Abstract
Powdery mildews, obligate biotrophic fungal parasites on a wide range of important crops, can be controlled by plant resistance (R) genes, but these are rapidly overcome by parasite mutants evading recognition. It is unknown how this rapid evolution occurs without apparent loss of parasite fitness. R proteins recognize avirulence (AVR) molecules from parasites in a gene-for-gene manner and trigger defense responses. We identify AVRa10 and AVRk1 of barley powdery mildew fungus, Blumeria graminis f sp hordei (Bgh), and show that they induce both cell death and inaccessibility when transiently expressed in Mla10 and Mlk1 barley (Hordeum vulgare) varieties, respectively. In contrast with other reported fungal AVR genes, AVRa10 and AVRk1 encode proteins that lack secretion signal peptides and enhance infection success on susceptible host plant cells. AVRa10 and AVRk1 belong to a large family with >30 paralogues in the genome of Bgh, and homologous sequences are present in other formae speciales of the fungus infecting other grasses. Our findings imply that the mildew fungus has a repertoire of AVR genes, which may function as effectors and contribute to parasite virulence. Multiple copies of related but distinct AVR effector paralogues might enable populations of Bgh to rapidly overcome host R genes while maintaining virulence.
INTRODUCTION
Obligate biotrophic parasites cause the most serious and widespread diseases of crop plants but are challenging to investigate because they cannot grow outside their host. The three major groups of biotrophic filamentous parasites are the powdery mildew and rust fungi and the downy mildews, which are oomycetes. Powdery mildews cause economic losses to most crops in temperate areas, infecting >9000 dicot and >650 monocot plant species (Chaure et al., 2000). Infection of host plants during the growing season results from wind-dispersed conidiospores that are the asexual (haploid) state of the fungus. Many powdery mildews also have a sexual phase resulting in the formation of ascospores (Figure 1A). Most powdery mildews show a high degree of host specialization, a feature well exemplified in Blumeria graminis f sp hordei (Bgh), which displays gene-for-gene interactions with its host plant, barley (Hordeum vulgare) (Schulze-Lefert and Panstruga, 2003).
Figure 1.
Lifecycle of Bgh and the Isolation of AVRk1 and AVRa10 by Map-Based Cloning.
(A) Compatible isolates of opposite mating types can be crossed to produce single ascospore cultures. These can be maintained as individual cultures in the haploid asexual phase on detached barley leaves, and DNA for mapping can be obtained from conidiospores.
(B) Genetic and physical map of the genomic region of Bgh containing AVRk1 and AVRa10. Genetic mapping data for two crosses is shown in blue, and recombinant progeny isolate numbers are in parentheses. Primer sequences for PCR markers are shown in Methods. M is a microsatellite mapped with marker CJR3. Marker CJR1 is a size polymorphism (541 bp inserted in DH14) and defines the left border of the region cosegregating with AVRk1. The right border of the AVRk1 cosegregating region was defined by sequencing PCR amplicons at CJR2. Homologies were established by BLASTX (Altschul et al., 1997) against ESTs in the NCBI and COGEME databases. LINE and SINE elements are as previously described (He et al., 1996; Wei et al., 1996). Numbers in brackets are COGEME accession numbers.
In gene-for-gene interactions (Flor, 1971), the recognition of an avirulence (AVR) molecule by a host resistance (R) protein triggers a localized cell death, known as the hypersensitive response, and other defense responses that prevent further parasite growth (Greenberg and Yao, 2004; Skamnioti and Ridout, 2005). Bacterial AVR proteins are introduced into host cells by the type-three secretion system and can contribute to successful infection in susceptible host varieties (Alfano and Collmer, 2004; Janjusevic et al., 2006). Thus, bacterial AVR proteins are often described as effectors since they have both elicitor (avirulence) and virulence activities. Some fungal AVR proteins could potentially function as effectors. For example, Avr4 from Cladosporium fulvum binds to chitin, so it may function to protect the fungus from plant chitinases during infection (van den Burg et al., 2003). Avr2, also from C. fulvum, binds to Rcr3, a Cys protease required specifically for the function of the resistance gene Cf-2 (Rooney et al., 2005). However, there is no proof that these or any other fungal AVR proteins contribute directly to infection success.
More than 25 independent AVR genes have been described in Bgh isolates (Brown and Jessop, 1995; Jensen et al., 1995), but none are yet isolated. Although AVR genes are distributed throughout the Bgh genome, a cluster containing AVRk1, AVRa10, and AVRa22 is linked by 1 to 2 centimorgans (cM) (Brown and Jessop, 1995; Jensen et al., 1995; Caffier et al., 1996). More than 85 barley R genes, each conferring resistance to specific Bgh AVR elicitors, have been described, including Mlk1 and 28 alleles at the Mla locus on barley chromosome 5 (Jørgensen, 1994). The six isolated Mla alleles (Mla1, Mla6, Mla7, Mla10, Mla12, and Mla13) are predicted to share >90% amino acid sequence identity (Zhou et al., 2001; Halterman et al., 2003; Shen et al., 2003; Halterman and Wise, 2004). Mla proteins have conserved coiled-coil N-terminal regions, nucleotide binding (NB) sites, variable leucine-rich repeat (LRR), and C-terminal regions that can confer recognition specificity for different Bgh AVR gene products (Shen et al., 2003). Mla proteins are related to other NB-LRR proteins, such as leaf rust and powdery mildew resistance genes Lr10 and Pm3b in wheat (Triticum aestivum), and to RPM1 in Arabidopsis thaliana (Feuillet et al., 2003; Yahiaoui et al., 2004). Mla and these related NB-LRR resistance proteins are likely to be located within the cytoplasm, where they recognize pathogen AVR molecules that enter the host cell (Schulze-Lefert and Panstruga, 2003). The powdery mildew resistance gene Mlk1 is also located on chromosome 5, ∼5 cM from the Mla locus (Maroof et al., 1994).
AVR genes have now been isolated from the two other groups of obligate biotrophs, represented by the flax rust fungus Melampsora lini and the oomycetes Hyaloperonospora parasitica, Phytophthora infestans, and Phytophthora sojae (Allen et al., 2004; Dodds et al., 2004; Shan et al., 2004; Armstrong et al., 2005; Rehmany et al., 2005; Catanzariti et al., 2006). Although nothing is known about the function of these AVR genes, all of them are subject to diversifying selection, which implies that they are maintained in populations because they play a role in pathogenicity. All biotrophs penetrate host plant cells and establish within them haustoria, specialized feeding structures for nutrient uptake. Some of the AVR genes identified in rust fungi are upregulated within the haustoria, suggesting a role in the establishment of biotrophy (Catanzariti et al., 2006).
Our objective was to isolate AVR genes from Bgh, a representative of the powdery mildews as the third major group of obligate biotrophic parasites. Isolation of AVR genes is a vital step in understanding the biology of Bgh in at least two respects. First, the large repertoire of AVR and R genes makes Bgh and barley excellent models for investigating how variation in molecular structures can account for specificity of parasite recognition. To understand the function of barley R genes in eliciting plant defenses, it is also essential to investigate the function of AVR genes corresponding to isolated R genes (Schulze-Lefert and Panstruga, 2003). Second, Bgh is an important model for research on the population biology of host–parasite coevolution in agriculture (Brown and Høvmoller, 2002). Cereal powdery mildews can rapidly evolve to overcome host R genes without apparent loss of fitness (Bronson and Ellingboe, 1986; Brown, 2003). The isolation of Bgh AVR genes would therefore permit investigation into the molecular basis for host adaptation and parasite evolution.
RESULTS
Genetic Delimitation of a Candidate AVRk1 Gene
To clone AVR genes from Bgh, we targeted the cluster containing AVRk1, AVRa10, and AVRa22, which are linked within 1 to 2 cM of each other. Two genetic crosses were made: one between Bgh isolates DH14 and CC52 and another between DH14 and CC148 (see Methods). The infection types of AVR genes segregating in these crosses are shown in Table 1. DNA from progeny and parental isolates was used for genetic mapping by amplified fragment length polymorphisms (AFLPs) (Vos et al., 1995), and a BAC library was made using DNA from the common parent isolate DH14. An AFLP marker PAAMCAC-365 present in DH14 DNA and mapping 2.4 cM (two recombinants out of 83) from the AVRa22 locus in the cross between Bgh isolates DH14 and CC52 was identified, providing a starting point for physical mapping at the AVRk1/AVRa10/AVRa22 cluster. PAAMCAC-365 was used to isolate three BACs by hybridization (Figure 1B). BAC 1817D was sequenced by shotgun cloning and primer walking. Comparative sequencing of BAC 1817D and parental DNA by long-range PCR identified a microsatellite (M) that consisted of 26 repeats of the DNA sequence (ATGTGG) in DH14 and 4.5 repeats in CC52 and CC148. M was therefore mapped in both DH14 × CC52 and DH14 × CC148 as PCR marker CJR3 with primers PCR3F and PCJR3R. In the cross DH14 × CC148, there were two recombinants between the AVRk1 and M loci, providing additional confirmation that BAC 1817D was located close to the AVRk1/AVRa10/AVRa22 cluster. Further sequence comparisons between DH14 and CC148 DNA near the M locus were made by PCR with primers designed from the sequence of BAC 1817D. Two further polymorphisms were identified at PCR loci CJR1 and CJR2 (Figure 1B), enabling a 5102-bp region cosegregating with the AVRk1 gene to be delimited. The entire cosegregating region was sequenced in both DH14 and CC148 by long-range PCR.
Table 1.
Infection Types of Bgh Isolates
| Infection Typea
|
||||
|---|---|---|---|---|
| Barley Variety | R Geneb | CC52 | CC148 | DH14 |
| Pallas or Siri | Mla8 | 4 | 4 | 4 |
| Midas and P03 | Mla6 | 0 | 4 | 0 |
| P09 and S09c | Mla10 | 4 | 0 | 4 |
| P10 | Mla12 | 4 | 4 | 0 |
| P17 and S17 | Mlk1 | 4 | 1 | 4 |
| Hordeum 1063 | Mlk1 | 4 | 1–2 | 4 |
| W37/136 | Mlh | 1–2 | 4 | 0 |
| P12 | Mla22 | 0 | 4 | 4 |
| Rupal | Mla13 | 4 | 0 | 0 |
| P08B | Mla9 | 4 | 0 | 0 |
| P06 | Mla7 | 4 | 1–2 | 1–2 |
Infection types are based on Brown and Wolfe (1990): 0, fully resistant, no visible symptoms; 1, necrotic flecks, no sporulation; 2, heavy necrotic flecks, scarce sporulation; 3, light necrotic flecks, moderate sporulation; and 4, fully susceptible, no necrosis or chlorosis.
R gene designations as in Jørgensen (1994).
P and S series of near-isogenic lines derived from Pallas and Siri, respectively (Kølster et al., 1986; Kølster and Stølen, 1987).
Within the cosegregating region, an open reading frame (ORF) was identified that was complete in three Bgh isolates with the phenotype Ak1 (avirulent on Mlk1 plants). In four Vk1 isolates (Vk1, virulent on Mlk1 plants), however, the amino acid sequence contained either stop codons or a frame shift (Figure 2A). This DNA sequence for this ORF was therefore a candidate for the AVRk1 gene (AVRk1-CAND). A retrotransposon sequence with homology to Cg T1 from Colletotrichum gloeosporoides (He et al., 1996) was located immediately 3′ to AVRk1-CAND. Other sequences with homology to Bgh ESTs were located within the cosegregating region but were discounted as candidates for the AVRk1 gene because no DNA sequence polymorphism between Ak1 and Vk1 isolates was detected.
Figure 2.
AVRk1 and AVRa10 Sequence Variants and Transcripts.
(A) Alignment of amino acid residues of AVRk1 isolated from Akl (CC148, GF13, and HLA) and Vk1 (CC52, DH14, GF4, and C24R2) isolates. The most conserved residues are boxed in black, and less conserved residues are in lighter gray. The sequence of isolate HLA is the same as CC148. The stop codon at position 14 in the amino acid alignment of DH14 is denoted by an X. The sequence of isolate CC52, which is the same as all the other Vk1 isolates, is extensively different to that of the Ak1 isolate CC148. This is caused by nucleotide polymorphisms that result in a frame shift and fusion with the downstream Cg T1 retrotransposon. Underlined residues are those characteristic of the NB domain of retrotransposons related to Cg T1 (He et al., 1996).
(B) Diagrams illustrating cDNA transcripts of AVRk1 and AVRa10 obtained by RACE-PCR. Numbers are the position of features in base pairs from the 5′ mRNA cap. A is ATG start, S is stop codon, and P is the location of the polyadenylation site. Two transcripts with the same stop codon and terminating at P1 and P2 were obtained for AVRk1.
(C) The nucleotide and translated amino acid sequence of AVRa10 is the same in isolates CC148 (Aa10) and DH14 and CC52 (both Va10), except at the C terminus, where variant nucleotides are shown in bold. The insertion of cytosine (arrowhead) in Va10 isolates results in a frame shift and fusion with a Cg T1 retrotransposon.
(D) Comparison of sequences of AVRa10 in isolate CC148 (Aa10) with the Vk1 isolate obtained from CC148 after EMS mutagenesis.
The AVRk1-CAND sequence is transcribed in Ak1 isolates of Bgh. cDNA transcripts containing the same stop codon were isolated by rapid amplification of cDNA ends (RACE)-PCR from barley leaf epidermis infected with the Ak1 Bgh isolate CC148. In one transcript, the polyadenylation site occurred shortly after the stop codon, whereas in the other, this occurred after the 3′ Cg T1 homologue sequence (Figure 2B).
AVRk1-CAND Causes Mlk1-Specific Cell Death
An AVR protein is predicted to elicit host cell death specifically in plants with the corresponding R gene. As no reliable transformation procedure is available for Bgh, we tested Mlk1-specific recognition of AVRk1-CAND by a cell death assay after transient expression in planta. This biolistic method has been used to verify other fungal, oomycete, and bacterial AVR genes (Leister et al., 1996; Jia et al., 2000; Allen et al., 2004; Rehmany et al., 2005). The method relies on R gene–dependent cell death causing a reduction in reporter protein accumulation when reporter and candidate AVR genes are coexpressed in cells of host plant leaves. We cobombarded a green fluorescent protein (GFP)–expressing reporter plasmid with an AVRk1-CAND–expressing plasmid and compared the results to a null plasmid containing a nonfunctional allele of AVRk1-CAND isolated from the Vk1 strain DH14. Near-isogenic barley lines with specific resistance genes backcrossed into varieties Pallas (P series) or Siri (S series) (Kølster et al., 1986; Kølster and Stølen, 1987) were used. A GFP index was measured as the proportion of GFP-expressing cells visible in Mlk1-resistant barley lines (P17 and S17) or lines with other resistance genes (P09, Mla10; S09, Mla10; P22, Mla22) relative to mlk1-susceptible lines (Pallas or Siri).
Using this method, we demonstrated that the AVRk1-CAND–expressing plasmid caused a highly significant (P > 0.001) Mlk1-dependent reduction in the GFP index to 59% (Figure 3A). This result is consistent with the recognition of AVRk1-CAND by Mlk1 causing cell death and a consequent reduction in GFP protein. No significant reduction in the GFP index was detected when AVRk1-CAND was cobombarded into leaves containing other Mla resistance alleles, confirming that recognition of AVRk1-CAND is specific to Mlk1. As expected, the GFP index was not reduced when the null plasmid was used on Mlk1 leaves.
Figure 3.
AVR Recognition Measured with a Cell Death Assay.
GFP index measured in barley leaves with Mla10, Mlk1, and Mla22 alleles compared with Pallas or Siri near-isogenic parental lines after expression of AVRk1 or the null plasmid (A) and AVRa10 or the null plasmid (B). Each data point summarizes five experiments with two replicates of each lisolate + plasmid combination per experiment and seven leaves of each variety per variety + plasmid replicate. Predicted means were calculated by generalized linear modeling of the logarithm of GFP index with the model experiment + plasmid × near-isogenic series. The plasmid effect was highly significant (F-test, P < 0.001). As there was no significant effect of the interaction between plasmid and near-isogenic series (Pallas or Siri), means were predicted for varieties and plasmids pooled across near-isogenic series. Significance test for difference of GFP index from 1: ***, P < 0.001; ns, P > 0.05. Bars indicate mean ± 1 se.
AVRk1-CAND Induces Inaccessibility in Mlk1 Leaves
We further characterized the function of AVRk1-CAND by exploiting a feature of powdery mildew gene-for-gene interactions known as induced inaccessibility, whereby virulent isolates can no longer infect living host cells first attacked by avirulent or nonhost isolates (Prats et al., 2006). We developed an infection assay based on the facts that GFP accumulated in 59% of AVRk1-CAND cobombarded Mlk1 cells (Figure 3A) and that live infection attempts on those cells could be viewed by microscopy. Leaves were first cobombarded with the GFP-expressing reporter plasmid and either the AVRk1-CAND–expressing or the null plasmid and then infected with either of two Vk1 isolates of Bgh, DH14 and CC52. Infection success or failure was determined by microscopy examination to establish whether GFP-expressing cells were penetrated by the fungus and whether such cells remained alive (Figures 4A to 4C).
Figure 4.
Scoring Criteria and Results of Infection Assay.
(A) to (C) Photomicrographs of GFP-expressing barley epidermal cells infected with Bgh, photographed rapidly to minimize tissue damage by UV irradiation.
(A) Germinated spore (sp) of Bgh showing successful infection on a living cell with secondary hyphae (sh), haustorium (h), and plant cell nucleus (n).
(B) Failed infection on a living cell. The fungus has attempted to penetrate the cell with an appressorium (ap), a specialized infection structure, but this has withered and lost turgor. The streaming cytoplasmic threads (ct) indicate that the cell is still alive.
(C) Failed infection with a dead cell and static granular cytoplasm.
(D) Percentage of infection success of Bgh isolate CC52 or DH14 (both Vk1 and Va10) in barley cells following cobombardment of plasmids expressing AVRk1 or AVRa10 with a GFP-expressing plasmid. Each data point summarizes three experiments (AVRk1) or two experiments (AVRa10), with seven replicate leaves per experiment on average of each variety + isolate + plasmid combination. Predicted means were calculated by generalized linear modeling of the number of successful infections. The model was experiment + variety × isolate × plasmid, and a logit link function was used to analyze binomial data of successful infections as a proportion of total infections. The variety × plasmid interaction was highly significant (P < 0.001). As there was no significant effect of isolate (DH14 or CC52) on the variety × plasmid interaction, means were predicted for varieties and plasmids pooled across isolates. Significance test for difference between null and test plasmids: **, P < 0.01; ***, P < 0.001.
There was a highly significant reduction in the number of AVRk1-CAND–expressing host cells successfully penetrated by the parasite on Mlk1 leaves compared with those expressing the null plasmid (Figure 4D). At failed infection sites, cytoplasmic aggregation typical of hypersensitive response–induced cell death, and live host cells on which the fungus had failed to penetrate, were observed. Our results show that transiently expressed AVRk1-CAND induces inaccessibility to virulent isolates attacking cells on Mlk1 lines and that this effect is specific to the AVRk1-Mlk1 interaction. Ten days after infection, necrotic lesions were observed on Mlk1 leaves cobombarded with GFP and AVRk1-CAND and subsequently infected by either of the two Vk1 isolates (Figure 5). The lesions resulted in diminished fungal growth, which was measured as a significant reduction in spore production.
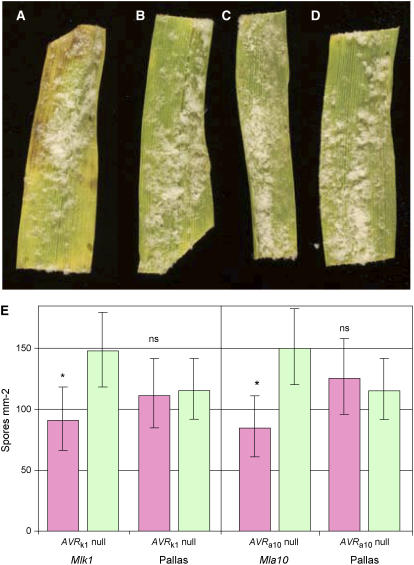
Figure 5.
AVRk1 and AVRa10 Induce Delayed Necrosis after Infection.
(A) Necrosis phenotype observed on P17 (Mlk1) leaves bombarded with AVRk1.
(B) P17 leaves bombarded with null plasmid.
(C) Pallas (mlk1) leaves with AVRk1.
(D) Pallas leaves with null plasmid. All leaves in (A) to (D) are shown 10 d after infection with DH14.
(E) Spore production (conidia mm−2) determined 10 d after infection following bombardment of AVRk1 or AVRa10 or the null plasmid as for (D) measured on a hemocytometer slide. Each data point summarizes three experiments with four replicates of each isolate + plasmid combination per experiment and seven leaves of each variety per variety + plasmid replicate. Predicted means were calculated by residual maximum likelihood analysis of the square root of the number of spores produced per leaf. The random effects model was experiment/isolate/replicate, and the fixed effects model was variety × plasmid. The variety × plasmid interaction was highly significant (Wald test, P < 0.001). As there was no significant effect of isolate (DH14 or CC52) on the variety × plasmid interaction, means were predicted for varieties and plasmids pooled across isolates. Significance test for difference between differential variety and Pallas: *, P < 0.05; ns, P > 0.05. Bars indicate mean ± 1 se.
AVRk1-CAND Enhances Infection on Susceptible Varieties
The ability to examine single, infected, living epidermal cells expressing AVRk1- CAND enabled us to make another important and unexpected observation. When expressed in the susceptible cultivar Pallas, AVRk1-CAND significantly increased the number of cells successfully infected by the Vk1 isolates (Figure 4D). This suggests that AVRk1-CAND has an effector function that enhances infection in compatible interactions. The number of spores produced on Pallas leaves was similar after bombardment with either AVRk1-CAND or the null plasmid (Figure 5). Infection is therefore probably enhanced only in the very small number of individual AVRk1-CAND–expressing cells obtained after the biolistic treatment, the rest of the leaf remaining unaffected.
In summary, AVRk1-CAND cosegregates with the AVRk1 phenotype, has a functional ORF sequence in Ak1 but not Vk1 isolates, is expressed by Ak1 isolates, elicits (1) host cell death, (2) induced inaccessibility, and (3) reduced fungal sporulation specifically in Mlk1 plants, and has an effector function in promoting susceptibility to disease in plants lacking Mlk1. Together, these seven points confirm that AVRk1-CAND is indeed AVRk1 with dual AVR and disease effector functions.
AVRa10 Is a Paralogue of AVRk1
We identified a paralogue of AVRk1 located 7.5 kb from AVRk1 and reasoned that this may encode another AVR gene within the AVRk1/AVRa10/AVRa22 cluster (Figure 1B). In the cross between Bgh isolates CC148 (Aa10, avirulent on Mla10 plants) and DH14 (Va10, virulent on Mla10 plants), this paralogue cosegregated with the Aa10 phenotype and so was a candidate for the gene (AVRa10-CAND). Sequences of AVRa10-CAND from natural Va10 isolates revealed mutations within the gene (Figure 2C). Furthermore, when CC148 was mutagenized with ethyl methanesulfonate (EMS) and selected for growth on Mla10 leaves, a derivative Va10 mutant isolate had a mutation in this nucleotide sequence resulting in a premature stop codon in the AVRa10-CAND protein (Figure 2D). In the cell death assay, we measured an AVRa10-CAND–dependent reduction in the GFP index to 73% on Mla10 leaves (Figure 3B). No reduction in the GFP index was measured on leaves containing Mlk1 or Mla22 genes or with the null plasmid on Mla10 leaves. In the infection assay, inaccessibility to DH14 and another Va10 isolate, CC52, was induced in an AVRa10-CAND–dependent, Mla10-specific manner (Figure 4D). Reduced infection was associated with both failed penetration and cell death. We also measured reduced spore production, consistent with diminished fungal growth, with both of the Va10 isolates 10 d after infecting Mla10 plants bombarded with AVRa10-CAND (Figure 5). The number of successful infections increased after expression of AVRa10 in Pallas cells (Figure 4D), indicating that AVRa10-CAND has an effector function of enhancing infection in susceptible lines (Pallas has Mla8, an ineffective Mla10 allele).
Our results confirm that AVRa10-CAND is indeed AVRa10. The seven lines of evidence adduced for the identity of AVRk1 and for the dual elicitor/effector function of that gene apply equally to AVRa10. In addition, EMS-induced point mutation of AVRa10 to produce premature termination of the coding sequence caused loss of AVRa10 activity.
AVRk1 and AVRa10 Are Members of a Gene Family in Powdery Mildew Fungi of Grasses
Six paralogues of AVRk1, with an E value <10−7, were identified by BLAST searching in GenBank as ESTs from Bgh (accession numbers BQ283794 and BQ283703) and from Bgh-infected barley (accession numbers DN83308, CK567720, CK568673, and CK568570). Three complete paralogous cDNAs were also amplified by RACE-PCR from barley leaves infected with Bgh isolate CC148 (Figure 6A). DNA gel blot hybridization experiments revealed that sequences homologous to AVRk1 are present in other formae speciales of B. graminis isolated from the Graminae hosts, wheat, rye (Secale cereale), oat (Avena sativa), and Agropyron sp (Figure 6B). PCR with degenerate primers also amplified bands from the DNA of these formae speciales. The AVRk1 sequence hybridized to 98 of 12,288 clones in the Bgh BAC library (Figure 6C). Taken together, the DNA gel blot and BAC filter hybridizations indicate that there are likely to be >30 paralogues within the genomes of Bgh and each of the other B. graminis formae speciales. In all, 87% (85/98) of the BAC clones hybridized to both AVRk1 and a Cg T1 sequence, revealing a close association between this AVR gene family and this class of retrotransposons. The AVRk1 sequence also weakly hybridized to DNA from Erysiphe cichoracearum, which infects Arabidopsis (Figure 6D), but the degenerate PCR primers failed to amplify a product indicating that more distantly related sequences may be present in this parasite. Apart from the Bgh and Bgh-infected barley ESTs, there was no significant (E value <10−5) homology to the sequence or structure of AVRk1 paralogues in any nucleotide or protein database or in the protein homology/analogy recognition engine (http://www.sbg.bio.ic.ac.uk/phyre).
Figure 6.
AVRk1and AVRa10 Belong to a Gene Family in Cereal Powdery Mildews.
(A) Sequences paralogous to AVRk1 were isolated by RACE-PCR from Bgh isolate CC148. The most conserved residues are boxed in black, and less conserved residues are in lighter gray. The conserved core region is underlined and contains a motif (between arrowheads) resembling that found in oomycetes and implicated in targeting avirulence proteins into host cells (Armstrong et al., 2005; Rehmany et al., 2005; Bhattacharjee et al., 2006).
(B) DNA gel blot hybridization of the conserved core sequence of AVRk1 (shown in [A]) against DNA (10 μg), restricted with EcoRI, obtained from B. graminis formae speciales isolated from grasses: Agropyron repens isolate Agr2 (lane 1), oat isolates M089-12 (lane 2) and OFR2 (lane 3), rye isolates RESP4 (lane 4) and PMAS (lane 5), wheat isolates P4RC (lane 6) and PIRIB (lane 7), and barley isolates CC52 (lane 8) and CC148 (lane 9). The approximate size of restriction fragments is shown.
(C) Hybridization of the AVRk1 core probe against a one genome equivalent of the Bgh BAC library. Hybridizing clones are visible as two replicate dots at each location on the filter. The DNA gel blot and BAC library hybridizations indicate >30 paralogues present in the genomes of B. graminis formae speciales.
(D) Dot blot of whole genome amplifications of (1) wash from noninfected Arabidopsis leaves, (2) DNA from noninfected Arabidopsis leaves, and (3) DNA from conidiospores from E. cichoracearum infecting Arabidopsis (all 10 μg of amplified DNA).
The central core of AVRk1, AVRa10, and the three complete RACE-PCR paralogues were highly conserved in terms of the transcribed amino acid sequence (Figure 6A). There was a deficit of nonsynonymous nucleotide changes (Ka, leading to amino acid changes) over synonymous nucleotide changes (Ks), implying that purifying selection has maintained the amino acid sequence, possibly owing to functional constraints on the protein. The ratio Ka:Ks within the core region ranged from 0.08 to 0.49 in pairs of comparisons between the five sequences. Ka:Ks = 1 would imply an absence of either purifying or diversifying selection. By contrast, the amino acid sequences outside the core and toward the N- and C-terminal regions of the AVR proteins and the RT-PCR paralogues were highly diverse. It is not possible to calculate Ka:Ks reliably for such diverse sequences, as its value depends on the alignment chosen; indeed, the process of sequence alignment causes a downwards bias in the estimate of Ka:Ks. Nevertheless, it is evident that great sequence diversity has evolved at the N- and C-terminal parts of the protein.
DISCUSSION
We have identified avirulence genes AVRk1 and AVRa10 and demonstrated that they belong to a gene family present in Bgh and other formae speciales of the grass powdery mildew fungi. It is possible that other paralogues may be different AVR genes; this could be tested by mapping and cell death assays in host plants with corresponding R genes, if these are available. We show that the AVRk1 paralogues are located near retrotransposons. Close physical association with retrotransposons and other repetitive sequences in both bacterial and fungal genomes may contribute to effector and AVR gene expansion and diversification (Rohmer et al., 2004; Skamnioti and Ridout, 2005; Gout et al., 2006).
Since powdery mildews infect single host epidermal cells, we were able to directly measure the effect of transiently expressing AVRk1 and AVRa10 on penetration success. Our results indicate that the AVR genes can enhance infection in susceptible varieties, indicating that they may function as effectors like some AVR proteins of bacterial pathogens (Alfano and Collmer, 2004). Further experiments will be required to validate these initial observations, for example, by localizing the AVRk1 proteins during infection and identifying potential host virulence targets. Effectors of Pseudomonas syringae and other pathogenic bacteria show considerable interstrain variation, implying that they play a role in host specialization (Guttman et al., 2002; Roden et al., 2004). If this also applies to powdery mildew fungi, the polymorphism between avirulence gene paralogues may be involved in host specialization and the origin of formae speciales.
When AVRk1 was expressed in the resistant barley varieties containing Mlk1, GFP reporter expression was reduced to 59% compared with the control plasmid. This value is comparable to the ∼50% reduction in reporter protein when avrRpt2 from P. syringae was expressed in Arabidopsis leaves containing the corresponding RPS2 resistance gene (Leister et al., 1996). Up to ∼95% reduction in reporter protein accumulation has been measured using similar methods with other AVR genes (Leister et al., 1996; Allen et al., 2004; Rehmany et al., 2005). These results indicate that cell death can proceed more rapidly in some AVR/R gene combinations. We were also able to measure induced inaccessibility in GFP-expressing cells that remained alive. This indicated that cell death does not always occur in cells expressing the AVR gene and that other changes have prevented the establishment of a successful infection. Race-specific resistance that does not result in cell death has been reported previously in both wheat and barley powdery mildew (Schiffer et al., 1997; Li et al., 2005).
Our results show that AVRk1 and AVRa10 are recognized when transiently expressed within host cells, which is consistent with the predicted cytoplasmic location of Mla proteins (Schulze-Lefert and Panstruga, 2003). Bgh AVR proteins must therefore be able to enter the host cell during infection, and we are currently developing immunolocalization procedures to investigate these predictions. The Uromyces fabae rust protein Uf RTP1p is known to enter host cells and localize to the nucleus (Kemen et al., 2005). However, the function of Uf RTP1p and the mechanism of its entry into the host cell are not known. Other fungal and oomycete AVR proteins are also predicted to enter the host cell, where they are recognized by cytoplasmic R proteins. A conserved amino acid motif Gx1x2R (where x1 is a hydrophobic or aromatic amino acid) common to four flax rust AVR proteins predicted to enter host cells has been identified (Catanzariti et al., 2006), but this sequence was not present in the Bgh AVR proteins. A conserved motif (RXLR, where X is any amino acid) has been implicated in the transport into host cells of AVR proteins and effectors from the oomycetes H. parasitica and P. infestans (Armstrong et al., 2005; Rehmany et al., 2005). This implication is based on amino acid sequence similarity to the PEXEL element, which directs exported Plasmodium virulence proteins into host cell erythrocytes (Marti et al., 2004). Indeed, the RXLR motif can substitute for the PEXEL element and drive the export and host targeting of GFP chimera proteins in Plasmodium sp (Bhattacharjee et al., 2006). However, there is no experimental proof that the RXLR motif can direct virulence proteins from oomycete parasites into host plant cells. Although AVRk1 and its paralogues do not contain the RXLR motif, we noted a similar sequence ([R/K]VY[L/I]R) within the conserved central core of the proteins (Figure 6A). However, searches of sequenced fungal genomes with this motif did not identify any proteins known to be involved in pathogenicity. Intriguingly, a motif (RMLLR), which closely resembles that found in the AVRk1 paralogues, has been identified as a small subset of RXLR proteins from P. sojae (R.H.Y. Jiang and B.M. Tyler, personal communication).
The AVRk1 paralogues are not predicted to contain signal peptides for secretion via the endoplasmic reticulum (ER). AVR-ACE1 isolated from Magnaporthe grisea is also not predicted to be secreted, but this intracellular protein is involved in the formation of a secondary metabolite that is the presumed avirulence elicitor (Böhnert et al., 2004). All other fungal proteins isolated so far function directly as AVR elicitors and are predicted to contain signal peptides for secretion via the ER. The cell death and infection assays demonstrate that AVRk1 and AVRa10 are functional inside the host cell, suggesting that an alternative route for secretion of these proteins may exist in Bgh. Increasing numbers of virulence and other proteins exported by non-ER-dependent routes have been identified in animal parasites, including Candida albicans, Aspergillus fumigatus, and Leishmania sp (Denny et al., 2000; Denikus et al., 2005; Nombela et al., 2006). Specialized delivery mechanisms may have evolved in these parasites, or the virulence proteins may contain features required for secretion and targeting to host cells. Our results indicate that searches for AVR proteins and effectors in other fungal plant parasites should not be restricted to those that contain signal peptides.
Powdery mildew fungi are vigorous parasites that rapidly adapt to overcome host recognition. We have shown that AVRk1 and AVRa10 are avirulence genes encoding proteins that elicit host defenses and contribute to successful infection. If Bgh AVR proteins are effectors that can functionally substitute for each other, parasite vigor and host compatibility could be maintained by the combined action and constant selection of multiple AVRk1 paralogues. This would explain why individual AVR genes can be lost without a fitness penalty (Bronson and Ellingboe, 1986; Brown and Wolfe, 1990) and why R genes are so easily defeated.
METHODS
Fungal Isolates and Genetic Analysis
Cultures of Bgh were grown on barley leaf segments (∼2-cm long) kept on agar supplemented with benzimidazole (0.1 gL−1) in plastic boxes (120 mm × 80 mm × 15 mm) (Brown and Wolfe, 1990). Bgh isolates CC52 and CC148 were crossed individually with DH14, which has the opposite mating type, resulting in the formation of cleistothecia. Single ascospore cultures were obtained from the cleistothecia (Brown and Wolfe, 1990), producing 83 progeny isolates from the cross CC52 × DH14 and 51 progeny from CC148xDH14. The parental and progeny cultures were scored for their avirulence phenotypes on Hordeum 1063 and P17 (Mlk1), P09 (Mla10), and P12 (Mla22). DNA was also extracted from the parental and progeny cultures as previously described (Robinson et al., 2002). Genetic mapping was performed using AFLPs (Vos et al., 1995). Parent isolates were screened with AFLP primer combinations to identify polymorphic sequences, which were then mapped with the progeny set. A selected AFLP marker band (PAAMCAC-365) was used for probing a BAC library.
Construction and Probing of a BAC Library
DNA from 7-d-old conidia (10 g) of DH14 was extracted in 10 mM Tris, pH 8.0, 100 mM EDTA, RNase (50 μg/mL), SDS (0.5% [w/v]), and proteinase K (to 100 μg/mL) and purified by ultracentrifugation on a 10 to 40% sucrose gradient. The DNA was restricted with Sau3a, centrifuged at 40,000 rpm for 2 h to purify high molecular weight DNA, ligated into SACBII vector (Bendahmane, 1999), and transformed into Electromax (Invitrogen) competent cells. The 12,288 clones were organized into 384 well plates and gridded onto Hybond nylon membranes for probing. A selected AFLP marker band (PAAMCAC-365) was excised from a polyacrylamide gel, rehydrated in water for 16 h at 4°C, reamplified with primers P11 (5′-GACTGCGTACATGCAGAA-3′) and M48 (5′-GATGAGTCCTGAGTAACAC-3′), labeled, and used for probing. Probes were labeled with the kit Rediprime (Amersham Biosciences) using 20 μCi [32P]dCTP. Hybridization was performed overnight at 65°C (Sambrook et al., 1989). Probes were also prepared from the AVRk1 and Bgh homologues of Cg T1 (5′-ATCCAACGAAACCGCCCTCCCATACAGCAATGTAGGCGATGTCTCGGTTTCCACGCCACCCGCGGATGTTCTCGCGCACCTGCCTGCTGGAACTGTGGATCCACCATGCACTCAGCCTTCGAGTGTAAAGCTCCAACCAAGTGCCGAAACTGCGGAGGGCCCCACCAATCTGGCAGTCGAG-3′).
Hybridization Experiments with Other Formae Speciales of B. graminis and Erysiphe cichoracearum
DNA was extracted from conidiospores of B. graminis f sp tritici (isolated from wheat [Triticum aestivum]), f sp secalis (isolated from rye [Secale cereale]), f sp avenae (isolated from oat, [Avena sativa]), and an isolate from Agropyron sp (Wyand and Brown, 2003). DNA from conidiospores of E. cichoracearum infecting Arabidopsis thaliana was extracted using the Bio-Rad Chelex 100 method, developed for the isolation of DNA from minute quantities of tissue (Hirata and Takamatsu, 1996), and amplified by the GenomePhi DNA amplification kit (Amersham Biosciences). As negative control for the amplification of DNA from other microorganisms, the same procedure was performed with the wash of noninfected Arabidopsis leaves. Samples were denatured and blotted onto a Hybond membrane (Amersham Biosciences) using a Biodot apparatus (Bio-Rad) and hybridized with an AVRk1 probe as described above. PCR with primers AVRDEGF (5′-GTCGARGCMRCCCTTCWWCC-3′, where R = A+G, M = A+C, and W = A+T) and AVRDEGR (5′-GTGGCMCSWGTGCTTYTGAG-3′, where Y = C+T and S = G+C) was performed with DNA from the B. graminis formae speciales and E. cichoracearum.
Analysis of Polymorphism and Transcripts
Following sequencing, primer pairs from regions within the BAC 1817D were designed and used to amplify sequences from the isolates DH14, CC52, and CC148 with Expand long template (Roche Diagnostics) or Herculase (Stratagene) DNA polymerases. Nucleotide polymorphisms were mapped at loci CJR1-CJR3 by PCR with primers CJR1F (5′-ATATTGCCTTAATTGGTATG-3′), CJR1R (5′-TTCCAACCAGCATCATCTAC-3′), CJR2F (5′-ATTCGAAGATGACTCGAGTTTGA-3′), CJR2R (5′-GTCCAAGTGATCTATTGAATTCT-3′), CJR3F (5′-GTGAATTGTAGATGTGGATGTGG-3′), and CJR3R (5′-CCCTATTCTTGGAGGTGTTTG-3′). For analysis of polymorphism, DNA was amplified from the additional isolates GF13 and HLA (both Ak1) and GF4 and C24R9 (both Vk1) with primers CJR2F and CJR2R, which produces a 1775-bp amplicon spanning the AVRk1 gene. RNA was extracted from leaves of barley cultivar Golden Promise with an RNAeasy kit (Qiagen) 3 d after inoculation with CC148 and cDNA prepared with a SMART RACE cDNA kit (BD Biosciences). Nested primers to amplify AVRk1 by RT-PCR were R1 first, then R2 (for 5′ RACE-PCR) and R3 then R4 (for 3′ RACE-PCR). Nested primers for amplification of AVRa10 were R5 then R6 (5′ RACE-PCR) and R7 then R8 (3′ RACE-PCR). Sequences of these primers were as follows: R1 (5′-ACGGCGGGAATTTGTATGCTCCT-3′), R2 (5′-AGGAGCCCTTGGGAGAGGGTT-3′), R3 (5′-CTATACAACAACGCGCCGCCA-3′), R4 (5′-GCGTCG AAGCCACCCTTCTTT-3′), R5 (5′-GCCGAAACCGAGGTGATATTTG-3′) R6 (5′-GGACATCGTTCTCCTTCGCTTG-3′), R7 (5′-ATCCTCCAGCCCAGAAGATGCA-3′), R8 (5′-AAGGCAGCAGGAGCCGAAAAC-3′).
Nucleotide and Protein Characterization and Analysis
Nucleotide sequence analysis and contig assembly were performed with the STADEN package (http://www.mrc-lmb.cam.ac.uk/pubseq). Homologies were detected by probing nucleotide or protein sequences against the NCBI and EMBL databases (http://www.ncbi.nlm.nih.gov/BLAST/) and fungal sequence databases at COGEME (http://cogeme.ex.ac.uk/blast.html), the Broad Institute (http://www.broad.mit.edu/), and The Institute for Genomic Research (http://www.tigr.org/). Searches for the short amino acid motif [RK]VY[IL]R were performed with MOTIF search (http://motif.genome.jp/MOTIF2.html) using the COGEME EST database and the Broad Institute fungal sequences. Potential ORFs were detected by ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Sequence alignment was performed with MUSCLE (Edgar, 2004) and edited with Genedoc (distributed by K.B. Nicholas, H.B. Nicholas, and D.W. Deerfield, http://www.psc.edu/biomed/genedoc/gdfeedb.htm). The relative rates of nonsynonymous and synonymous nucleotide substitutions (Ka:Ks) were calculated by the program K-Estimator 6.0 (Comeron, 1999).
EMS Mutagenesis
Isolate CC148 was grown on barley (cv Golden Promise) leaf segments for 3 d, and the leaves were transferred onto agar containing 0.2% EMS for 16 h. In each experiment, 20 plastic boxes were used, each containing ∼50 leaf segments. Conidia were collected after 10 d and inoculated onto leaves of barley cv P09 (Mla10). A single colony was isolated after performing the experiment eight times. The colony was tested on P09 to confirm that it had the phenotype VIRa10 and on a set of 13 other differential varieties (Brown and Wolfe, 1990) to confirm that it had the same avirulence determinants as the original isolate, CC148.
Cell Death Assay Procedures
The cell death assay was performed by cobombarding two plasmids into plant cells, one expressing the reporter gene GFP under the control of maize (Zea mays) ubiquitin promoter (Pu hGFP-C3-N; Neilsen et al., 1999) and another expressing the test AVR gene. To prepare the test plasmids, AVRk1 and AVRa10 coding sequences were first amplified from genomic DNA of Bgh isolate CC148 by PCR with primers containing PstI and SacI restriction sites at the N and C termini, respectively. The amplification products were cloned into pGEMTeasy (Promega) and the sequences checked. After removal of GFP coding sequence by PstI and SacI, pUbi-GFP-Nos (for maize ubiqitin 1 promoter-GFP-Nos, obtained from K. Shirasu; Shen et al., 2003) was used as a backbone for cloning candidate AVR genes. AVRk1 and AVRra10 were removed from the pGEMTeasy vector by the restriction enzymes PstI and SacI and inserted into pUbi-GFP-Nos to create plasmids pCJR37 (AVRk1) and pPS1 (AVRra10). A null AVR plasmid containing the virulence (DH14) allele of AVRk1 with the mutation L14stop was also prepared (pCJR17). The AVR constructs were applied to gold particles (8.33 μg/mg gold) with Pu hGFP-C3-N reporter (1.8 μg/mg gold). This ratio of AVR to reporter plasmid was selected after preliminary trials to achieve the maximum number of GFP-expressing cells without diluting too far the concentration of AVR-expressing plasmid. The particles were bombarded onto leaves of the barley cultivars Pallas and Siri (both susceptible to all isolates; Kølster et al., 1986; Kølster and Stølen, 1987) and the near-isogenic resistant lines P17 and S17 (both Mlk1), P09 and S09 (both Mla10), P08B (Mla9), and P12 (Mla22) with a particle gun (Bio-Rad). Pairs of leaves (∼2 cm each in length) comprising Pallas or Siri and the test variety from the same near-isogenic series (P series is derived from Pallas, for example P09; S series is derived from Siri) were placed in each of seven replicate positions under a hepta manifold such that both varieties received a similar quantity of gold particles. The position of all leaves was marked before bombardment, and the number of cells expressing GFP in resistant varieties relative to susceptible varieties (GFP index) was determined after 44 h by viewing through a UV dissecting microscope (Leica). Statistical analysis was done by generalized linear modeling, and each experiment was performed five times.
Infection Assays
To determine the effect of AVR expression on infection, resistant and susceptible leaves were cobombarded at the same time as for cell death assay, with pCJR37 (AVRk1), pPS1 (AVRa10), or the null plasmid (pCJR17). The bombarded leaves were inoculated 24 h later with the isolates DH14 or CC52 (both Vk1 and Va10). Cells expressing GFP were scored for successful or failed infection after a further 40 h using the criteria described in Figures 4A to 4C using a UV microscope (Leica). These illustrative photomicrographs of living tissue were taken rapidly to minimize deterioration of the structures by UV light. The experiment was performed three times for AVRk1 and two times for AVRa10. Since there was no significant difference between results for the two isolates, means were predicted for varieties and plasmids pooled across isolates. The viability of fungal colonies was also determined by a spore-counting procedure. The infection assay was performed without the inclusion of the GFP-expressing plasmid (since no UV microscopy was involved). Ten days after infection, the concentration of conidiospores on the leaf was determined by tapping them onto a hemocytometer slide with the aid of a tube (1.5-cm diameter) positioned over the counting grid. The number of spores/mm2 in the hemocytometer grid directly represents the spores produced on the leaf. The experiment was performed three times.
Accession Numbers
Nucleotide and protein sequences of AVRk1 and AVRa10 mRNA have been deposited in the GenBank data library under accession numbers DQ679912 and DQ679913, respectively, and the genomic region cosegregating with AVRk1 under accession number DQ679914.
Acknowledgments
This research was supported by the Biotechnology and Biological Sciences Research Council, the Department for Environment, Food, and Rural Affairs, the Gatsby Charitable Trust, the European Union Framework Six Program (BIOEXPLOIT), the Hellenic Republic Studentships Foundation (I.K.Y.), and the intra-European Marie Curie fellowship program. We thank David Studholme for protein structure predictions and Marielle Vigouroux for nucleotide and protein sequence analysis.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christopher J. Ridout (christopher.ridout@bbsrc.ac.uk).
References
- Alfano, J.R., and Collmer, A. (2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42 385–414. [DOI] [PubMed] [Google Scholar]
- Allen, R.L., Bittner-Eddy, P.D., Grenvitte-Briggs, L.J., Meitz, J.C., Rehmany, A.P., Rose, L.E., and Beynon, J.L. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306 1957–1960. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, M.R., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognised in the host cytoplasm. Proc. Natl. Acad. Sci. USA 102 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A. (1999). Zero-background plasmid vector for BAC library construction. Biotechniques 26 228–232. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S., Hiller, N.L., Liolios, K., Win, J., Kanneganti, T., Young, C., Kamoun, S., and Halder, K. (2006). The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog 2 e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnert, H.U., Fudal, I., Dioh, W., Tharreau, D., Nottenghem, J., and Lebrun, M. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson, C.R., and Ellingboe, A.H. (1986). The influence of four unnecessary genes for virulence on the fitness of Erysiphe graminis f. sp tritici. Phytopathology 76 154–158. [Google Scholar]
- Brown, J.K.M. (2003). Little else but parasites. Science 299 1680–1681. [DOI] [PubMed] [Google Scholar]
- Brown, J.K.M., and Høvmoller, M.S. (2002). Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297 537–541. [DOI] [PubMed] [Google Scholar]
- Brown, J.K.M., and Jessop, A.C. (1995). Genetics of avirulences in Erysiphe graminis f. sp. hordei. Plant Pathol. 44 1039–1049. [Google Scholar]
- Brown, J.K.M., and Wolfe, M.S. (1990). Structure and evolution of a population of Erysiphe graminis f. sp hordei. Plant Pathol. 39 376–390. [Google Scholar]
- Caffier, V., de Vallavieille-Pope, C., and Brown, J.K.M. (1996). Segregation of avirulences and genetic basis of infection types in Erysiphe graminis f. sp. hordei. Phytopathology 86 1112–1121. [Google Scholar]
- Catanzariti, A., Dodds, P.N., Lawrence, G.J., Ayliffe, M.A., and Ellis, J.G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaure, P., Gurr, S.J., and Spanu, P. (2000). Stable transformation of Erysiphe graminis, an obligate biotrophic pathogen of barley. Nat. Biotechnol. 18 205–207. [DOI] [PubMed] [Google Scholar]
- Comeron, J.M. (1999). K-estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15 763–764. [DOI] [PubMed] [Google Scholar]
- Denikus, N., Orfaniotou, F., Wulf, G., Lehmann, P.F., Monod, M., and Reichard, U. (2005). Fungal antigens expressed during invasive aspergillosis. Infect. Immun. 73 4704–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny, P.W., Gokool, S., Russell, D.G., Field, M.C., and Smith, D.F. (2000). Acylation dependendent protein export in Leishmania. J. Biol. Chem. 275 11017–11025. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A.M., Ayliffe, M.A., and Ellis, J.G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced space and time complexity. BMC Bioinformatics 5 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., and Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 100 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of gene for gene concept. Annu. Rev. Phytopathol. 9 275–296. [Google Scholar]
- Gout, L., Fudal, I., Kuhn, M., Blaise, F., Eckert, M., Cattolico, L., Balesdent, M., and Rouxel, T. (2006). Lost in the middle of nowhere: The AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol. Microbiol. 60 67–80. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6 201–211. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S., Vinatzer, B.A., Sarker, S.F., Ranall, M.V., Kettler, G., and Greenberg, J.T. (2002). A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295 1722–1726. [DOI] [PubMed] [Google Scholar]
- Halterman, D.A., Wei, F.S., and Wise, R.P. (2003). Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 131 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman, D.A., and Wise, R.P. (2004). A single-amino acid substitution in the sixth leucine-rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 38 215–226. [DOI] [PubMed] [Google Scholar]
- He, C., Nourse, J.P., Kelemu, S., Irwin, J.A.G., and Manners, J.M. (1996). CgT1: A non-LTR retrotransposon with restricted distribution in the fungal phytopathogen Colletotrichum gloeosporioides. Mol. Gen. Genet. 252 320–331. [DOI] [PubMed] [Google Scholar]
- Hirata, T., and Takamatsu, S. (1996). Nucleotide sequence diversity of rDNA internal transcribed spacers extracted from conidia and cleistothecia of several powdery mildew fungi. Mycoscience 37 283–288. [Google Scholar]
- Janjusevic, R., Abramovitch, R.B., Martin, G.B., and Stebbins, C.E. (2006). A bacterial inhibitor of host defenses is an E3 ubiquitin ligase. Science 311 212–225. [DOI] [PubMed] [Google Scholar]
- Jensen, J., Jensen, H.P., and Jørgensen, J.H. (1995). Linkage studies of barley powdery mildew virulence loci. Hereditas 122 197–209. [Google Scholar]
- Jia, Y., McAdams, S.S., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1994). Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13 97–119. [Google Scholar]
- Kemen, E., Kemen, A.C., Rafiqui, M., Hempel, U., Mendgen, K., Hahn, M., and Voegele, R.T. (2005). Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant Microbe Interact. 18 1130–1139. [DOI] [PubMed] [Google Scholar]
- Kølster, P., Munk, L., Stølen, O., and Løhde, J. (1986). Near isogenic barley lines with genes for resistance to powdery mildew. Crop Sci. 26 903–907. [Google Scholar]
- Kølster, P., and Stølen, O. (1987). Barley isolines with genes for resistance to Erysiphe graminis f. sp. hordei in the recurrent parent Siri. Plant Breed. 98 79–82. [Google Scholar]
- Leister, R.T., Ausubel, F.M., and Katagiri, F. (1996). Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis resistance genes RPS2 and RPM1. Proc. Natl. Acad. Sci. USA 93 15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A.L., Wang, M.L., Zhou, R.H., Kong, X.Y., Huo, N.X., Wang, W.S., and Jia, J.Z. (2005). Comparative analysis of early H2O2 accumulation in compatible and incompatible wheat powdery mildew interactions. Plant Pathol. 54 308–316. [Google Scholar]
- Maroof, M.A.S., Zhang, Q., and Biyashev, R.M. (1994). Molecular marker analyses of powdery mildew resistance in barley. Theor. Appl. Genet. 88 733–740. [DOI] [PubMed] [Google Scholar]
- Marti, M., Good, R.T., Rug, M., Knuepfer, E., and Cowman, A.F. (2004). Targeting malaria virulence and remodelling proteins to the host erythrocyte. Science 306 1930–1933. [DOI] [PubMed] [Google Scholar]
- Neilsen, K., Olsen, O., and Oliver, R. (1999). A transient expression system to assay putative antifungal genes on powdery mildew infected barley. Physiol. Mol. Plant Pathol. 54 1–12. [Google Scholar]
- Nombela, C., Gil, C., and Chaffin, L. (2006). Non-conventional protein secretion in yeast. Trends Microbiol. 14 15–21. [DOI] [PubMed] [Google Scholar]
- Prats, E., Carver, T.L.W., Lyngkjaer, M.F., Roberts, P.C., and Zeyen, R.J. (2006). Induced inaccessibility and accessibility in the oat powdery mildew system: Insights gained from use of metabolic inhibitors and silicon nutrition. Mol. Plant Pathol. 7 47–59. [DOI] [PubMed] [Google Scholar]
- Rehmany, A.P., Gordon, A., Rose, L.E., Allen, R.L., Armstrong, M.R., Whisson, S.C., Kamoun, S., Tyler, B.M., Birch, P.R.J., and Beynon, J.L. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, H.L., Ridout, C.J., Sierotzki, H., Gisi, U., and Brown, J.K.M. (2002). Isogamous, hermaphroditic inheritance of mitochondrion-encoded resistance to Qo inhibitor fungicides in Blumeria graminis f. sp. tritici. Fungal Genet. Biol. 36 98–106. [DOI] [PubMed] [Google Scholar]
- Roden, J.A., Belt, B., Ross, J.B., Tachibana, T., Vargas, J., and Mudgett, M.B. (2004). A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA 101 16624–16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer, L., Guttman, D.S., and Dangl, J.L. (2004). Diverse evolutionary mechanisms shape the type III effector virulence factor repertiore in the plant pathogen Pseudomonas syringae. Genetics 167 1341–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, H.C., van't Klooster, J.W., van der Hoorn, R.A., Joosten, M.H., Jones, J. D., and de Wit, P.J. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786.Erratum. Science 310, 54. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schiffer, R., Gorg, R., Jarosch, B., Beckhove, U., Bahrenberg, G., Kogel, K.H., and Schulze-Lefert, P. (1997). Tissue dependence and differential cordyceptin sensitivity of race-specific resistance responses in the barley powdery mildew interaction. Mol. Plant Microbe Interact. 10 830–839. [Google Scholar]
- Schulze-Lefert, P., and Panstruga, R. (2003). Establishment of biotrophy by parasitic fungi and reprogramming of host cells for disease resistance. Annu. Rev. Phytopathol. 41 641–667. [DOI] [PubMed] [Google Scholar]
- Shan, W.X., Cao, M., Dan, L.U., and Tyler, B.M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant Microbe Interact. 17 394–403. [DOI] [PubMed] [Google Scholar]
- Shen, Q.H., Zhou, F.S., Bieri, S., Haizel, T., Shirasu, K., and Schulze-Lefert, P. (2003). Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skamnioti, P., and Ridout, C.J. (2005). Microbial avirulence determinants: Guided missiles or antigenic flak. Mol. Plant Pathol. 6 551–559. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A., Westerink, N., Francoijs, K.J., Roth, R., Woesterenenk, E., Boeren, S., de Wit, P.J.G.M., Joosten, M.H.A.J., and Vervoot, J. (2003). Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278 27340–27346. [DOI] [PubMed] [Google Scholar]
- Vos, P., Hogers, R., Bleeker, M., Reijans, M., van der Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., and Zabeau, M. (1995). AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 23 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y.D., Collinge, D.B., Smedegaard-Petersen, V., and Thordal-Christensen, H. (1996). Characterization of the transcript of a new class of retroposon-type repetitive element cloned from the powdery mildew fungus, Erysiphe graminis. Mol. Gen. Genet. 250 477–482. [DOI] [PubMed] [Google Scholar]
- Wyand, R.A., and Brown, J.K.M. (2003). Genetic and forma specialis diversity in Blumeria graminis of cereals and its implications for host-pathogen co-evolution. Mol. Plant Pathol. 4 187–198. [DOI] [PubMed] [Google Scholar]
- Yahiaoui, N., Srichumpa, P., Dudler, R., and Keller, B. (2004). Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37 528–538. [DOI] [PubMed] [Google Scholar]
- Zhou, F.S., Kurth, J.C., Wei, F.S., Elliott, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R., and Schulze-Lefert, P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signalling pathway. Plant Cell 13 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]