Abstract
Few biomedical subjects of study are as resource-intensive to teach as gross anatomy. Medical education stands to benefit greatly from applications which deliver virtual representations of human anatomical structures. While many applications have been created to achieve this goal, their utility to the student is limited because of a lack of interactivity or customizability by expert authors. Here we describe the first version of the Biolucida system, which allows an expert anatomist author to create knowledge-based, customized, and fully interactive scenes and lessons for students of human macroscopic anatomy. Implemented in Java and VRML, Biolucida allows the sharing of these instructional 3D environments over the internet. The system simplifies the process of authoring immersive content while preserving its flexibility and expressivity.
Keywords: Virtual Reality, Anatomy, Education, Knowledgebase
Introduction
Few subjects are as foundational to the practice of medicine as gross anatomy, yet few subjects are as resource intensive to teach. Both the availability of cadavers and the expertise required to teach the subject are prohibitive barriers which medical education must overcome.
Paper-based, electronic static illustrations, video, and interactive computer applications have attempted to relieve our dependence on expert-led, cadaver-based instruction. However, these materials suffer shortfalls in conveying the important three-dimensional spatial relationships of anatomical structures. Computer-generated 3-D animations and atlases are available to convey these relationships, but these systems are customized for specific areas, and are not easily modified by the individual anatomy instructor, who may have little or no knowledge of visualization techniques.
We have previously described the Digital Anatomist Dynamic Scene Generator [7] (DSG) that addresses some of these limitations. Using a web interface a user requests scenes showing objects and relationships encoded in an ontology of anatomy. The web application retrieves the models for these objects, constructs a scene, renders it, and sends a snapshot to the web client. However, this system is written in a non-portable language, employs a nonintuitive forms-based interface, and only outputs what we call “prosaic” scenes – scenes that are passive and do not actively demonstrate a concept. While the DSG is capable of producing virtual reality modeling language (VRML) for the viewing of a static set of objects, these scenes are of only limited utility.
VRML is a standardized declarative scene description language. The latest standard, VRML97, describes the geometries, behavior, and ambient characteristics of scene and their constituent three-dimensional objects. VRML files can be readily viewed over the web via a number of freely available browser plug-ins, or by using the open source toolkit, XJ3D.
In the present paper we describe a second generation Dynamic Scene Generator (called Biolucida) that greatly improves the previous version by employing a more modern portable architecture, and by allowing the creation of “expressive” 3-D scenes - those that contain animations and other interactive features that permit active demonstration of anatomical concepts. Unlike the DSG, which primarily outputs static 2-D snapshots of the 3-D scene over the web, and which does not separate the authoring interface from the end-user interface. Biolucida is an authoring application that outputs a VRML world that can be delivered intact as an end-user standalone lesson for inclusion in an anatomy curriculum. In addition, the architecture of this first version of Biolucida forms the basis for future work in visual access, not only to 3-D anatomical models, but also to biomedical data that can be related to structure.
Functionality
The Biolucida system allows an author to create a virtual reality based 3D scene complete with animation, object behaviors, and audio narration. While there are a number of systems which render 3D representations of anatomical structures, such as those using the visible human dataset [1], Biolucida offers four unique features: (1) it uses an ontology to organize individual model “primitives”, thereby allowing intelligent generation of scenes, (2) it has the capability of integrating models from heterogeneous specimens in cases where simple transforms will suffice to achieve proper registration, (3) it allows authors to add new models to their heterogeneous datasets, and (4) it lends authors the ability of creating truly custom scenes and interactive content where other systems may limit such material to a prepackaged library of animations authored by developers.
The author has control over various aspects of the scene, conveying a variety of new concepts to the student who subsequently views it. Once the prosaic scene is constructed, modifications of objects’ appearances (such as highlighting, or increasing transparency) serve as illustrative tools. An author may also change the position and orientation of any of the models in the scene. The alterations may express concepts such as anatomical abnormalities, pathology, or possibly even the results of surgical procedures. Moreover, the author may make these alterations, along with various camera movements, text displays, and audio narration, as part of a cohesive animation.
System Description
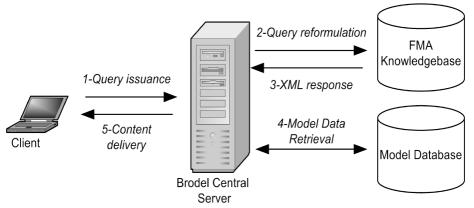
The Biolucida architecture (Fig 1) has four main components: 1) an anatomical knowledge base, 2) a 3-D model database, 3) a central server, and 4) an authoring and viewing client. All components cooperate to assemble and render a virtual scene according to an anatomist author’s design.
Figure 1.
Schematic of DSG2’s architecture and scene creation process.
1. Anatomical Knowledge Base: Foundational Model of Anatomy (FMA) [5]
The FMA is an ontology, represented in Protégé [3], that consists of over 70,000 concepts and 110,000 anatomical terms. Its 168 relationship types yield over 1.5 million relations between its concepts. The FMA is made available to other programs by means of the OQAFMA [4] server, which receives queries in the STRuQL [2] query language, and provides XML in response.
2. Three Dimensional Model Database
The 3D model database is implemented in PostgreSQL, with pointers to 3-D model files. The database and associated files store spatial information regarding the surface mesh representations of anatomical structures corresponding to leaf nodes in the FMA. Such information includes vertices and faces, along with transform and “bounding box” data. The database currently contains 167 anatomical models, representing structures of the skull and thorax. The database is meant to eventually serve as a 3D model repository, where the public can both submit and retrieve anatomical models in a variety of formats.
3. Brodel Central Server
The Brodel server is implemented in Java and manages communication between the client, the FMA, and the 3D Model Database. The Brodel server also manages the scene graph in an abstract form, by enforcing a part-of structure. It manages additions and deletions to the scene, determines appearances, and processes the abstract scene graph into format-specific content which is sent to the client to be rendered.
4. Client
The client is a Java application, which is deployed via an Applet or by Webstart. The client is responsible for both rendering the scene, and for allowing the author to manipulate the scene via a set of Java Swing control panels. From the user’s point of view, the authoring interface is composed of two distinct panels (Figs 2,3). The panel on the right serves as a canvas on which to render and interact with the scene. The panel on the left represents a display area for a variety of control modules, offering a level of interactivity and control over the scene that is best managed by two dimensional widgets and text. Selecting from the menus, a user can control a variety of modules which create animations, alter the appearance of objects in the scene, add annotations, and execute many other tasks.
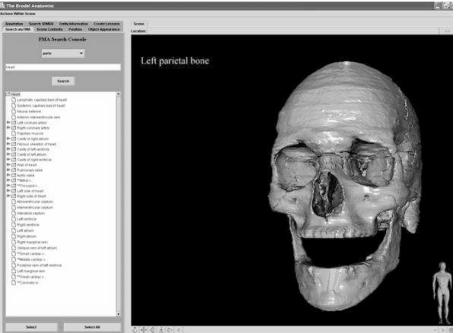
Figure 2.
Construction of a prosaic scene. The control panel on the left is used to intelligently query the FMA. The previously posed query was “all the parts of the skull”.
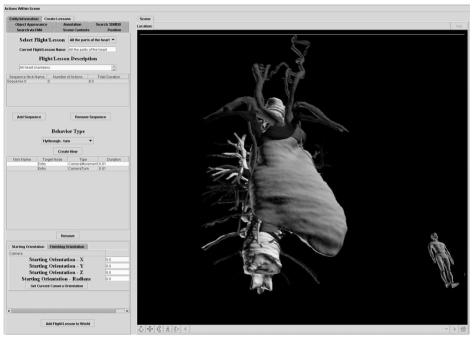
Figure 3.
User Interface. Adding animation to a prosaic scene (this scene depicts the mediastinum)
Both the server and the viewing client are implemented in Java because of its platform-independent nature, and because of the utility of the EAI (External Authoring Interface) of VRML97, which allows external Java applications to easily communicate with the VRML scene. The scene itself is rendered using the open source XJ3D library provided by Yumetech [8].
Workflow
Creating a Scene
In order to illustrate how the Biolucida application works, it is best to use an example. In this case, an anatomist is using the system to create a scene containing the basic parts of the skull (Fig 2).
First, a query is issued from the client (Fig 1), requesting the addition of “all structures which are part of the skull”. Next, the query is received by the central server, re-formulated as StruQL, and reissued to the FMA. An XML reply, denoting the anatomical structures which satisfy the query (all the parts of the skull) is subsequently processed. Structures for which the 3D model database does not contain representations are then eliminated. Models for the remaining structures are then retrieved, converted into the Brodel server’s native data structures, and placed into the abstract scene graph. The abstract scene graph is then given to a translator object, which converts its contents into format-specific syntax (i.e. VRML, X3D) fro transmission to the client. The client renders the scene for the author/user to view (fig. 2).
Adding Interactivity: Animations and Behavior
The creation of expressive (active) scenes requires more input from the user than was needed to construct the original prosaic scene. Creation of animations, the changing of transform data, and the editing of object behaviors requires an authoring system that allows real-time interaction with the scene.
As another example, the anatomist uses the interface in Fig 2 to create a scene showing the contents of the mediastinum. The anatomist then opts to make this scene expressive by authoring an lesson within the animation module (Fig. 3). This module contains a table that will be filled with sequences of actions in the new animation. The author creates a new sequence, adding to it several camera movements and turns, followed by sequential highlighting of various structures in the mediastinum. The author may add several audio voice-overs to serve as narration for the lesson which will play in parallel with the highlighting of the structures. The animation is reviewed, edited as needed, and then the entire scene, and its animations, is saved as a VRML file. This self-contained VRML file is subsequently made available to students via a web server.
Preliminary Evaluation
The Biolucida authoring system has been used by its developer to create more than 30 scenes to date, using the available models found in the 3D model database. Since each model contains a variable number of polygons, the time to create a basic prosaic scene depends on the complexity of the models chosen. A scene which includes all models found in the database takes 18 minutes for the system to assemble. Such a scene includes just over one million polygons.
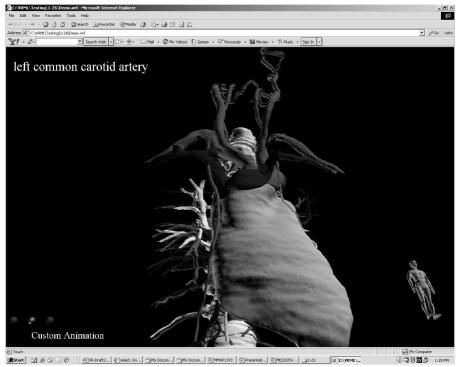
A variety of animations have been produced and tested, each of which could be constructed quickly. Fig 4 shows a screenshot of one such animation. A demonstration scene, representing a subset of Biolucida’s capabilities, can be accessed at http://sig.biostr.washington.edu/projects/biolucida. This VRML97 world is best viewed using the Blaxxun Contact 5.1 VRML plug-in (http://www.blaxxun.com).
Figure 4.
Output. Typical .wrl file viewed by a student, complete with HUD controls
As Biolucida is currently in a prototype stage, it will be necessary to compare its current functionality with user requirements solicited from anatomy education experts. Such an assessment is planned to coincide with the beta-release of Biolucida. We believe that Biolucida’s high functionality and generalized approach with regard to the design of behaviors and animations will result in refinements to its interface, but preserve basic design and functionality.
Discussion
In order to maximize the functionality and flexibility of the Biolucida system, a few novel design choices were made which are worth noting:
Content Independence through Implementation of an Abstract Scene Graph
Biolucida implements a format-independent abstract scene graph in memory. This approach allows us to simply implement one translator to which the data structures are passed to produce a particular scene description language (i.e. VRML97, X3D). Using this approach allows Biolucida to be easily adaptable to any future output formats.
The Mediator Method
The system’s simplicity is reinforced by the use of a mediator [6], which enables messaging between the many Java-based control modules and the virtual scene. Implementing a single message broker between the VRML scene and any control modules simplifies the system’s design. The mediator implementation is specific to the display technology, yet presents a standard interface to any Java modules which need to manipulate the scene. Should we choose to interact with a different rendering window, which may rely on different technology (i.e. OpenGL), a new mediator implementation will require no reworking of the system.
Leveraging 2D and 3D capabilities together
Although “a picture is worth a thousand words”, there are some functions that require the pragmatic simplicity of 2D displays. For example, querying the FMA by selecting “parts of” and typing “heart” is considerably more efficient than using a visual control to achieve the same end. Therefore, the functionalities of both 2D text-based modules and 3D visualization are combined in Biolucida to maximize its flexibility. Other developers may extend Biolucida’s functionality or to provide some type of external analytical functions by extending these 2D panel objects.
Scenes and Programmatic Prototyping
Prototyping is a familiar concept in VRML and X3D. It is the extension of the language by the declarative specification of a custom node, defining its accessible fields and its standard node implementation. This practice allows the simplification of complex nodes by abstraction. Biolucida builds on this concept by saving its scenes in an XML format similar to a prototyped node. However, the standard node implementation rests not in an external file, but in the program logic of Biolucida itself, and in the references to entries in the 3D model database.
There are strong similarities between standard prototyping, and Biolucida’s programmatic prototyping. Biolucida’s format-specific wrappers output a predefined set of nested nodes for every anatomical structure inserted into the scene. Scenes are saved in an XML format, where the small set of user-customized attributes are noted for each anatomical structure. When loaded into the authoring system, these XML structures are parsed and returned to standard form. Biolucida’s storage format method reduces file size by eliminating references to vertices and faces – the most storage intensive portion of the file. Instead, only the anatomical structures name is needed, and the model database is queried for these data. Of course, these data must be written explicitly if the file is to be made available to viewers as a standard VRML world.
Sharing Interactive Content
Custom behaviors, sequences, and animations created by authors are stored in a format that emulates our X3D programmatic prototyping approach. By accessing this content database, users can share animation sequences, referenced by unique names (i.e. “spin and flip”, “flash”) and extend them by compounding their content (i.e. “spin, flip, and flash”). This saves an author from having to “reinvent the wheel”, and allows for the creation of increasingly complex behaviors.
The sharing of custom content between authors is Biolucida’s most promising educational asset. As creating such content can be time and resource intensive, such a mechanism promises to be a strong impetus for the adoption of Biolucida as an educational tool. We hope that distributable lessons in virtual environments will have an impact on the way medical education is practiced in institutions lacking available resources, or expertise.
The Economical Storage of Declarative Animations
The declarative nature of animations in VRML97 are a highly efficient way to store long interactive lessons. The declarative syntax describing a lesson several hours in duration may require only a few thousand lines of text. Other media formats, such as AVI movies, store each graphical frame explicitly leading to storage requirements of many orders of magnitude higher than VRML.
Future Work
Immediate plans for the Biolucida system include improving its efficiency in creating scenes and augmenting the number of models found in the 3D model database. Investigations have revealed that minor refinements to the FMA query engine will result in faster scene construction. Augmenting the number of models in the 3D model database will greatly increase the utility of the Biolucida system. We will begin to search for freely available surface models of anatomical structures in the near future. With more models, a wider array of lessons can be created using the system.
There is tremendous potential to leverage the FMA knowledgebase in a variety of ways based on the feedback we receive. The inclusion of the FMA knowledgebase increases the potential functionality of Biolucida from a content authoring and presentation tool to possibly a fully functional computer-administered exam system, study aid, or an injury propagation modeling environment (utilizing VRML’s collision detection mechanisms). Biolucida’s implementation suggests that it could also be used for haptic applications, such as surgery simulation.
We will also investigate Biolucida’s uses for visualization and information access. With many freely available biomedical data sources in existence and more being created every day, interesting possibilities exist for including external data sources to create many other kinds of scenes. Scenes ranging from our current macroscopic anatomy views, to molecular and cellular scenes, can be used to visualize and access molecular, physiological, and possibly patient record data.
Acknowledgements
NIH Health Informatics Training Grant Program Number: 1 T15 LM07442 and BISTI Planning grant LM07714.
References
- 1.Ackerman MJ. The Visible Human Project: a resource for anatomical visualization. Medinfo. 1998;9(Pt 2):1030–2. [PubMed] [Google Scholar]
- 2.M. Fernandez, D. Florescu, A. Levy, D. Suciu, A Query Language for a Web-Site Management System,, SIGMOD Record, vol. 26, no. 3, pp. 4–11, 9/1997.
- 3.Gennari J, Musen MA, Fergerson RW, et al. The Evolution of Protégé: An Environment for Knowledge- Based System Development. International Journal of Human-Computer Studies. 58;2003 ;(1):89–123. http://protege.stanford.edu. [Google Scholar]
- 4.Mork P, Brinkley JF, Rosse C. OQAFMA Querying Agent for the Foundational Model of Anatomy: a Prototype for Providing Flexible and Efficient Access to Large Semantic Networks. Journal of Biomedical Informatics. 2003;36:501–517. doi: 10.1016/j.jbi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Rosse C, Mejino JL, Modayur BR, Jakobovits RM, Hinshaw KP, Brinkley JF. Motivation and Organizational Principles for Anatomical Knowledge Representation: The Digital Anatomist Symbolic Knowledge Base. Journal of the American Medical Informatics Association. 1998;5(1):17–40. doi: 10.1136/jamia.1998.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan, K.J., Mediators: Easing the Design and Evolution of Integrated Systems, Ph.D. Dissertation, University of Washington Department of Computer Science and Engineering, Technical Report UW-CSE-94-08-01, August, 1994
- 7.Wong, B. A. and Rosse, C. and Brinkley, J. F. (1999) Semi-Automatic Scene Generation Using the Digital Anatomist Foundational Model. In Proceedings, American Medical Informatics Association Fall Symposium, pages 637–641 [PMC free article] [PubMed]
- 8.XJ3D API. http://www.xj3d.org/javadoc/index.html