Abstract
In DNA-dependent RNA polymerases, reactions of RNA synthesis and degradation are performed by the same active center (in contrast to DNA polymerases in which they are separate). We propose a unified catalytic mechanism for multisubunit RNA polymerases based on the analysis of its 3′–5′ exonuclease reaction in the context of crystal structure. The active center involves a symmetrical pair of Mg2+ ions that switch roles in synthesis and degradation. One ion is retained permanently and the other is recruited ad hoc for each act of catalysis. The weakly bound Mg2+ is stabilized in the active center in different modes depending on the type of reaction: during synthesis by the β,γ-phosphates of the incoming substrate; and during hydrolysis by the phosphates of a non-base-paired nucleoside triphosphate. The latter mode defines a transient, non-specific nucleoside triphosphate-binding site adjacent to the active center, which may serve as a gateway for polymerization of substrates.
Keywords: catalytic magnesium/exonuclease/RNA polymerase/stimulating NTP/two-metal mechanism
Introduction
The multisubunit cellular DNA-dependent RNA polymerase (RNAP) is a molecular machine that carries out the fundamental reaction of DNA transcription and serves as the focus for genetic regulation. The machine advances in elementary steps, in which the catalytic reaction in the active center is coupled to the translocation of the RNAP protein relative to DNA/RNA in the ternary elongation complex (TEC). Thus, a structure–functional description of the catalytic reaction is a prerequisite for understanding how the transcription complex moves and how it is regulated.
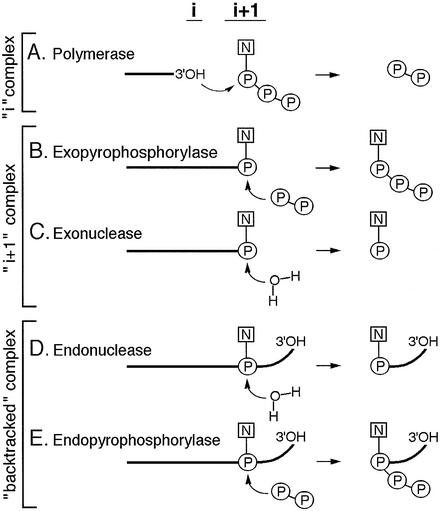
The principal enzymatic activity of RNAP is the transfer of a nucleotidyl moiety from the NTP substrate to the 3′-hydroxyl on the RNA terminus (Figure 1A). Thus, the RNAP active center should a priori include the binding sites for the incoming NTP (the ‘i + 1’ site) and for the RNA terminus (the ‘i’ site). The resultant formation of the phoshodiester bond is followed by translocation of the newly formed terminus from the ‘i + 1’ to the ‘i’ site. The reaction and the movement are reversible: pyrophosphate stimulates processive degradation of RNA with the release of 3′-terminal NTPs (Rozovskaya et al., 1984). Prior to pyrophosphorolysis, the 3′-terminus should go back into the ‘i + 1’ site (Figure 1B). Thus, at any point during elongation, the TEC exists in equilibrium between the ‘i’ and ‘i + 1’ complexes (Figure 1A and B), leading to the forward and reverse reactions, respectively.
Fig. 1. Enzymatic activities of RNAP. Schematic representation of reactions performed by RNAP in various types of TEC. ‘i’ and ‘i + 1’ denote the two sites of the active center. Bold lines represent RNA. Bent arrows show the direction of nucleophilic attack.
The RNAP active center can also perform hydrolysis of the phosphodiester bond. In the ‘backtracked’ complex (Figure 1D), RNA is threaded through the active center yielding a protruding 3′ terminus (Komissarova and Kashlev, 1997a,b; Nudler et al., 1997). The 3′ fragment can be removed by an intrinsic endonuclease activity (Orlova et al., 1995), which is stimulated by the prokaryotic GreA and GreB proteins (Borukhov et al., 1993) and by eukaryotic SII factor (Reines, 1992; Izban and Luse, 1993). Pyrophosphate can also stimulate RNA cleavage in backtracked complexes of eukaryotic Pol II, releasing an RNA fragment with 5′-triphosphate (Rudd et al., 1994) (Figure 1E).
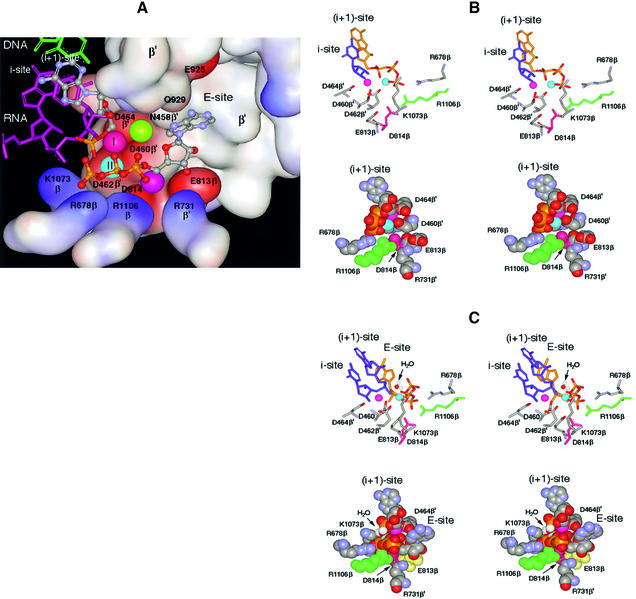
Elucidation of the RNAP X-ray crystal structure rendered a three-dimensional view of the active center (Figure 2A) (Zhang et al., 1999; Korzheva et al., 2000; Cramer et al., 2001; Gnatt et al., 2001; Vassylyev et al., 2002). Its principal component is a patch of three evolutionarily invariant aspartate residues, D460β′, D462β′ and D464β′, of the β′-subunit (hereafter, the amino acid nomenclature used is that of Escherichia coli), which coordinate a tightly bound Mg2+ ion involved in catalysis (Zaychikov et al., 1996). This Mg2+ is referred to hereafter as Mg-I (represented by a red ball in Figure 2A). Mg-I is suspended in the aperture joining two principal channels in the RNAP molecule: the main channel accommodating DNA and RNA (upper left in Figure 2A) and the secondary channel for the entry of NTP substrates (center right in Figure 2A), which also serves to accommodate the protruding 3′ end of RNA during backtracking (Epshtein et al., 2002).
Fig. 2. Molecular model of the RNAP active center. The model combines prior knowledge and present results, as explained in the text. (A) The active center in surface rendition performing the 3′–5′ exonuclease reaction. Basic and acidic amino acid residues are represented by the blue and red surfaces, respectively. In the top left of the panel, the 3′-terminal nucleoside (gray–blue metallic) in the i + 1 site is paired with the DNA base (green). The penultimate RNA nucleotide in the i site is depicted in purple. Of the four phosphates shown (yellow–red), one belongs to the 3′-terminal NMP in the i + 1 site and three belong to the non-complementary NTP in the E-site. Balls marked I and II represent catalytic Mg-I and Mg-II. Unmarked balls indicate the positions of Mg2+ ions from the published structures [green from Vassylyev et al. (2002); purple from Cramer et al. (2001)]. The water molecule participating in the exonuclease reaction is omitted. (B) Stereoscopic rendition (for crossed eyes) of the active center performing synthesis. Crucial amino acid residues are indicated. The purple and blue balls represent Mg-I and Mg-II, respectively. Top panel: the 3′-terminal nucleotide in the i site is shown in blue and the incoming NTP in the i + 1 site in yellow; its phosphates coordinate Mg-II. Bottom panel: space-filled model of the same reaction from a different angle. The 3′-terminal nucleotide is not shown; the incoming NTP in the i + 1 site is shown. (C) Stereo view of the active center performing the 3′–5 exonuclease reaction from two angles. The arrow points to the water molecule that serves as acceptor for nucleotidyl transfer. Top panel: the two 3′-terminal nucleotides (blue) are shown occupying the i and i + 1 sites, while the yellow NTP in the E-site coordinates Mg-II. Bottom panel: space-filled model from a different angle. Only the 3′-terminal nucleotide is shown in the i + 1 site.
In addition to the three above-mentioned aspartates, the aperture is built up of several β-subunit residues (bottom in Figure 2A), including two acidic residues (E813β and D814β) and a basic cluster (K1073β, R678β, R1106β and R731β′), and the ‘wall structure’ of the β′-subunit (center top in Figure 2A).
In addition to Mg-I, a second Mg2+ ion has been postulated to take part in the active center of large multisubunit RNA polymerases, by analogy with small single-subunit enzymes (Steitz, 1998). However, the three reported crystal structures of RNAP provide conflicting data about the second catalytic Mg2+: in the bacterial core RNAP structure, it is not seen at all (Zhang et al., 1999) while in the yeast Pol I structure (Cramer et al., 2001) and in the RNAP holoenzyme from Thermus thermophilus (Vassylyev et al., 2002), it has been positioned in different locations (represented by unmarked purple and green balls in Figure 2A, respectively).
In this investigation, we took advantage of a fortuitous Mg2+-dependent 3′–5′ exonuclease activity in RNAP of E.coli (Figure 1C) to explore the structure–functional aspects of the two Mg2+ ions in the active center. The results, in combination with molecular modeling, lead to unequivocal positioning of the second catalytic Mg2+ (hereafter referred to as Mg-II, represented by the blue ball in Figure 2A) and reveal a functional architecture of the active center that explains all of the RNAP reactions depicted in Figure 1 by a single unified mechanism.
Results
Non-complementary NTPs stimulate exonuclease in the TEC
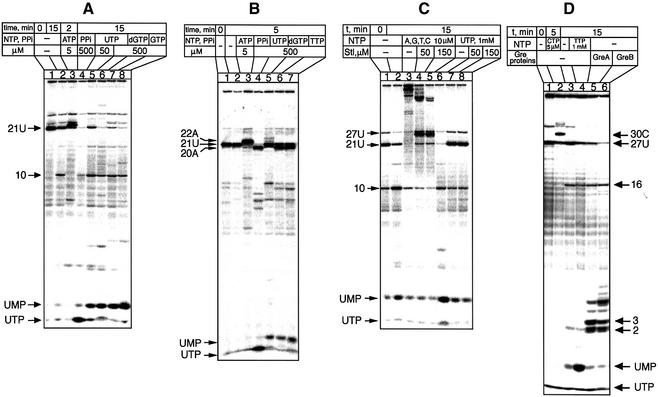
As the experimental system to study degradation of RNA in the TEC, we used defined complexes obtained by RNAP ‘walking’ on the T7A1 promoter fragment, which were stalled due to the lack of the next nucleotide. The complexes are designated by the length of RNA and by the terminal nucleotide, which is labeled with 32P; e.g. TEC-21U carries the 21mer RNA transcript with a radiolabeled UMP residue at the 3′ terminus (Figure 3A, lane 1). Incubation of TEC-21U without nucleotides yielded three principal slowly accumulating products: UMP, UTP and the 10mer 3′-terminal oligonucleotide (lane 2). The two latter products could be explained by pyrophosphorolysis caused by traces of PPi and by internal cleavage in backtracked TEC, respectively. The generation of UMP, which was the main focus of this investigation, apparently was caused by hydrolysis of the 3′-terminal nucleotide.
Fig. 3. Characterization of the 3′–5′ exonuclease of RNAP. Autoradiograms show the effect of incubation of defined TECs carrying radiolabeled transcripts (lanes 1) under the indicated conditions. The RNA fragments are identified by their length (numbers) and 3′-terminal nucleotide (letters). The starting complexes in (A), (C) and (D) carry a 32P label only in the 3′-terminal nucleotide, while in (B) RNA was labeled both at the 3′-terminus and internally.
The origin of the three degradation products was confirmed by control experiments (Figure 3A, lanes 3 and 4). The addition of ATP, which is the 22nd nucleotide in the sequence, resulted in the extension of 21U to 22A and suppression of all three products of degradation (lane 3), whereas pyrophosphate converted all radioactivity to UTP through pyrophosphorolysis (lane 4). A crucial observation of this investigation is the stimulation of UMP production by non-complementary nucleotides, UTP (lanes 5 and 6), dGTP (lane 7) or GTP (lane 8). The same effect was observed with all non-complementary ribo- and deoxyribo-NTPs, and with the triphosphate derivative of 4-methylumbelliferone, but not with mono- or diphosphates (data not shown). The dependence of the cleavage rate on the concentration of stimulatory NTP gives a value of 0.8 ± 0.1 mM for the apparent binding constant. The release of the terminal NMP was not nucleotide specific; it was observed in TEC-21U, TEC-27U, TEC-12C and TEC-14C (data not shown).
Nonetheless, the exonuclease reaction displayed some sequence specificity: it was not observed in TEC-11A and TEC-20A. These two complexes were also not susceptible to pyrophosphorolysis, as is illustrated for TEC-20A in Figure 3B. It can be seen that the starting transcript 21U (lane 1), which, in this case, was radiolabeled both at the 3′ teminus and internally, was readily degradable to 20A, which was resistant to further degradation induced by either pyrophosphate (lane 4) or non-complementary nucleotides (lanes 6 and 7). Such a correlation between hydrolysis and pyrophosphorolysis suggests that hydrolysis occurs from the i + 1 site.
It should be noted that a non-complementary NTP that is identical to the 3′-terminal nucleotide, e.g. UTP in the case of TEC-21U (Figure 3A, lane 5), has a different effect from that of a non-complementary NTP that is not the one at the terminus, e.g. dGTP (lane 6). dGTP stimulated conversion of 21U to 20A (lane 6), whereas UTP only generates UMP without the disappearance of the 21U band. This apparently reflects the exchange of the terminal nucleotide whereby UTP first effects the removal of UMP, and then is incorporated in its place. Indeed, UTP caused the 21U band to disappear when it was 32P-radiolabeled terminally (Figure 3A, lane 6) but not when the radioactive 32P was present both at the 3′ terminus and in the body of RNA (Figure 3B, lane 5).
We next tested the response of the exonuclease activity to streptolydigin (Stl), which blocks elongation by interfering with the functioning of the RNAP active center (McClure, 1980). Controls in Figure 3C demonstrate that the transcript in TEC-21U (lanes 1 and 2) can be chased to the run-off product by adding the four NTPs (lane 3) or degraded to UMP upon addition of non-complementary UTP (lane 6). Increasing concentrations of Stl inhibited both reactions (lanes 4 and 5, and 7 and 8, respectively), suggesting that they are mechanistically related.
The exonuclease reaction was not due to the action of Gre proteins. This is evident from Figure 3D, in which the starting material was TEC-27U labeled with 32P at the 3′-terminal UMP (lane 1). The next three nucleotides in the sequence are C, so the addition of CTP led to the product extension to 30C (lane 2). The addition of GreA (lane 5) and GreB (lane 6) stimulated extensive degradation of 27U through internal cleavage, generating predominantly di-, tri- and longer 3′-terminal fragments, whereas UMP was only a minor product. Under the same conditions, the non-complementary nucleotide TTP stimulated predominantly 3′-terminal UMP production (lane 4).
Non-complementary NTP recruits Me2+ for the exonuclease
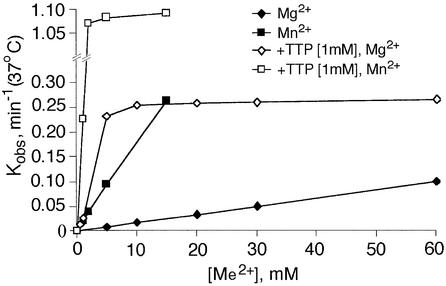
The exonuclease activity requires divalent metal ions. Figure 4 shows the dependence of the initial rate of UMP release from TEC-21U on the concentration of Mg2+ (diamonds) or Mn2+ (squares) in the absence (closed symbols) or presence of an excess of TTP. The apparent dissociation constant of Mg2+ without TTP cannot be derived reliably from the concentration dependence, which remains linear in the range 0–100 mM; it is certainly higher than 100 mM (Table I). Mn2+ stimulates the exonuclease much more efficiently than Mg2+, but the dissociation constant also could not be determined. These values are at least three orders of magnitude higher than the respective constants for Mg-I; the principal Me2+ ion coordinated in the RNAP active center (Koren and Mildvan, 1977; Mustaev et al., 1997). Thus, the stimulation was due to Mg-II—the second, weakly bound Me2+ ion rather than the principal strongly bound Mg-I.
Fig. 4. Dependence of the 3′–5′ exonuclease reaction on Me2+ and TTP. The curves show the initial rate of UMP cleavage from 3′-terminally labeled TEC-21U as a function of Mg2+ or Mn2+ concentration in the presence or absence of 1 mM dTTP.
Table I. Influence of mutations, pH and TTP on the apparent RNAP–Mg-II binding constants (mM).
| RNAP | pH 7.9 |
pH 10 |
|
|---|---|---|---|
| –TTP | +TTP, 1 mM | –TTP | |
| Wild type | >100 | 2–3 | 50–60 |
| ED813,814AA | >100 | 2–3 | >100 |
| R1106A | ∼10 | ∼10 | ∼10 |
When TTP was added (Figure 4, open symbols), the affinity for Me2+ rose dramatically, with apparent association constants increasing at least 30-fold (Table I). In other words, the stimulation of the exonuclease by non-complementary NTP consists of the recruitment of Me2+, apparently through coordination involving β,γ-phosphates, since only triphosphates were effective (see previous section).
A mutation in the active center enhances exonuclease activity
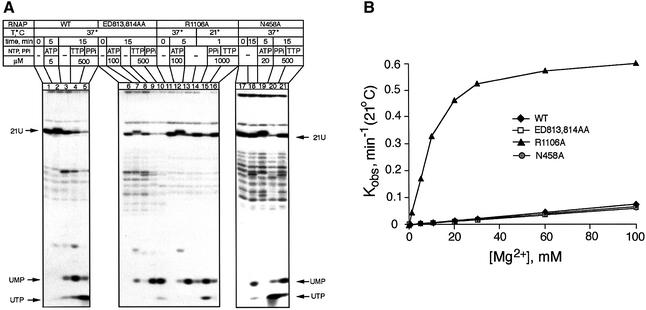
Our initial strategy for a mutational study of the exonuclease activity was based on the report by Cramer et al. (2001) that in the X-ray crystal structure of Pol II, the presumed second catalytic Mg2+ ion is held by two invariant residues, E813β and D814β (purple ball in Figure 2A). In the yeast crystal structure, D814β forms a salt bridge with R1106β. Another study (Vassylyev et al., 2002) revealed the second Mg2+ ion held by D460β′, D464β′ and N458β′. Earlier it was shown that E813K substitution in E.coli, as well as E813A, D814A or a double mutant in the archaeon Methanococcus janaschii, inhibited the polymerase activity of RNAP (Lee et al., 1991; Werner et al., 2002), which pointed to their possible role in Mg-II holding. To investigate the issue further, we engineered a double alanine substitution of E813β, D814β (designated ED/AA) as well as the substitution mutations R1106A and N458A. To study their biochemical effects, recombinant RNAP was reconstituted from individually expressed subunits (Borukhov and Goldfarb, 1993). All mutant polymerases exhibited significantly decreased elongation rates, which could be partially compensated by elevated concentrations of NTP (not shown). However, contrary to the initial expectation, the results demonstrated that the double substitution ED/AA as well as N458A did not affect Mg2+-mediated stimulation, whereas R1106A displayed a surprising effect of enhancing the exonuclease activity. These results are presented in Figure 5.
Fig. 5. Effect of mutations on the 3′–5′ exonuclease activity. (A) Autoradiograms show degradation of the terminally labeled transcript in TEC-21U by the wild-type and mutant RNAPs. (B) Dependence of the rate of UMP cleavage on Mg2+ concentration.
Lanes 1–5 of Figure 5A represent the wild-type control demonstrating extension (lane 3), pyrophosphorolysis (lane 5) and hydrolysis (lane 4) of the 3′ terminus in TEC-21U.
The double substitution ED/AA (Figure 5A, lanes 6–10) did not change the qualitative cleavage pattern (lanes 8 and 9), nor did it affect the rate of cleavage (Figure 5B). Surprisingly, the mutation changed the effect of pyrophosphate, whose addition stimulated the production of UMP through hydrolysis rather than of UTP through pyrophosphorolysis (compare lanes 5 and 10 in Figure 5A). The stimulation of the exonuclease activity in the ED/AA polymerase by TTP was the same as for wild-type enzyme (compare lanes 4 and 9; see Table I). Moreover, the absolute TTP-dependent cleavage rate in the mutant was 2–3 times greater (data not shown), which can be accounted for by improved binding of the stimulatory NTP (0.35 ± 0.1 mM). Clearly, the double substitution ED/AA does not inhibit the exonuclease reaction.
The substitution N458A (Figure 5A, lanes 17–21) did not affect the rate of cleavage (Figure 5B) or response to pyrophosphate (lane 20). However, the stimulatory effect of NTP on the cleavage was substantially reduced (lane 21).
The R1106A substitution (Figure 5A, lanes 11–16) displayed a significant increase in the intrinsic exonuclease activity in the absence of TTP, as is evidenced by the prominent UMP spot in lane 13 and the disappearance of the radioactive label from the original 21-U band. This effect is illustrated further in Figure 5B, which shows the dependence of the initial rate of exonuclease reaction on the concentration of Mg2+. Non-complementary TTP did not accelerate the rate of cleavage further (Figure 5A, lane 16). In other words, the R1106A mutation apparently made the 3′-exonuclease activity independent of TTP. Similarly to ED/AA, the R1106A mutation changed the response of TEC to pyrophosphate: instead of causing pyrophosphorolysis (production of UTP), it stimulated the exonuclease reaction (lane 15).
In conclusion, these experiments indicate that E813β, D814β and N458β′ are not involved in Mg-II binding, whereas the removal of the R1106β residue appears to improve the binding of the stimulatory Mg2+ (Table I).
Two modes of pyrophosphate action
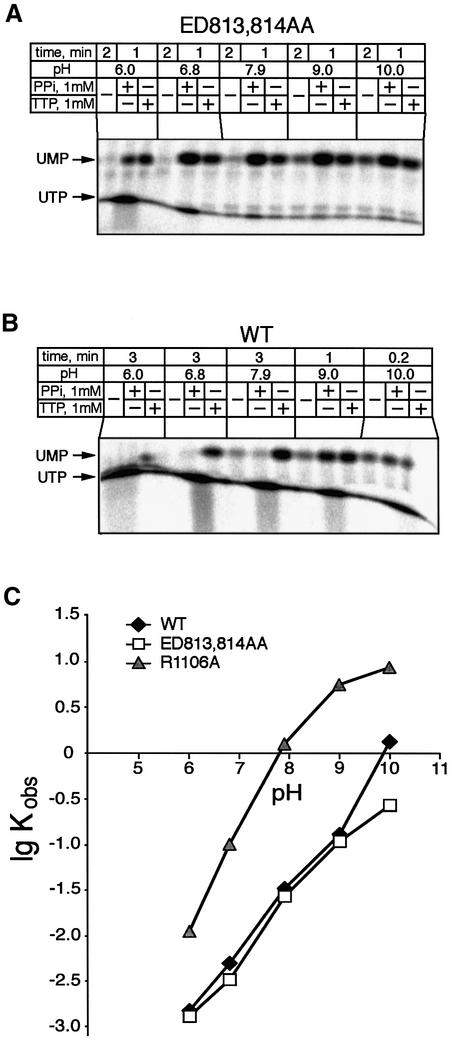
From the above experiments, it appears that pyrophosphorolysis and 3′-terminal hydrolysis are two alternative effects of PPi, discernible by the mutations ED/AA and R1106A. This phenomenon was investigated further in the experiment of Figure 6, in which the effect of PPi was determined for the ED/AA (Figure 6A) and wild-type RNAP (Figure 6B) over a range of pH values.
Fig. 6. Effect of pH on the 3′–5′ exonuclease and the pyrophosphorylase activities. (A and B) Autoradiograms show the release of terminal nucleotide from TEC-21U by the ED/AA mutant and wild-type RNAPs. (C) Effect of pH on the initial rate of terminal UMP release by the wild-type and mutant RNAPs at 10 mM MgCl2.
As can be seen, in the ED/AA TEC (Figure 6A) at pH 6, the PPi-dependent pyrophosphorolysis (UTP production) proceeded much faster than exonuclease cleavage, while at pH >7, PPi induced exclusively 3′-terminal hydrolysis. In the case of wild-type RNAP (Figure 6B), PPi also stimulated significant 3′-terminal hydrolysis, but only in the pH range above 9. The same effect was observed for N458A substitution (data not shown). Thus, the two reactions caused by PPi are intrinsic to the wild-type enzyme, and appear to be competitive. Alkaline pH appears to favor the hydrolytic effect of PPi over pyrophosphorolysis, and the mutations enhance this effect. The simplest interpretation of these results is to assume that PPi can bind in the active center in two alternative modes, depending on the microenvironment affected by the pH or minor structural distortions caused by mutations.
Stimulation of exonuclease by alkaline pH
The notion of transitions in the active center affected by pH has found support in the study of the effect of pH on the Mg2+-dependent exonuclease (Figure 6C). It should be noted that elevation of pH should be expected to stimulate hydrolysis unspecifically, due to increased deproton ation of the active water molecule. Normally, such stimulation should plateau out as the hydrogen ion concentration approaches the system’s pK value. However, in the case of wild-type (as well as for N458A) RNAP, the pH dependence curve displayed an abrupt upturn in the range of 9–10 (diamonds), reflecting an additional effect. The upturn was not seen in the case of the ED/AA double substitution (squares) and in the R1106A mutant (triangles), even though the latter performed hydrolysis at a much higher rate. This allele-specific upsurge could reflect a transition in the wild-type protein structure at pH 9–10 leading to better retention of Mg2+.
This interpretation is consistent with the observation that the apparent dissociation constant of the stimulatory Mg2+ for the wild-type RNAP decreased in the pH interval 9–10, whereas in the case of R1106A and ED/AA mutants, it remained at the constant level of ∼10 mM and >100 mM, respectively (Table I).
Modeling of the active center
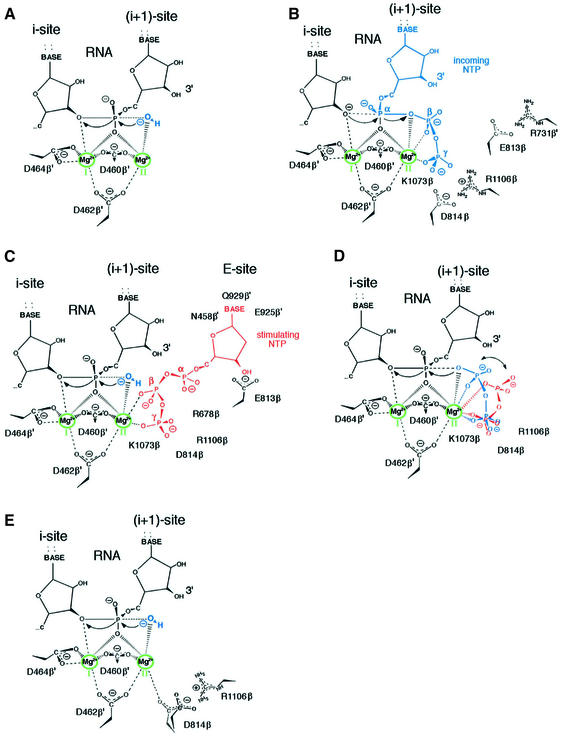
To explain the above results, we built a working model of the RNAP active center that was based on the following premises: (i) the polymerase and the nuclease (hydrolysis or pyrophosphorolysis) reactions are performed by the same active site; (ii) there are two Mg2+ ions involved— one of which is bound strongly (Mg-I) and the other weakly (Mg-II); (iii) in the polymerase reaction, Mg-II is stabilized additionally by the β,γ-phosphates of the incoming NTP; (iv) both reactions are based on the substitution nucleophilic bimolecular (Sn2) mechanism (Yee et al., 1979) operating in opposite directions; and (v) the reactions’ geometry requires that the two Mg2+ ions be situated at equal distances from the non-bridging oxygen of the scissile phosphate collinearly to the axis of the attack by an activated water molecule.
As noted in the Introduction, only Mg-I has been detected unequivocally in the catalytic center of RNAP on the basis of X-ray crystal structure. Thus, the modeling, in essence, consisted of fitting of Mg-II into the structure of the catalytic center so that it would participate in both the nuclease and the polymerase reactions in accord with the above-listed requirements.
The molecular coordinates for the modeling were those of the yeast RNAP ternary complex, in which the 3′ terminus of RNA is positioned in the i + 1 site (Gnatt et al., 2001). The yeast structure is preferable to the bacterial RNAP structure (Zhang et al., 1999) because the latter reflects the non-productive conformation of the active center (Epshtein et al., 2002). The coordinates were combined with the parameters of the nuclease reaction as it occurs in the exonuclease center of the Klenow fragment of DNA polymerase I (Brautigam and Steitz, 1998). As required by the reaction geometry, Mg-I and Mg-II were placed in the model parallel to the line of attack by the activated water, symmetrically with regard to the non-bridging oxygen (Figure 7A). The position of Mg-I is from the X-ray crystal structure. The metal is coordinated by the three conserved aspartates from the NADFDGD motif of the β′-subunit (D460, D462 and D464). The position of Mg-II is unequivocal. Remarkably, it fits into the structure so that Mg-II is coordinated by two of the three aspartates of the NADFDGD motif (D460 and D462); this explains why it is bound more weakly than Mg-I (Figure 7A). To allow for Mg-II to be stabilized by the phosphate of the incoming NTP, the γ-phosphate should fit into the positively charged pocket formed by the evolutionarily invariant side chains of K1073, R678 and R1106 of the β-subunit (Figures 7B, and 2A and B).
Fig. 7. Modes of the action of RNAP active center. The schemes present the results of molecular modeling and proposed reaction mechanisms when coordination of Mg-II is effected in different modes. (A) The 3′–5′ exonuclease in the absence of additional stabilization of Mg-II. (B) The polymerase with Mg-II stabilized by the incoming NTP (shown in blue). (C) The 3′–5′ exonuclease with Mg-II stabilized by non-complementary NTP (red). (D) The pyrophosphorylase with stabilizing pyrophosphate bound in two alternative modes (red or blue). (E) The 3′–5′ exonuclease with Mg-II stabilized by D814 residue shift resulting from disrupted interaction with R1106.
The model answers the bidirectionality requirement. In the nuclease reaction, Mg-I ligates the leaving group, facilitating the bond breakage and leaving of the 3′-oxyanion by neutralizing the developing negative charge on the oxygen (Figure 7A). Both Mg-I and Mg-II ligate the non-bridging oxygen to stabilize the pentacovalent transition state. The attacking water molecule is activated by Mg-II. In the polymerase reaction, the roles of Mg-I and Mg-II are reversed (Figure 7B). The attacking 3-hydroxyl is coordinated by Mg-I, whereas Mg-II ligates the leaving group, facilitating the breakage of the bond between the α- and the β-phosphates of the NTP substrate.
A binding site for non-base-paired NTP
Our results attribute stimulation of the exonuclease reaction by non-complementary NTPs to stabilization of Mg-II in the active center. To exert this effect, the stimulatory NTP should bind in the bottom of the substrate pore with its β,γ-phosphates facing Mg-II (site E in Figures 7C, 2A and C). In fitting the stimulatory NTP into the structure, the requirements were to ensure coordination of Mg-II by the β,γ-phosphates while leaving space for the activated water molecule. These structural requirements uniquely define the position of the β,γ-phosphates.
It should be emphasized that the γ-phosphate of the stimulatory non-base-paired NTP in the nuclease reaction (Figure 7C) and of the substrate NTP in the polymerase reaction (Figure 7B) occupy nearly identical positions and are engaged in the same set of ionic interactions, with residues K1073, R678 and R1106. However, the positions of the β-phosphate in these two situations are substantially different, i.e. the stimulatory NTP allows an exogenous water molecule to take the attacking position, whereas in the case of the NTP substrate this position is taken by the oxygen between the α- and β-phosphates. The notion of two modes of binding of β,γ-phosphates in the active center is strongly supported by the observation of two modes of action of PPi: phosphorolytic and hydrolytic, as shown in Figure 7D.
For positioning of the nucleoside moiety of the stimulatory NTP (Figure 7C), we envisaged a hydrogen bond between the 3′-hydroxyl of ribose and the carboxyl group of E813 (see Figure 2A). While this is a speculation, it is supported by the observation that 3′-amino derivatives of NTPs, which should be electrostatically attracted to the side chain of E813, were extremely potent stimulators of the exonuclease activity (data not shown). Anchoring of the stimulatory NTP through its β,γ-phosphates and the 3′-hydroxyl of ribose conveniently positions the nucleotide base facing the residue N458β′. This is consistent with the reduced stimulatory effect of NTP caused by N458A substitution. Besides residue N458β′, two other highly conserved residues (E925β′ and Q929β′) within the secondary channel in the wall structure contact the NTP base (Figure 2A).
Breaking of a salt bridge causes exonuclease enhancement
The crucial aspect of the proposed model is that it explains two allele-specific effects: (i) stimulation of the exonuclease activity by substitution R1106A; and (ii) the upsurge of the exonuclease activity in the pH range 9–10 that is seen only in wild-type RNAP. Our model attributes these effects to the destruction of the salt bridge between residues R1106 and D814. The removal of R1106 would destroy the bridge and make the negative charge of the D814 carboxyl available to form an additional coordination bond with Mg-II—an effect that would require slight shifting of the D814 side chain (Figure 7E). Thus, the R1106A mutation would lift the requirement for a stimulatory NTP for stabilization of Mg-II. Elevation of pH would make the exonuclease activity nucleotide independent by a similar mechanism, by destroying the R1106–D814 salt bridge through titration of the positive charge of the R1106 side chain. Obviously, elevated pH would not have a significant effect in the case of mutations removing either D814 or R1106, although for different reasons. In the absence of D814 (ED/AA mutant), the reaction cannot be stimulated due to the lack of the side chain to coordinate Mg-II. In the case of R1106A, the intrinsic exonuclease level would already be maximal since the 1106–814 salt bridge would be destroyed in the first place, even without alkaline pH.
Discussion
The main experimental result of this work is the elucidation of the role and stuctural localization of the second Mg2+ ion (Mg-II) involved in the 3′–5′ exonuclease reaction, which, admittedly, is not the principal biological function of RNAP. However, the model of the active center derived from these data explains all RNAP activities, including biologically relevant polymerization of RNA, in terms of a logical mechanism assuming that Mg-II is retained in the same position and plays the same role in all other reactions. Of course, the latter statement remains to be confirmed by experiment.
The 3′–5′ exonuclease activity of RNAP was first noted in human RNA polymerase II (Wang and Hawley, 1993; Thomas et al., 1998). Here we characterize this reaction for RNAP of E.coli. Its biological significance remains unclear; it could have a role in error correction, similar to other RNA degradation reactions. From the standpoint of understanding the mechanism of RNAPs, this activity fits into a logical place in the repertoire of RNA degradation reactions associated with lateral movements of the transcript in the active center (Figure 1A–E).
Indeed, in the backtracked complex, RNAP can cleave an internal phosphodiester bond, through both hydrolysis (Figure 1D) and pyrophosphorolysis (Figure 1E). In the i + 1 complex, only the removal of the terminal nucleotide through pyrophosphorolysis had been characterized in some detail (Figure 1B). In other words, the 3′–5′ exonuclease activity (Figure 1C) completes the view of the RNAP active center as a non-specific nuclease capable of cleaving any phosphodiester bond using either water or pyrophosphate as the nucleotidyl acceptor.
On the basis of these results, we offer a unified structure–functional model of the active center, which fits the X-ray structure, explaining all of the activities of RNAP—the polymerase, the exopyrophosphorylase and the endopyrophosphorylase, the exonuclease and the endonuclease—by a single mechanism based on the known Sn2 reaction geometry (Figure 7). According to the model, the degradation and the synthesis of the phophodiester bond are performed by the same active site, being in effect mirror images of each other. In both reactions, the same symmetrical pair of Mg2+ ions is involved, which alternate their roles of coordinating the donor and acceptor oxygens during nucleotidyl transfer (compare Figure 7A and B).
The crucial result that led to the above model has been the structure–functional characterization of the weakly bound catalytic Mg2+ ion (Mg-II). We found that Mg-II is recruited for the exonuclease reaction by a non-complementary NTP, through its β,γ-phosphates. The β,γ-phos phates can be substituted by free pyrophosphate. A R1106A substitution, which distorts the active center in a particular manner, increases the binding of Mg-II, thus stimulating the exonuclease. A similar distortion can be caused by alkaline pH, in a highly localized effect abolished by another specific mutation, ED/AA.
These results led to the conclusion that Mg-II, while being an essential component of the active site, is non-resident in the enzyme and has to be recruited by an auxiliary helper. An ad hoc recruitment of a metal ion into the catalytic center is well documented in other systems, e.g. in the active site of DNA polymerase I (Li et al., 1998). What is notable in the present case is that the recruitment of Mg-II may occur in a variety of ways, with the aid of different helper molecules, leading to the same end result of symmetrical, two-Mg geometry of the active center. In the case of the polymerase reaction, Mg-II is recruited by the incoming substrate. In the case of pyrophosphorolysis, PPi recruits it. For hydrolysis, the recruiter can be either a non-complementary NTP or PPi. In all of these cases, the recruitment consists of coordination of Mg-II by a diphosphate, which can bind in two different modes (Figure 7D).
Alternatively, Mg-II may be stabilized through a structural transition in the protein, without the involvement of diphosphate, when the D814β residue shifts to establish the third coordination bond with Mg-II. For this to happen, the salt bridge between D814β and R1106β needs to be destroyed, either by a mutation or by high pH. The result, obtained here for the exonuclease activity, is consistent with the observation that internal cleavage of RNA in the backtracked complex can be stimulated by alkali in the absence of auxiliary proteins (Orlova et al., 1995). It is tempting to assume that the role of cleavage factors GreA and GreB consists of distorting the microenvironment of the active center so that D814β is made available for recruiting Mg-II.
The notion of ad hoc recruitment may explain the discrepancy between our positioning of Mg-II and the coordinate assignments given to the second catalytic Mg2+ ion in the two crystal structures (Cramer et al., 2001; Vassylyev et al., 2002) (unmarked balls in Figure 2A). In the absence of a recruiter, Mg-II is bound so weakly that it is not likely to be retained in the crystals in the first place. The lack of effect on the exonuclease reaction by the ED/AA substitution, which would disrupt the retention of Mg2+ in the model of Cramer et al. (2001), confirms this interpretation. Earlier observations that mutations in this site inhibited the polymerase activity (Lee et al., 1991; Werner et al., 2002) could reflect a decrease in nucleotide binding rather than in metal retention.
It should be emphasized that neither of the published assignments for the second Mg2+ fits the Sn2 reaction geometry, or explains the effect of the R1106A substitution. Since many Mg2+ ions are bound to RNAP protein (Vassylyev et al., 2002), those crystals could have retained one that is unrelated to catalysis. A way to test this explanation directly would be to obtain the crystal structure of the R1106A mutant RNAP.
A principal result of this study is the discovery of a non-specific binding site (E-site, for entry) for NTP at the bottom of the substrate pore (Figure 2A and B). The incoming nucleotide bound in the E-site would enter the i + 1 site by rotating around Mg-II so that its β,γ-phos phates shift position as shown in Figure 7D. Such rotation would entail only a slight shift in the position of the γ-phosphate, but a wide swing in the position of the base (compare the nucleotides in the E-site and the i + 1 site in Figure 2A). A direct demonstration of the existence of the E-site would require obtaining a cross-link of the bound nucleotide, and a way to distinguish it from the NTP bound in the i + 1 site, which is the goal of our future research.
It should be noted that the E-site is probably unrelated to the allosteric substrate-binding site suggested by Foster et al. (2001), since the latter involves base pairing and attracts nucleotides with much higher affinity. On the other hand, the notion of the unspecific NTP site is supported by the reports that non-complementary NTPs stimulate 3′-terminal degradation of RNA in yeast RNAP III (Whitehall et al., 1994) and in vaccinia virus RNAP (Hagler and Shuman, 1993).
The existence of a distinct nucleotide entry site is intuitively appealing if one imagines how the incoming substrates can be sorted efficiently for complementarity. A non-specific substrate site would increase the local concentration of the substrate for transient presentation into the active center. Considering that the occupancy of the i + 1 site by the 3′ terminus should compete with the substrate entry, and may be regulated additionally by a structural switch within the RNAP molecule (Epshtein et al., 2002), the strictly diffusion-driven delivery of substrates may not be sufficiently fast for shifting the kinetic equilibrium in TEC in the direction of the synthetic reaction.
Materials and methods
Engineering of rpoB and rpoC mutations
R1106A and ED813,814AA mutations were generated initially in pMKSe2 (Severinov et al., 1994), which carries the β-subunit gene (rpoB) under the lac promoter by two-round PCR using DeepVent DNA polymerase (New England Biolabs) and appropriate primers (the sequences and detailed protocols are available on request). Then, the EcoRI fragments carrying the corresponding mutations were recloned from this vector into a pET28 derivative coding for the β-subunit with a His6 tag on the C-terminal end. The N458A mutation was generated in the pET29 derivative containing the rpoC gene with the His6 tag at the 3′ terminus, using the QuikChange XLSite-Directed Mutagenesis Kit (Stratagene). The substitutions were confirmed by restriction analysis and DNA sequencing. Plasmids carrying the mutations were transformed into BL21 (DE3) E.coli strain, and the mutant β-subunits were overproduced, purified and used for reconstitution of RNAP as described (Borukhov and Goldfarb, 1993).
Transcriptional complexes
The 270 bp BamHI–HindIII DNA fragment containing the T7A1 promoter was cut from pTZ19 plasmid containing this fragment. Defined TECs were obtained by RNAP ‘walking’ on the T7A1 promoter fragment using RNA polymerases immobilized on Ni-NTA–agarose (Kashlev et al., 1996). To obtain 3′-labeled TEC21, 1 pmol of RNAP in 10 µl of transcription buffer (TB) containing 50 mM Tris–HCl pH 7.9, 100 mM KCl, 10 mM MgCl2, 1 mM 2-mercaptoethanol, was mixed with 4 µl of pre-equilibrated Ni-NTA–agarose suspension and incubated for 3 min at 25°C. Then 1 pmol of DNA template was added to the pellet RNAP beads in a 1 µl volume, incubated for 5 min at 37°C and washed with cold TB (4 × 1 ml). Transcription was initiated by the addition of the priming trinucleotide ApUpC (10 µM) and ATP, GTP and CTP (20 µM each) [in the case of the internal labeling of RNA, 0.3 µM [α-32P]ATP (3000 Ci/mmol) was used] to allow the formation of TEC-20 (5 min, 37°C), and then the RNAP beads were washed with cold TB, supplemented with 0.3 µM [α-32P]UTP (3000 Ci/mmol), incubated for 5 min at 25°C and washed with cold TB. TEC-11, TEC-12, TEC-14 and TEC-27 were obtained by the same protocol but with different combinations of unlabeled and [α-32P]NTPs.
Exonuclease cleavage
Exonuclease cleavage was usually performed at 37°C in TB, containing different MgCl2 concentrations. The reaction time varied from 10 s to 10 min to ensure that no more than ∼20% of RNA was shortened. In the case of fast reaction by the R1106A mutant RNAP, the rate was measured at 21°C and extrapolated to 37°C using coefficient 5.6 determined by measuring the 37°C/21°C rate ratio for the wild-type RNAP in separate experiments. The reaction was stopped by adding 1 vol. of the loading buffer (7 M urea, 20 mM EDTA, bromophenol blue and xylene cyanol FF, 0.25% each), and reaction products were resolved on a 20% polyacrylamide gel (19:3 acrylamide:bis-acrylamide, 1× TBE, 7 M urea). Details of reaction conditions with PPi and r/dNTP (Amersham) are described in the figures. Transcription buffers with different pH contained: 50 mM PIPES–HCl pH 6, 50 mM Tris–HCl pH 7–9, 50 mM CAPS–NaOH pH 10; other components were as in TB. GreA- and GreB-induced cleavage was performed using a 10-fold excess of the cleavage factors over RNAP.
The identity of NMP as a product of the exonuclease reaction was confirmed by TLC and PAGE, where unlabeled NMP was used as marker. The triphosphate derivative of 4-methylumbelliferone was synthesized from 4-methylumbelliferone-monophosphate (Sigma) as described (Hoard and Ott, 1965).
Modeling
Modeling was performed using the program WebLab ViewerLight 4.0 (Molecular Simulations Inc.).
Acknowledgments
Acknowledgements
We thank Sergey Borukhov for fruitful discussions. This work was supported by NIH grants GM49242 and GM30717 to A.G., and by the Russian Foundation for Basic Research, grant 02-04-48525, and Foundation for Support of Leading Scientific Schools, grant 00-15-97751.
References
- Borukhov S. and Goldfarb,A. (1993) Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr. Purif., 4, 503–511. [DOI] [PubMed] [Google Scholar]
- Borukhov S., Sagitov,V. and Goldfarb,A. (1993) Transcript cleavage factors from E.coli. Cell, 72, 459–466. [DOI] [PubMed] [Google Scholar]
- Brautigam C.A. and Steitz,T.A. (1998) Structural principles for the inhibition of the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol., 277, 363–377. [DOI] [PubMed] [Google Scholar]
- Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Epshtein V., Mustaev,A., Markovtsov,V., Bereshchenko,O., Nikiforov,V. and Goldfarb,A. (2002) Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol. Cell, 10, 623–634. [DOI] [PubMed] [Google Scholar]
- Foster J.E., Holmes,S.F. and Erie,D.A. (2001) Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell, 106, 243–252. [DOI] [PubMed] [Google Scholar]
- Gnatt A.L., Cramer,P., Fu,J., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science, 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- Hagler J. and Shuman,S. (1993) Nascent RNA cleavage by purified ternary complexes of vaccinia RNA polymerase. J. Biol. Chem., 268, 2166–2173. [PubMed] [Google Scholar]
- Hoard D.E. and Ott,D.G. (1965) Conversion of mono- and oligodeoxyribonucleotides to 5′-triphosphates. J. Am. Chem. Soc., 87, 1785–1789. [DOI] [PubMed] [Google Scholar]
- Izban M.G. and Luse,D.S. (1993) The increment of SII-facilitated transcript cleavage varies dramatically between elongation competent and incompetent RNA polymerase II ternary complexes. J. Biol. Chem., 268, 12874–128785. [PubMed] [Google Scholar]
- Kashlev M., Nudler,E., Severinov,K., Borukhov,S., Komissarova,N. and Goldfarb,A. (1996) Histidine-tagged RNA polymerase of Escherichia coli and transcription in solid phase. Methods Enzymol., 274, 326–34. [DOI] [PubMed] [Google Scholar]
- Komissarova N. and Kashlev,M. (1997a) Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA, 94, 1755–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N. and Kashlev,M. (1997b) RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem., 272, 15329–15338. [DOI] [PubMed] [Google Scholar]
- Koren R. and Mildvan,S. (1977) Magnetic resonance and kinetic studies of the role of the divalent cation activator of RNA polymerase from Escherichia coli. Biochemistry, 16, 241–249. [DOI] [PubMed] [Google Scholar]
- Korzheva N., Mustaev,A., Kozlov,M., Malhotra,A., Nikiforov,V., Goldfarb,A. and Darst,S.A. (2000) A structural model of transcription elongation. Science, 289, 619–625. [DOI] [PubMed] [Google Scholar]
- Lee J., Kashlev,M., Borukhov,S. and Goldfarb,A. (1991) A β subunit mutation disrupting the catalytic function of Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA, 88, 6018–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Korolev,S. and Waksman,G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J., 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W.R. (1980) On the mechanism of streptolydigin inhibition of Escherichia coli RNA polymerase. J. Biol. Chem., 255, 1610–1616. [PubMed] [Google Scholar]
- Mustaev A., Kozlov,M., Markovtsov,V., Zaychikov,E., Denissova,L. and Goldfarb,A. (1997) Modular organization of the catalytic center of RNA polymerase. Proc. Natl Acad. Sci. USA, 94, 6641–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E., Mustaev,A., Lukhtanov,E. and Goldfarb,A. (1997) The RNA–DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell, 89, 33–41. [DOI] [PubMed] [Google Scholar]
- Orlova M., Newlands,J., Das,A., Goldfarb,A. and Borukhov,S. (1995) Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl Acad. Sci. USA, 92, 4596–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D. (1992) Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J. Biol. Chem., 267, 3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Rozovskaya T.A., Rechinsky,V.O., Bibilashvili,R.S., Karpeisky,M.Ya., Tarusova,N.B., Khomutov,R.M. and Dixon,H.B. (1984) The mechanism of pyrophosphorolysis of RNA by RNA polymerase. Endowment of RNA polymerase with artificial exonuclease activity. Biochem. J., 224, 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd M.D., Izban,M.G. and Luse,D.S. (1994) The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc. Natl Acad. Sci. USA, 91, 8057–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinov K., Kashlev,M., Severinova,E., Bass,I., McWilliams,K., Kutter,E., Nikiforov,V., Snyder,L. and Goldfarb,A. (1994) A non-essential domain of Escherichia coli RNA polymerase required for the action of the termination factor Alc. J. Biol. Chem., 269, 14254–14259 [PubMed] [Google Scholar]
- Steitz T.A. (1998) A mechanism for all polymerases. Nature, 391, 231–232. [DOI] [PubMed] [Google Scholar]
- Thomas M.J., Platas,A.A. and Hawley,D.K. (1998) Transcriptional fidelity and proofreading by RNA polymerase II. Cell, 93, 627–637. [DOI] [PubMed] [Google Scholar]
- Vassylyev D.G., Sekine,S., Laptenko,O., Lee,J., Vassylyeva,M.N., Borukhov,S. and Yokoyama,S. (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature, 417, 712–719. [DOI] [PubMed] [Google Scholar]
- Wang D. and Hawley,D.K. (1993) Identification of a 3′→5′ exonuclease activity associated with human RNA polymerase II. Proc. Natl Acad. Sci. USA, 90, 843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F. and Weinzierl,R. (2002) A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell, 10, 635–646. [DOI] [PubMed] [Google Scholar]
- Whitehall S.K., Bardeleben,C. and Kassavetis,G.A. (1994) Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J. Biol. Chem., 269, 2299–2306. [PubMed] [Google Scholar]
- Yee D., Armstrong,V.W. and Eckstein,F. (1979) Mechanistic studies on deoxyribonucleic acid dependent ribonucleic acid polymerase from Escherichia coli using phosphorothioate analogues. 1. Initiation and pyrophosphate exchange reactions. Biochemistry, 18, 4116–4120. [DOI] [PubMed] [Google Scholar]
- Zaychikov E. et al. (1996) Mapping of catalytic residues in the RNA polymerase active center. Science, 273, 107–109. [DOI] [PubMed] [Google Scholar]
- Zhang G., Campbell,E.A., Minakhin,L., Richter,C., Severinov,K. and Darst,S.A. (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell, 98, 811–824. [DOI] [PubMed] [Google Scholar]