang,Z. and Grierson,C.S. (2002) The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell, 14, 763–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baluska F., Salaj,J., Mathur,J., Braun,M., Jasper,F., Samaj,J., Chua,N.-H., Barlow,P.W. and Volkmann,D. (2000) Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol., 227, 618–632. [DOI] [PubMed] [Google Scholar]
- 10.Fu Y., Li,H. and Yang,Z. (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell, 14, 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H.-T. and Cosgrove,D.J. (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell, 14, 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran S., Christensen,H.E.M., Ishimaru,Y., Dong,C.-H., Wen,C.-M., Cleary,A.L. and Chua,N.-H. (2000) Profilin plays a role in cell elongation, cell shape maintenance and flowering in Arabidopsis. Plant Physiol., 124, 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosgrove D.J. (2000) Loosening of plant cell walls by expansins. Nature, 407, 321–326. [DOI] [PubMed] [Google Scholar]
- 14.Ji S.-J., Lu,Y.-C., Li,J., Wei,G., Liang,X. and Zhu,Y.-X. (2002) A β-tubulin-like cDNA expressed specifically in elongating cotton fibers induces longitudinal growth of fission yeast. Biochem. Biophys. Res. Commun., 296, 1245–1250. [DOI] [PubMed] [Google Scholar]
- 15.Wan C.-Y. and Wilkins,T.A. (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem., 223, 7–12. [DOI] [PubMed] [Google Scholar]
- 16.Ruan Y.-L. and Chourey,P.S. (1998) A fiberless seed mutation in cotton is associated with lack of fiber cell initiation in ovule epidermis and alterations in sucrose synthase expression and carbon partitioning in developing seeds. Plant Physiol., 118, 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpita N. and McCann,M. (2000) The cell wall. In Buchanan,R.B., Gruissem,W. and Jones,R.L. (eds), Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp. 52–108.
- 18.Zhang T. and Pan,J. (1991) Genetic analysis of a fuzzless-lintless mutant in Gossypium hirsutum L. Jiangsu. J. Agric. Sci., 7, 13–16. [Google Scholar]
- 19.Whittaker D.J. and Triplett,B.A. (1999) Gene-specific changes in alpha-tubulin transcript accumulation in developing cotton fibers. Plant Physiol., 121, 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy A.S.N. and Day,I.S. (2000) The role of the cytoskeleton and a molecular motor in trichome morphogenesis. Trends Plant Sci., 5, 503–505. [DOI] [PubMed] [Google Scholar]
- 21.Mathur J. and Chua,N.-H. (2000) Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell, 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romagnoli S., Cai,G. and Cresti,M. (2003) In vitro assays demonstrate that pollen tube organelles use kinesin-related motor proteins to move along microtubules. Plant Cell, 15, 251–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thines E., Weber,R.W.S. and Talbot,N.J. (2000) MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell, 12, 1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weston C.R., Lambright,D.G. and Davis,R.J. (2002) MAP kinase signaling specificity. Science, 296, 2345–2346. [DOI] [PubMed] [Google Scholar]
- 25.Samaj J., Ovecka,M., Hlavacka,A., Lecourieux,F., Meskiene,I., Lichtscheidl,I., Lenart,P., Salaj,J., Volkmann,D., Bogre,L. et al. (2002) Involvement of the mitogen-activated protein kinase SIMK in regulation of root hair tip growth. EMBO J., 21, 3296–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLong A., Mockaitis,K. and Christensen,S. (2002) Protein phosphorylation in the delivery of and response to auxin signals. Plant Mol. Biol., 49, 285–303. [PubMed] [Google Scholar]
- 27.Jones A.M., Im,K.-H., Savka,M.A., Wu,M.-J., DeWitt,N.G., Shillito,R. and Binns,A.N. (1998) Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science, 282, 1114–1117. [DOI] [PubMed] [Google Scholar]
- 28.Napier R.M., David,K.M. and Perrot-Rechenmann,C. (2002) A short history of auxin-binding proteins. Plant Mol. Biol., 49, 339–348. [PubMed] [Google Scholar]
- 29.Nishi T. and Forgac,M. (2002) The vacuolar (H+)-ATPases—nature’s most versatile proton pumps. Nature Rev. Mol. Cell Biol., 3, 94–103. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher K., Vafeados,D., McCarthy,M., Sze,H., Wilkins,T. and Chory,J. (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes Dev., 13, 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz C.J., Johnson,K.L., Currie,G. and Bacic,A. (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell, 12, 1751–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Hengel A.J. and Roberts,K. (2002) Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J., 32, 105–113. [DOI] [PubMed] [Google Scholar]
- 33.Shi H., Kim,Y., Guo,Y., Stevenson,B. and Zhu,J.-K. (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell, 15, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vissenberg K., Martinez-Vilchez,I.M., Verbelen,J.-P., Miller,J.G. and Fry,S.C. (2000) In vivo colocalization of xyloglucan endotransglycosylase activity and its donor substrate in the elongation zone of Arabidopsis roots. Plant Cell, 12, 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vissenberg K., Fry,S.C. and Verbelen,J.-P. (2001) Root hair initiation is coupled to a highly localized increase of xyloglucan endotransglycosylase action in Arabidopsis roots. Plant Physiol., 127, 1125–1135. [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y., Choi,D. and Kende,H. (2001) Expansins: ever-expanding numbers and functions. Curr. Opin. Plant Biol., 4, 527–532. [DOI] [PubMed] [Google Scholar]
- 37.Rose J.K.C., Lee,H.H. and Bennett,A.B. (1997) Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc. Natl Acad. Sci. USA, 94, 5955–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L., Li,J., Qu,L., Hager,J., Chen,Z., Zhao,H. and Deng,X.W. (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell, 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.