Abstract
In the phytopathogenic fungus Ustilago maydis, pathogenic development is controlled by a heterodimer of the two homeodomain proteins bE and bW, encoded by the b-mating-type locus. We have identified a b-dependently induced gene, clampless1 (clp1), that is required for the proliferation of dikaryotic filaments in planta. We show that U. maydis hyphae develop structures functionally equivalent to clamp cells that participate in the distribution of nuclei during cell division. In clp1 mutant strains, dikaryotic filaments penetrate the plant cuticle, but development is stalled before the first mitotic division, and the clamp-like structures are not formed. Although clp1 is immediately activated upon b-induction on the transcriptional level, nuclear-localized Clp1 protein is first observed at the stage of plant penetration prior to the first cell division. Induced expression of clp1 strongly interferes with b-dependent gene regulation and blocks b-dependent filament formation and b-dependent cell cycle arrest. We speculate that the Clp1 protein inhibits the activity of the bE/bW heterodimer to facilitate the cell cycle progression during hyphal growth.
INTRODUCTION
The phytopathogenic Basidiomycete Ustilago maydis is the causal agent of the smut disease on maize (Zea mays). Similar to other smut fungi, U. maydis displays a dimorphic life style and grows in its haploid phase as a saprophytic yeast. Sexual development is initiated by fusion of two haploid sporidia. The resulting dikaryon is filamentous; however, at the initial stage only the apical cell of the hyphae is filled with cytoplasm, while the distal part consists of empty sections. Cell division is stalled until the dikaryon invades the plant via a specialized infection structure, the appressorium, and only after plant penetration is the true filament with multiple septated compartments formed. At the initial stage of infection, the hyphae traverse plant cells without an apparent host defense response. Plant cells remain alive until late in the infection process. At later stages, plant tumors are induced by the fungus, followed by massive proliferation of fungal hyphae, nuclear fusion, and the formation of diploid teliospores. Meiosis occurs upon spore germination, and a septated basidia is formed, from which haploid sporidia are released by successive budding events (for review, see Banuett, 1995; Kahmann and Kämper, 2004).
The crucial step for the infection process is the formation of the dikaryon. This process is controlled by the two mating type loci, termed a and b (Rowell and DeVay, 1954; Rowell, 1955; Holliday, 1961; Puhalla, 1970; Day et al., 1971). The biallelic a-locus encodes a pheromone/receptor system that is responsible for cell recognition and cell fusion of the sporidia (Bölker et al., 1992; Spellig et al., 1994). After formation of the dikaryon, different alleles of the multiallelic b-mating type are the prerequisite to initiate the biotrophic stage (Holliday, 1961; Puhalla, 1968; Banuett and Herskowitz, 1989). The b-locus encodes two distinct homeodomain proteins, termed bE and bW, that form a heterodimer, but only if they derive from different alleles (Schulz et al., 1990; Gillissen et al., 1992; Kämper et al., 1995). The central role of the bE/bW heterodimer for pathogenic development was demonstrated by the construction of haploid strains with a hybrid b-locus expressing different alleles of bE and bW (Bölker et al., 1995). Such strains are solopathogenic (i.e., they are able to infect the host plant without a mating partner). The bE/bW heterodimer has been shown to bind elements (b-binding sites) in the promoter region of b-responsive genes (Romeis et al., 2000; Brachmann et al., 2001). It is conceivable that genes that are either directly regulated by the bE/bW heterodimer or that are indirectly regulated via a b-dependent regulatory cascade are involved in the establishment of the biotrophic phase. In several attempts, 21 b-regulated genes have been identified (Bohlmann et al., 1994; Schauwecker et al., 1995; Urban et al., 1996; Wösten et al., 1996; Brachmann et al., 2001, 2003), but with the exception of the mitogen-activated protein kinase kpp6, which is involved in appressorium formation (Brachmann et al., 2003), none of the genes tested so far have been linked to pathogenic development. A more detailed insight into the b-dependent processes was achieved using DNA arrays that allow monitoring of gene expression of ∼90% of all U. maydis genes (M. Scherer and J. Kämper, unpublished data). Array analysis identified ∼350 b-responsive genes that change their expression within a 12-h time course after formation of an active bE/bW heterodimer. Several of these b-regulated genes are involved in cell cycle control, mitosis, and DNA replication, consistent with the observed cell cycle arrest after b-induction that is released only upon plant infection (M. Scherer and J. Kämper, unpublished data).
In most dikaryotic Basidiomycetes, the proper distribution of the two genetically distinct nuclei during mitotic cell divisions is ensured by the formation of clamps. Clamp cells develop at the most apical cell of the dikaryotic hyphae at the position where the future septum will be formed. The process starts with the formation of a backward-directed, hook-formed cell. Subsequently, the two nuclei in the lateral cell undergo a synchronous division. From the first dividing pair of nuclei, one nucleus migrates into the clamp primordium, while the other moves toward the apex of the hyphae. From the second dividing pair, one nucleus moves backward to the distal part of the hypha, while the other moves to the apical part. A septum formed at the base of the clamp cell entraps one nucleus within the clamp cell; a second septum is formed subapical to the clamp cell, creating an apical compartment with two nuclei and a distal compartment with one nucleus. Subsequently, the clamp cell fuses with the subapical compartment to reconstitute its dikaryotic status. In Coprinopsis cinerea and Schizophyllum commune, clamp cell initiation and nuclear division are controlled by the A-mating type, while clamp cell fusion is controlled by the B-mating type (Swiezynski and Day, 1960a, 1960b; Raper, 1966). The A-locus encodes two classes of homeodomain transcription factors that function in a similar way as the bE/bW proteins in U. maydis (reviewed in Casselton and Olesnicky, 1998; Hiscock and Kües, 1999; reviewed in Casselton, 2002). The B-locus encodes pheromones and the corresponding receptors that are discussed to mediate the fusion of the clamp via an extracellular communication of the genetically diverse cells (Kües, 2000; Brown and Casselton, 2001; Kothe, 2001).
In U. maydis, clamp-like structures are present in the dikaryotic stage; however, it has not been observed that these structures are involved in the distribution of nuclei. In particular, the fact that a fusion of the clamp-like cells to the distal hyphal compartment has not been observed prompted the conclusion that these structures are not the functional homologues of clamps (Snetselaar and Mims, 1994; Banuett and Herskowitz, 1996).
Here, we show that the clamp-like structures are involved in the distribution of nuclei in the hyphae of U. maydis. We have identified a b-dependently expressed gene encoding a protein with similarities to the Clampless1 (Clp1) protein from C. cinerea. In this higher Basidiomycete, it is necessary and sufficient to induce clamp formation (Inada et al., 2001). In U. maydis, clp1 is required for the formation of the clamp-like structures. Strains deleted for clp1 are still able to penetrate the plant cuticle; however, the tip cells of the filaments are unable to undergo mitotic divisions, resulting in a block at the earliest stage of biotrophic development. Induced expression of clp1 leads to a block of b-dependent filament formation and b-dependent cell cycle arrest. Concomitantly, b-dependent genes are downregulated. Most likely, the Clp1 protein is needed to stall the b-induced cell cycle control as a prerequisite for mitotic divisions.
RESULTS
U. maydis clp1 Is b-Dependently Regulated
To identify genes that are regulated in dependency of the bE/bW heterodimer, we have performed microarray experiments with custom Affymetrix arrays that cover ∼90% of the predicted 6902 U. maydis genes. In the haploid strains AB31 and AB33, the bE1 and bW2 genes are under the control of the arabinose-inducible crg1 promoter and the nitrate-inducible nar1 promoter, respectively (Brachmann et al., 2001), allowing the initiation of b-dependent development in axenic culture. The change in expression upon bE1/bW2 gene induction was monitored independently in both strains during a 12-h time course using strains AB32 and AB34, which harbor the incompatible combination bE2 and bW2, as controls. We identified ∼350 genes that were induced or repressed more than twofold at least at one time point during the 12-h time course in both AB31 and AB33. A detailed analysis of the DNA array experiments will be published elsewhere.
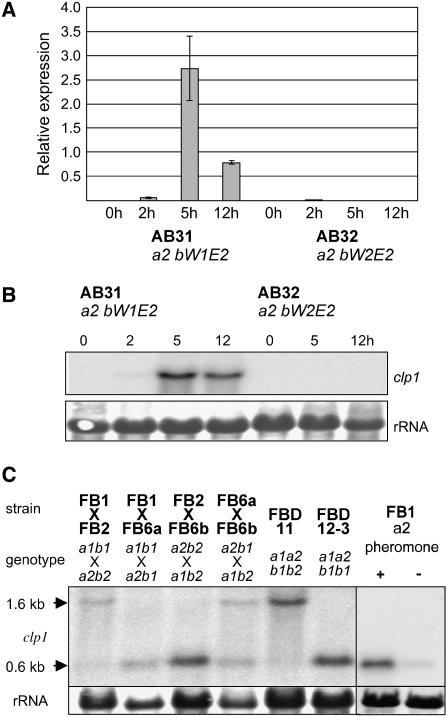
One of the genes identified as b-dependently regulated is clp1 (annotated as um02438 at the MIPS Ustilago maydis DataBase; http://mips.gsf.de/genre/proj/ustilago). The array experiment suggests that expression of clp1 is induced early after formation of the bE1/bW2 heterodimer (data not shown). In strain AB31 (a1 bE1crg1P/bW2crg1P), clp1 expression can be detected 2 h after b-induction by real-time RT PCR and peaks after 5 h. In the control strain AB32 (a1 bE2crg1P/bW2crg1P), clp1 expression is not detectable (Figure 1A). The b-dependent expression profile of clp1 in AB31 was confirmed by RNA gel blot analysis (Figure 1B). Similarly, when an active bE/bW heterodimer is formed upon mating of the two compatible haploid wild-type strains FB1 (a1b1) and FB2 (a2b2), a 1.6-kb clp1 transcript is induced. When strains with compatible a but incompatible b alleles are mated, this transcript cannot be detected (Figure 1C). Surprisingly, RNA gel blot analysis revealed the presence of a second, smaller clp1 transcript of ∼0.6 kb in size. The small transcript was detected (1) in mating reactions of strains with compatible a loci, (2) in the diploid strain FBD12-3 (a1a2b1b1), and (3) upon stimulation of FB1 (a1b1) with the a2 pheromone, indicating an a-dependent regulation (Figure 1C).
Figure 1.
a- and b-Dependent Expression of clp1.
(A) and (B) Analysis of clp1 expression after induction of compatible (strain AB31) and incompatible (strain AB32) combinations of bE and bW. Samples were taken before and 2, 5, and 12 h after induction with arabinose.
(A) Real-time quantitative RT PCR analysis. clp1 expression was measured relative to the constitutively expressed gene for peptidylprolyl isomerase (ppi; um03726). Shown are the mean values and standard deviation of four technical replicates.
(B) RNA gel blot analysis; 10 μg of total RNA was loaded per lane. As a loading control, the filter was stained with methylene blue to visualize blotted rRNA.
(C) Wild-type strains FB1, FB2, FB6a, and FB6b and the diploid strains FBD11 and FBD12-3 were grown for 48 h on complete medium containing charcoal as indicated, prior to RNA isolation. FB1 was treated with pheromone a2 for 75 min. The probe used for hybridization covered 744 bp of the 5′-region of the clp1 ORF. Note that only in combinations with an active b1/b2 combination is the 1.6-kb clp1 transcript visible; the smaller 0.6-kb transcript is formed in combinations with an active a1/a2 combination.
U. maydis Clp1 Is Similar to the Clp1 Protein from C. cinerea
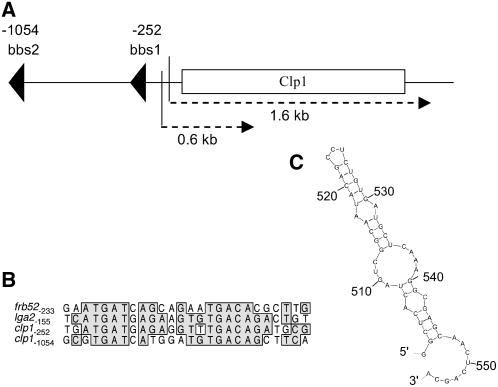
Inspection of the clp1 cDNA obtained by 5′- and 3′-rapid amplification of cDNA ends did not reveal the presence of introns. clp1 has 5′- and 3′-untranslated regions of 88 and 94 bp, respectively, and the gene encodes a potential protein of 444 amino acids. Two sequence motifs with high similarity to the b-binding sites in the promoter regions of the directly b-regulated genes lga2 (Romeis et al., 2000) and frb52 (Brachmann et al., 2001) at positions –252 and –1054 suggest that clp1 is a direct b-target gene (Figures 2A and 2B). The deduced protein sequence of clp1 shows weak similarity to Clp1 of C. cinerea (23% identities and 41% positives in a window of 282 amino acids at the C terminus). Clp1 in C. cinerea was found to be essential for A-dependent clamp formation, and forced expression of Clp1 bypassed the requirement for an active A-pathway for clamp formation (Inada et al., 2001). The genome of C. cinerea contains one additional predicted gene with similarity to clp1. Likewise, the genome of the second sequenced Homobasidiomycete, Phanerochaete chrysosporium, contains two sequences with clp1 similarity, while that of Cryptococcus neoformans (belonging to the Filobasidiales) contains only one. Comparison of the deduced proteins did not reveal any conserved regions shared among all Clp1-related proteins nor the presence of any known structural motifs (see Supplemental Figure 1 online). Outside the Basidiomycete phylum, no sequences with similarities to Clp1 were found.
Figure 2.
Structure of the clp1 Gene.
(A) Schematic presentation of the clp1 locus. Two potential b-binding sites and the two different transcripts (1.6 and 0.6 kb) are indicated.
(B) Alignment of the two potential b-binding sites within the clp1 promoter to the binding sites within the lga2 and frb52 promoters (Romeis et al., 2000; Brachmann et al., 2001). Numbers indicate the position of the sequences with respect to the ATG start codon of the genes. Identical nucleotides are boxed.
(C) Potential secondary structure at the 3′-end of the small 0.6-kb transcript. The secondary structure was predicted using the mfold program (Zuker, 2003). The calculated initial ΔG for the structure is −16.2 kcal/mol.
To elucidate whether the a-dependent transcript has the capacity to encode a protein, we aimed to isolate the corresponding cDNA from pheromone-treated FB1 cells; under this condition, only the small transcript is detectable. Because all attempts using poly(dT) primers for cDNA synthesis failed, we used circular RT-PCR (cRT-PCR), a method that is suitable to generate cDNA from nonpolyadenylated RNAs (Couttet et al., 1997). We obtained a cDNA of 555 bp that covers 437 bp of the clp1 open reading frame (ORF) and an additional 118 bp upstream region (Figure 2A). Sequence inspection revealed that the a-dependent transcript lacks a poly(A) tail; however, the 3′-end of the mRNA is predicted to form a stable secondary structure (Figure 2C). The ORF deduced from the short transcript lacks an in-frame stop codon and thus has no coding potential.
clp1 Is Required for Pathogenic Development
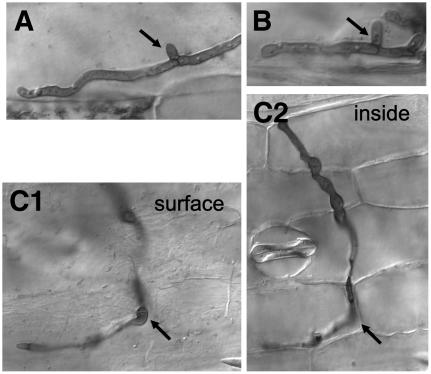
The clp1 ORF was replaced by a hygromycin resistance cassette in U. maydis wild-type stains FB1 (a1b1) and FB2 (a2b2) and the haploid solopathogenic strain SG200 (a1mfa2bE1bW2). Deletion of clp1 did not cause any obvious phenotype in haploid cells growing in axenic culture. The growth rate in minimal or complete media was not altered, and the mating reaction of the FB1Δclp1 and FB2Δclp1 (UVO161 and UVO162, respectively) strains was indistinguishable from that of the respective wild-type strains (data not shown). However, pathogenic development was strongly affected. Maize plants infected with a mixture of the strains FB1Δclp1 and FB2Δclp1 still showed chlorosis, but formation of tumors was not observed (Table 1). Similar results were obtained for the clp1 deletion in the solopathogenic strain SG200, indicating (1) that the pathogenicity defect is not due to a defect in cell recognition or cell fusion and (2) that the requirement for clp1 during pathogenic development is not connected to the dikaryotic stage per se. To determine at which stage the clp1 mutants are blocked during pathogenic development, fungal cells in infected leaves were visualized by staining with Chlorazole Black E. As control for the infection experiments, we used the solopathogenic strain UMS71, a SG200 derivative expressing enhanced green fluorescent protein (eGFP) linked to a nuclear localization sequence (see below). Three days after inoculation, UMS71 had entered the plant via appressoria-like penetration structures, and massive proliferation of fungal material was observed in the leaf tissue. Typical clamp-like structures with Y-shaped septa (Snetselaar and Mims, 1994; Banuett and Herskowitz, 1996) were frequently found (Figure 3A). The clp1 deletion strain UMS71Δclp1 (UMS67) showed no defect in appressoria formation and was able to invade the leaf tissue (Figure 3C1). However, no proliferation of fungal cells could be observed. Mostly, invading hyphae were short and restricted to single plant cells of the outer epidermal layer. Only in few cases did we observe that the hyphae surpassed more than one plant cell (Figure 3C2). Clamp-like structures were completely absent in the clp1 mutant strain (Figure 3C2).
Table 1.
Pathogenicity of clp1 Deletion Strains
| Strains Inoculated | No. of Plants | Tumor Formation (%) |
|---|---|---|
| FB1 × FB2 | 61 | 96 |
| FB1Δclp1 × FB2Δclp1 | 62 | 0 |
| SG200 | 18 | 94 |
| SG200Δclp1 | 38 | 0 |
Six-day-old maize plants were inoculated with the strains indicated, and tumor formation was scored 10 to 14 d after infection.
Figure 3.
clp1 Is Required for Proliferation in Planta during Pathogenic Development.
Infected leaves were stained with Chlorazole Black E 3 d after inoculation. Hyphae of the solopathogenic strains UMS71 (a1mfa2bE1/bW2:NLS-3xeGFP) (A) and CL13 (a1bE1/bW2) (B) form clamp-like structures in planta (arrows). Hyphae of strain UMS71Δclp1 can form appressoria ([C1], arrow; focus on the leaf surface) and penetrate, but clamp-like structures are not observed. Fungal proliferation stops at latest when the hyphae have grown through two to three plant cells ([C2]; focus within the epidermal cell layer). The site of penetration is marked with an arrow.
Clamp-Like Structures in U. maydis Participate in Nuclear Distribution in Heterodikaryotic Hyphae
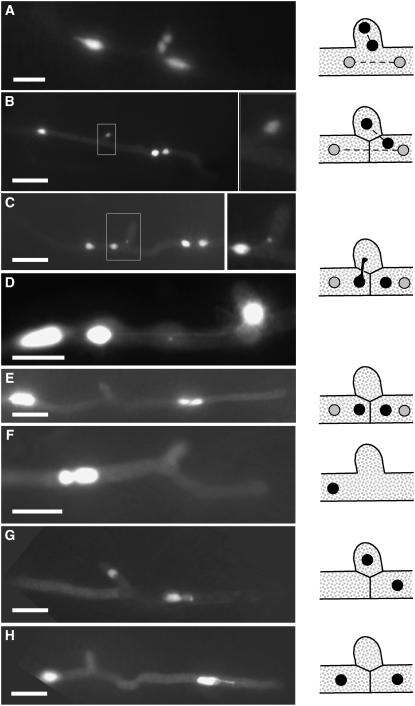
Although clamp-like structures formed by U. maydis during proliferative growth in planta have been described previously, a function with respect to nuclear distribution during cell divisions of the heterodikaryon has been excluded because the clamp cells do not fuse with the distal hyphal compartment (Snetselaar and Mims, 1994; Banuett and Herskowitz, 1996). To allow live imaging of nuclei in the dikaryotic hyphae of U. maydis during plant infection, we fused the ORF of eGFP arranged triple in tandem orientation (3xeGFP) to the nuclear localization signal (NLS) from VP16 that has been proven to be functional in U. maydis (Quadbeck-Seeger et al., 2000). NLS-3xeGFP was expressed under control of the mig2-5 promoter, which is strongly induced in planta (Farfsing et al., 2005; Basse and Farfsing, 2006). When a mixture of strains FB1 and FB2, both expressing NLS-3xeGFP (UMS73 and UMS74, respectively), were coinjected into maize plants, nuclei showed strong fluorescence after the heterodikaryotic hyphae had penetrated the plant. At 48 h after inoculation, the clamp-like structures were observed at various stages of development. Initially, a branch-like structure was formed; upon mitotic cell division, (1) either two nuclei were found within the branch, and the other two nuclei were positioned distal and proximal to the position of the branch (Figure 4A), or (2) only one nucleus was localized in the branch, one distal to the branch and two in the proximal part of the hypha (Figure 4B). Surprisingly, the nucleus located within the branch remained at this position in situations where the cell of the clamp-like structure was separated from the main hypha by a Y-shaped septum. By that, the nucleus appears to be trapped in the clamp-like structure (Figure 4B). In other Basidiomycetes, the subapical cell and the clamp fuse to release the trapped nucleus. Concomitantly with the observations of others, we did not observe a visible dissolution of the cell wall between the cell of the clamp-like structure and the subapical cell in U. maydis. Nevertheless, the nucleus moved out of the clamp-like structure into the subapical hyphal compartment. During this process, the nucleus often appeared as an elongated structure, of which parts still resided in the clamp-like structure, while the other part could be localized within the distal cell compartment; both parts of the nucleus were connected via thin projections (Figures 4C and 4D). Finally, the apical and subapical cell contained one pair of nuclei each (Figure 4E). Our data clearly demonstrate that clamp-like structures of U. maydis participate in nuclear distribution during division of heterodikaryotic hyphae. The solopathogenic strain UMS71 (SG200:NLS-3xeGFP) formed clamp-like structures as well (Figures 4F to 4H), although it proliferates in planta as a haploid monokaryon. During proliferation in planta, we observed that upon nuclear division one of the two daughter nuclei migrates into the clamp-like structure, while the second nucleus is positioned in the apical part of the hypha (Figure 4G). After formation of the Y-shaped septum, the nucleus migrates from the clamp-like structure to the distal cell, similar to the situation found in the dikaryon (Figure 4H).
Figure 4.
Clamp-Like Structures Participate in Nuclear Distribution.
(A) to (E) Dikaryotic hyphae of crosses of strains UMS73 (a1b1) and UMS74 (a2b2).
(F) to (H) Hyphae of the haploid solopathogenic strain UMS71 (a1mfa2bE1/bW2); all strains express a nuclear-localized NLS-3xeGFP; nuclei are visualized by fluorescence microscopy. Samples were taken 48 h after inoculation of maize plants.
Insets in (B) and (C) give an enlarged view of the clamp cell shown. The distribution of the nuclei with respect to the clamp cell is given in the illustrations at the right. For a detailed description, see text. Bars = 5 μm in (A) and (D) and 10 μm in all other panels.
In the Basidiomycetes C. cinerea and S. commune, it is known that the formation of the clamp is dependent on the A-mating type locus, while fusion of the clamp cell with the distal cell compartment is dependent on the pheromone receptor system encoded by the B-locus (Kües, 2000; Brown and Casselton, 2001; Kothe, 2001). In U. maydis, we observed the formation of clamp-like structures in the solopathogenic strain CL13 (a1bE1bW2); since this strain is harboring only a single a-locus, we must assume that in contrast with other Basidiomycetes, the pheromone/receptor system is dispensable for the function of the clamp cells in U. maydis (Figure 3B).
Clp1 Is Localized in the Nucleus and Is Posttranscriptionally Regulated
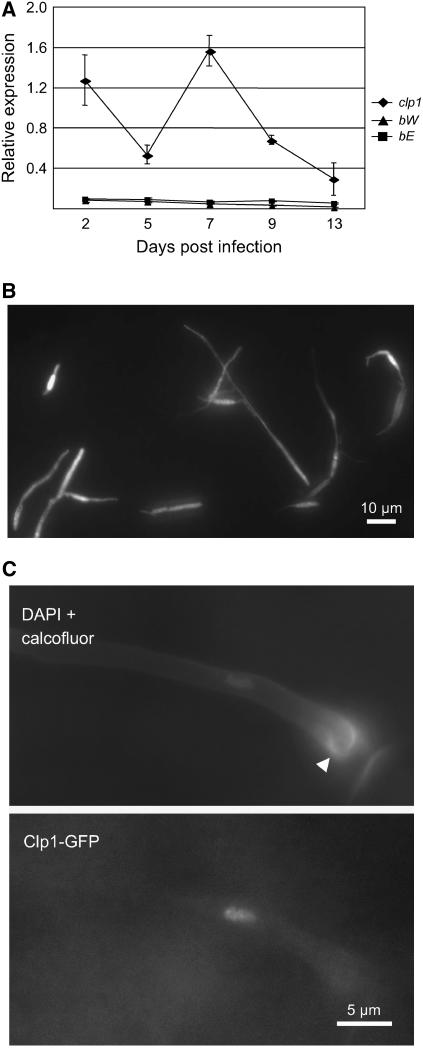
The expression of clp1 during pathogenic development was analyzed by real-time RT-PCR using samples of tumor material collected 2, 5, 7, 9, and 13 d after infection with a mixture of the compatible wild-type strains FB1 (a1b1) and FB2 (a2b2). clp1 expression was detectable at all time points, with a peak at 7 d after inoculation (Figure 5A). To address clp1 expression on the plant surface prior to infection, the ORF was replaced with the ORF of the eGFP gene in the solopathogenic strain SG200. In the resulting strain, UMS76, we observed a bright GFP fluorescence in all hyphae on the leaf surface, indicating that clp1 is expressed before hyphae penetrate the plant cuticle (Figure 5B).
Figure 5.
clp1 Expression during Pathogenic Development.
(A) Real-time quantitative RT-PCR analysis of clp1, bW, and bE expression in planta. Samples from maize plants were taken 2, 5, 7, 9, and 13 d after infection with a mixture of compatible wild-type cells (FB1 × FB2). Expression was measured in relation to the constitutively expressed ppi gene. Shown are the mean values and standard deviation of four technical replicates.
(B) clp1 gene expression during the early infection phase was monitored using a clp1 promoter:eGFP reporter construct. The clp1 ORF was replaced by eGFP in the solopathogenic strain SG200 by homologous recombination to create strain UMS76. eGFP expression of cells on a leaf surface 24 h after inoculation was monitored by fluorescence microscopy.
(C) Clp1:3xeGFP localizes to the nucleus of cells with appressoria (arrowhead). A functional clp1:3xeGFP fusion construct was expressed under control of the native clp1 promoter in the strain SG200. Cells on the leaf surface were stained with 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor to visualize nuclei and appressoria (top panel). GFP fluorescence (bottom panel) shows the localization of the Clp1:3xeGFP fusion protein in the nucleus of a cell that has developed an appressorium.
To analyze the subcellular localization of the protein, the C terminus of Clp1 was fused to triple eGFP. The fusion construct was integrated into the clp1 locus of the solopathogenic strain SG200, replacing the native clp1 gene. The fusion protein was shown to be functional, since the resulting strain UMS48 infects maize plants with a rate similar to that of the respective unmodified strain SG200 (data not shown). When grown on the surface of maize leaves, the Clp1:3xeGFP fusion protein was localized in the nucleus of appressoria-like structures (Figure 5C); however, expression of the fusion protein was restricted to the stage of plant penetration. In no case did we detect the Clp1:eGFP signal in filaments prior to penetration (data not shown). Since clp1 appears to be transcribed at a similar level in all cells, as judged from the eGFP expression levels of the clp1-promoter:egfp fusion, these observations suggest a posttranscriptional regulation of clp1.
Overexpression of clp1 Inhibits b-Dependent Filament Formation
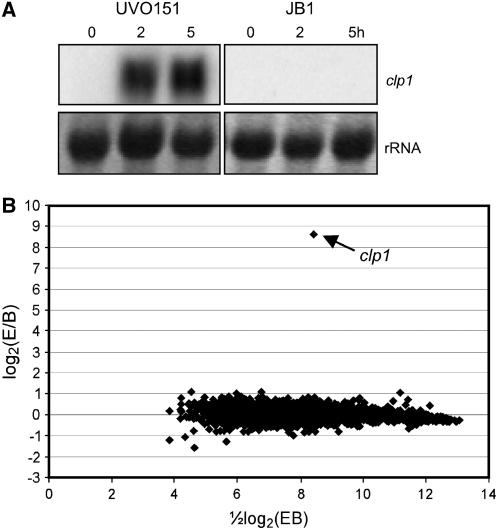
In C. cinerea, expression of clp1 is sufficient to initiate the formation of clamps in the absence of an active A-heterodimer, suggesting a regulatory function downstream of A (Inada et al., 2001; Kamada, 2002). To test whether U. maydis clp1 could have a similar function, we introduced an arabinose-inducible clp1 allele into strain JB1 (a1Δb) that carries a deletion of the entire b-locus to create strain UVO151. After 2 h of growth in arabinose-containing medium, clp1 expression was found to be drastically induced (Figure 6A). The induction of clp1 in UVO151 did not change the morphology of the sporidia (data not shown). To address the question whether Clp1 exhibits its function as a transcriptional regulator, expression profiles of UVO151 and JB1 were compared 2 h after arabinose-induced expression of clp1 by DNA microarray analysis. Data analysis revealed no significant changes in gene expression, with the exception of the 390-fold-induced expression level of clp1 (Figure 6B). Thus, it can be excluded that U. maydis Clp1 acts as a transcriptional regulator under these conditions.
Figure 6.
Clp1 Does Not Act as a Transcriptional Regulator by Itself.
(A) and (B) Strain UVO151 (a1Δb crg1P:clp1) is deleted for the entire b-locus and harbors an arabinose-inducible version of the clp1 gene integrated into the ip locus as described (Brachmann et al., 2001). The expression of clp1 was induced by 2 and 5 h of growth in arabinose-containing medium. As a control, strain JB1 (a1Δb) was grown under identical conditions.
(A) RNA gel blot analysis of clp1 induction; 5 μg of total RNA were loaded per lane. As a loading control, the filter was stained with methylene blue to visualize blotted rRNA. A 32P-labeled, 0.7-kb fragment of clp1 was used as probe.
(B) MA plot (Yang et al., 2002) illustrating Affymetrix GeneChip results. Results represent two independent experiments. Expression profiles of UVO151 (experiment [E]) and JB1 (baseline [B]) were compared. Ratios of expression values were plotted versus signal intensities. Genes that were not consistently detected in either JB1 or UVO151 are not shown. Data analysis revealed no significant changes in gene expression, with the exception of the 390-fold-induced expression level of clp1 (arrow). The entire data set can be viewed at http://www.ncbi.nlm.nih.gov/projects/geo/ (accession number GSE4689).
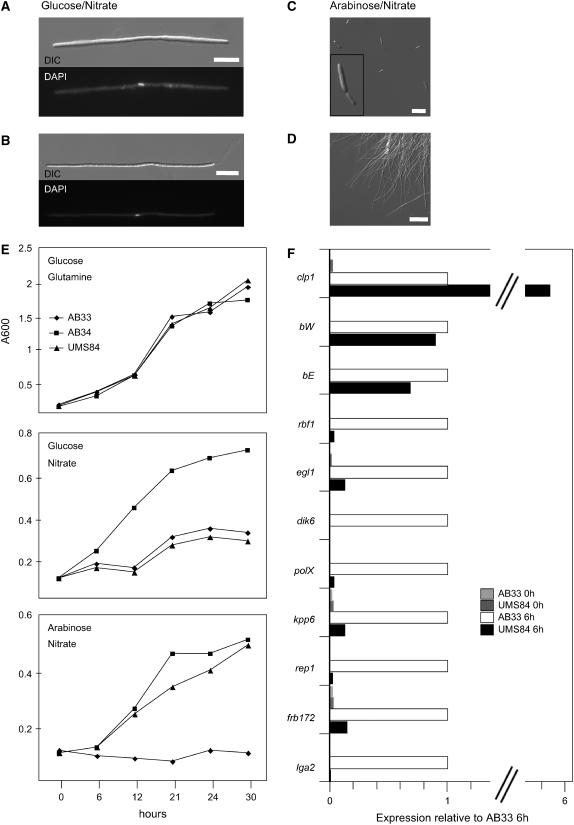
Next, we were interested in the effect of Clp1 expression on b-dependent processes. The arabinose-inducible clp1 allele was integrated into the ip locus of strain AB33, which carries the bE1 and bW2 genes under control of the nitrate-inducible nar1 promoter. The resulting strain, UMS84, allows independent induction of Clp1 (by arabinose) and of an active bE/bW heterodimer (by nitrate). When cells were transferred to medium containing nitrate and glucose, leading to induction of bE1/bW2 only, filament formation of UMS84 was comparable to that of AB33 (Figures 7A and 7B). Cell division of the filamentous cells was not observed, and nuclear staining with DAPI revealed that the filaments, comparable to AB33, contained a single nucleus, implying that the filaments are cell cycle arrested (Figures 7A and 7B). Simultaneous induction of b and clp1 in UMS84 by nitrate/arabinose medium led to a drastic reduction of filamentation (Figure 7C) compared with the filaments formed by AB33 upon growth in nitrate/arabinose medium (Figure 7D). Less than 2% of the UMS84 cells exhibited filamentous growth after 20 h of induction compared with >90% filaments formed in strain AB33 under identical conditions. In medium containing glucose and Gln (i.e., under conditions where neither b nor clp1 is induced), strains AB33, AB34 (harboring the inactive bE2/bW2 combination), and UMS84 display a similar growth curve (Figure 7E, top panel). In medium containing glucose and nitrate (induction of bE and bW), in both AB33 and UMS84 cell divisions are stalled caused by the b-mediated cell cycle arrest, while growth of AB34 is unaffected (Figure 7E, middle panel). However, the simultaneous induction of clp1 in arabinose/nitrate medium results in a growth rate of UMS84 that is comparable to that of AB34, indicating that the b-mediated cell cycle block must have been released (Figure 7E, bottom panel). Thus, both the change in cell morphology (sporidia instead of hyphae) and the cell cycle release indicate that Clp1 counteracts the function of the bE/bW heterodimer. Concomitantly, we observed a downregulation of b-dependent genes. The induction of clp1, driven by the crg1 promoter in strain UMS84, leads to an expression level that is ∼6 times higher than the expression from the native clp1 promoter after b-induction in strain AB33 (Figure 7F). The expression of the bE and bW genes, both under the control of the nar1 promoter, is insignificantly altered after clp1 induction (Figure 7F). However, under the same conditions, expression of the b-dependently expressed genes lga2 (Romeis et al., 2000), frb172 (Brachmann et al., 2001), rep1 (Wösten et al., 1996), kpp6 (Brachmann et al., 2003), polX (Brachmann et al., 2001), dik6 (Bohlmann et al., 1994; G. Weinzierl and J. Kämper, unpublished data), egl1 (Schauwecker et al., 1995), and rbf1 (M. Scherer and J. Kämper, unpublished data) is strongly reduced in UMS84 compared with AB33 6 h after b-induction (Figure 7F).
Figure 7.
clp1 Overexpression Inhibits b-Dependent Gene Regulation.
(A) to (D) Strains AB33 (a1 bE1nar1P/bW2nar1P), AB34 (a1 bE2nar1P/bW2nar1P), and UMS84 (a1 bE1nar1P/bW2nar1Pclp1crg1) were grown in minimal medium with Gln and glucose to an OD600 of 0.1 prior to induction. Expression of the b-genes was induced by nitrate, and clp1 expression was induced by arabinose. Induction of the active bE1/bW2 combination in strains UMS84 (A) and AB33 (B) leads to the formation of filaments with single nuclei (DAPI stain, bottom panel). Simultaneous induction of clp1 in strain UMS84 suppresses filamentation; the cells display a yeast-like phenotype (C). Strain AB33 is filamentous under identical conditions (D). DIC, differential interference contrast. Bars = 10 μm in (A), (B), and (D) and 20 μm in (C).
(E) Growth curves of strains AB33, AB34, and UMS84. Top panel: In medium containing glucose (clp1-off) and Gln (b-off), all strains display a similar growth curve. Middle panel: In medium containing nitrate (b-on) and glucose (clp1-off), strains AB33 and UMS84, both harboring the compatible bE1/bW2 combination, are drastically reduced in growth compared with strain AB34 harboring the incompatible bE2/bW2 combination. Bottom panel: In medium containing nitrate (b-on) and arabinose (clp1-on in strain UMS84), the growth rate of AB34 and UMS84 are comparable, despite the compatible bE1/bW2 combination in strain UMS84.
(F) Quantitative real-time PCR analysis of b-dependently expressed genes. Strains AB33 and UMS84 were shifted to arabinose/nitrate medium to induce the bE1/bW2 genes and additionally in UMS84 the clp1 gene. RNA was extracted before the shift (0 h) and 6 h after induction. Differences in expression of the monitored genes were calculated relative to the expression in AB33 6 h after induction of b (defined as 100%). Values were normalized to the expression of the constitutively expressed ppi gene. Given are the mean values of two technical replicates that gave similar results. The expression of the following b-induced genes was monitored: lga2, a gene with unknown function involved in mitochondrial fragmentation (Romeis et al., 2000; Bortfeld et al., 2004); frb172, encoding a protein with similarities to K+/H+ antiporters (Brachmann et al., 2001); rep1, encoding a hydrophobic surface protein (Wösten et al., 1996); kpp6, encoding a mitogen-activated protein kinase involved in appressoria formation (Brachmann et al., 2003); polX, encoding a protein with weak similarities to polymerase X (Brachmann et al., 2001); dik6, encoding a seven–transmembrane domain protein with unknown functions (Bohlmann et al., 1994; G. Weinzierl and J. Kämper, unpublished data); egl1, encoding a cellulase (Schauwecker et al., 1995); and rbf1, encoding a zinc finger transcription factor (M. Scherer and J. Kämper, unpublished data).
DISCUSSION
In U. maydis, the switch from saprophytic to biotrophic lifestyle and the morphological changes from budding to filamentous growth are closely interconnected. The dikaryotic hyphae formed upon fusion of two haploid sporidia initiate the biotrophic phase: without the host plant, cells do not divide any further. Only upon plant infection do the dikaryotic hyphae start to proliferate. In most Basidiomycetes, the proliferation of the dikaryon is accompanied by the formation of clamp cells that ensure the proper distribution of the genetically diverse nuclei. However, although widespread, clamps are not essential for the maintenance of a heterokaryotic mycelium; clampless dikaryons have been described in various cases (Swiezynski and Day, 1960a; Kemp, 1980; Salo et al., 1989), especially for various Uredinomycetes (for recent examples, see Frieders and McLaughlin, 2001; Ikeda et al., 2003). Similarly, it was generally believed that in U. maydis clamps are not required for dikaryotic growth. Although dikaryotic filaments develop hyphal projections located at the septa of adjacent hyphal cells directly after penetration of the plant cuticle, these structures were generally considered to be branch primordia rather than true clamp connections, in particular because a fusion with the subapical hyphal cell was never observed (Sleumer, 1931; Snetselaar and Mims, 1994; Banuett and Herskowitz, 1996). Here, we provide evidence for a function of these clamp-like structures in the orderly distribution of nuclei using live imaging of the nuclei in dikaryotic hyphae. The formation of clamp-like structures of U. maydis shares several features with the formation of clamp connections in other Basidiomycetes: (1) The clamp-like projection is formed at the most apical cell of a dikaryotic hypha when the nuclei undergo coordinated division. (2) It is placed at the position of future septum formation. (3) One of the four resulting nuclei becomes located in the clamp-like structure and (4) is trapped there after septa have been formed. However, there are major differences with respect to the way the trapped nucleus is released to the subapical hyphal cell. In most filamentous Basidiomycetes, this is achieved by the fusion of the backward-growing clamp with a peg formed by the subapical cell (Badalyan et al., 2004). By contrast, we observed neither changes in the shape of clamp-like structures during mitotic cell division nor a visible dissolution of the cell wall in U. maydis. Despite the integral appearance of the cell wall, the clamp-located nucleus migrates into the subapical hyphal cell, obviously through a narrow opening. One possibility is that the opening is the result of a locally restricted dissolution of the cell wall and by that resembling mechanistically a fusion event comparable to that of other Basidiomycetes. Another possibility is that the clamp cell is separated by formation of a septum that is similarly structured as the porated septae in between hyphal compartments. Since the pori (Bauer et al., 1997) facilitate nuclear migration in between the compartments (for example, see Giesy and Day, 1965), a fusion event would be expendable. Such an explanation would be in line with the finding that clamp formation in U. maydis is independent from the presence of an active pheromone pathway that, in higher Basidiomycetes, is required for cell fusion.
Clamp-like structures were also found in the solopathogenic strains SG200 and CL13, although their hyphae contain only single nuclei. Similar observations have been made for AmutBmut monokaryons of C. cinerea that, comparable to U. maydis SG200, harbor self-activating alleles of the A- and B-loci (Swamy et al., 1984; Kües, 2000). Obviously, in both C. cinerea and U. maydis, clamp formation is a program that is closely interconnected with the mitotic division program of hyphal cells.
Taken together, we find significant functional similarity between the formation of clamp-like structures in U. maydis and the formation of clamp connections in other Basidiomycetes; however, further ultrastructural studies are needed to provide evidence whether clamp formation in U. maydis differs mechanistically from that of higher Basidiomycetes.
Regulation of Clamp Formation by Clp1
We have identified the U. maydis clp1 gene to be essential for clamp formation and pathogenic development. clp1 expression is b-dependent, and the early induction of clp1 upon b-activation as well as the presence of two promoter regions with similarity to the known b-binding sites make it likely that the gene is directly regulated by the bE/bW heterodimer. The clp1 deletion does not affect the initial stages of b-mediated development; neither the formation of the dikaryotic filament nor the formation of appressoria is altered. The hyphae are capable of penetrating the plant cuticle, indicating that the b-dependent infection process per se is unaffected. In wild-type hyphae, the plant penetration marks the earliest time point for the first mitotic cell division, which is accompanied by the formation of clamp-like structures. In the clp1 mutant strains, however, we never observed clamp formation or cell divisions, and as a consequence, hyphal development was blocked directly after plant penetration. One of the obvious functions of the b-locus is the control of the mitotic cell cycle. Nuclear division is stalled in dikaryotic hyphae until the fungus penetrates the plant (Snetselaar and Mims, 1992), and induced expression of the bE1/bW2 genes in strains AB31 or AB33 is sufficient to induce a G2 cell cycle arrest (J. Pérez-Martín, unpublished data). However, bE and bW are expressed during the entire biotrophic growth phase, and recently we were able to show by means of a temperature-sensitive bE allele that b-function is required for pathogenic development subsequent to invasion (R. Wahl and J. Kämper, unpublished data). Thus, to release the cell cycle block and to allow proliferation, the b-dependent regulation has to be adapted after plant penetration. We favor that such an adaptation of b-function is mediated by the Clp1 protein. First, in clp1 deletion strains, the b-mediated block is obviously not released. In addition, our data show that induced expression of clp1 in a strain with an active bE1/bW2 heterodimer is sufficient to counteract b-dependent filamentation and to release the b-induced cell cycle block. How is this clp1-mediated regulation achieved? Because induction of clp1 in cells lacking an active bE/bW heterodimer causes no significant alterations of the transcription profile, it is improbable that Clp1 exerts its function as a transcriptional regulator. Second, in strain UMS84, the expression of various b-dependent genes is drastically reduced upon clp1 overexpression, although the bE and bW expression itself is only insignificantly reduced. Thus, we favor a regulation via Clp1 at the posttranscriptional level, possibly by inhibition of the transactivation activity of the b-heterodimer via protein–protein interaction. The inhibition of the yeast regulator Gal4p by Gal80p is a well-studied example for this mode of action (Lohr et al., 1995). Another level of complexity is added by the downregulation of the rbf1 gene. rbf1 is a directly b-regulated gene encoding a zinc finger transcription factor required for the expression of the majority of b-regulated genes (M. Scherer and J. Kämper, unpublished data). It is expected that by the inactivation of bE/bW function in combination with the downregulation of rbf1 the expression of most b-dependent genes is altered upon clp1 induction.
Similar to the situation in U. maydis, the C. cinerea clp1 gene is considered to be a direct target of the homeodomain proteins encoded by the A-locus. Clp1 is thought to act as a repressor of Pcc1, a potential HMG box transcription factor. A loss-of-function mutation in pcc1 produces clamp cells uncoupled from the presence of active A- and B-mating-type loci; Pcc1 is thus thought to act as a repressor of clamp cell formation. The current model suggests that in C. cinerea, clamp formation is initiated by the Clp1-mediated release of Pcc1 repression, consistent with the observation that forced clp1 expression leads to an initiation of clamp formation independent from the A-mating-type locus. As in U. maydis, the Clp1-mediated control of Pcc1 is proposed to appear at the posttranscriptional level, since the expression of pcc1 is not decreased upon clp1 induction (Murata et al., 1998; Inada et al., 2001; Kamada, 2002). Thus, although the Clp1 proteins from U. maydis and C. cinerea do not share known structural motifs and lack regions of highly similar amino acid sequence, they appear to have a conserved mechanism of altering the function of key regulators of development.
In U. maydis, the Clp1 protein itself appears to be the subject of posttranscriptional regulation. Although the gene is expressed during the entire biotrophic stage, the protein can be detected within the nucleus only when the appressorium is formed (i.e., at the earliest detectable time when the cell cycle block can be released). It is possible that Clp1 transport to the nucleus is regulated (and the protein present in the cytoplasm is not detectable due to the lower concentration); alternatively, the translation or the stability of Clp1 could be regulated. It is tempting to speculate that a cyclic transport of Clp1 to the nucleus could set the timing for the nuclear divisions during the entire biotrophic growth phase. Unfortunately, due to the weak signal of the Clp1:3xeGFP fusion protein, we were not able to detect any Clp1:3xeGFP protein signals after plant penetration.
In addition, clp1 transcription appears to be regulated via the a-mediated signaling cascade. a-mediated induction leads to a truncated transcript, lacking a poly(A) tail and devoid of any coding capacity. It is possible that the transcript is the result of a stalled RNA polymerase or that it is caused by a premature abortion of transcription, triggered by a potential stem-loop structure formed during transcription. Both the mechanism of how this short transcript is generated and its potential function have to await further studies for clarification. However, since the a-locus is not required for clamp formation, an essential function of the short transcript for this process can be excluded.
With our work, we have provided insights into the molecular processes associated with the onset of hyphal proliferation after plant penetration of U. maydis. The first step in the dimorphic switch of U. maydis cells is the initiation of polarized growth leading to filament formation; this can be achieved on artificial media and has been well studied (Steinberg et al., 1998, 2001; Weber et al., 2003, 2006; Fuchs et al., 2005; Schuchardt et al., 2005). For the true filamentous growth, however, a multitude of different processes have to be coordinated to achieve synchronized nuclear divisions, sorting of nuclei, and cell differentiation. With our work, we have paved the road for the detailed study of these processes that are expected to be closely interwoven with the potential of U. maydis to infect its host plant. It will be of great interest to elucidate the particular role of the b-mating-type locus, especially to dissect its function with respect to the establishment of the pathogenicity program (i.e., those processes that facilitate infection) and to cope with the host environment, and the morphological program required for hyphal proliferation. We anticipate that this work will be of relevance not only to U. maydis but also to other Basidiomycetes and will lead to a better understanding of dikaryotic hyphal growth.
METHODS
Strains and Growth Conditions
Escherichia coli strain TOP10 (Invitrogen) was used for cloning purposes. Ustilago maydis cells were grown at 28°C in YEPS (Tsukuda et al., 1988), potato dextrose medium (Difco), complete medium, or minimal medium (Holliday, 1974). Mating assays and plant infections were performed as described by Gillissen et al. (1992). U. maydis strains used in this study are listed in Table 2.
Table 2.
Strains of U. maydis Used in This Study
| Strain | Relevant Genotype | Reference |
|---|---|---|
| AB31 | a2 Pcrg:bW2,bE1 | Brachmann et al. (2001) |
| AB32 | a2 Pcrg:bW2,bE2 | Brachmann et al. (2001) |
| AB33 | a2 Pnar:bW2,bE1 | Brachmann et al. (2001) |
| AB34 | a2 Pnar:bW2,bE2 | Brachmann et al. (2001) |
| FB1 | a1 b1 | Banuett and Herskowitz (1989) |
| FB2 | a2 b2 | Banuett and Herskowitz (1989) |
| FB6a | a2 b1 | Banuett and Herskowitz (1989) |
| FB6b | a1 b2 | Banuett and Herskowitz (1989) |
| FBD11 | a1a2 b1b2 | Banuett and Herskowitz (1989) |
| FBD12-3 | a1a2 b1b1 | Banuett and Herskowitz (1989) |
| SG200 | a1mfa2 bW2bE1 | Bölker et al. (1995) |
| CL13 | a1 bW2bE1 | Bölker et al. (1995) |
| JB1 | a1 Δb∷hph | This study |
| UMS48 | a1mfa2 bW2bE1 clp1:3xeGFP | This study |
| UMS67 | a1mfa2 bW2bE1 Δclp1 ipr[Pmig2-5:NLS-3xeGFP]ips | This study |
| UMS71 | a1mfa2 bW2bE1 ipr[Pmig2-5:NLS-3xeGFP]ips | This study |
| UMS73 | a1 b1 ipr[Pmig2-5:NLS-3xeGFP]ips | This study |
| UMS74 | a2 b2 ipr[Pmig2-5:NLS-3xeGFP]ips | This study |
| UMS76 | a1mfa2 bW2bE1 Pclp1:eGFP | This study |
| UMS84 | a2 Pnar:bW2,bE1 ipr[Pcrg:clp1]ips | This study |
| UVO151 | a1 Δb ipr[Pcrg:clp1]ips | This study |
| UVO159 | a1mfa2 bW2bE1 Δclp1 | This study |
| UVO161 | a1 b1 Δclp1 | This study |
| UVO162 | a2 b2 Δclp1 | This study |
DNA and RNA Procedures
Molecular methods followed described protocols (Sambrook et al., 1989). DNA isolation from U. maydis and transformation procedures were performed as described (Schulz et al., 1990). Homologous integration of constructs was verified by gel blot analyses. Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples to be used for microarray analyses or real-time RT-PCR were further column purified (RNeasy; Qiagen) and the quality checked using a Bioanalyzer with an RNA 6000 Nano LabChip kit (Agilent). RNA ladder 3191 (Promega) was used as a size marker for RNA gel blot analyses. Membranes were stained for 5 min in 200 mg/mL of methylene blue in 300 mM sodium acetate to assess equal loading and transfer. Probes for radioactive labeling of gel blots were generated by random priming (New England Biolabs). Detection and quantification of radioactive signals were performed with a STORM 840 PhosphorImager and ImageQuant 5.2 software (Molecular Dynamics).
Real-Time RT-PCR
First-strand cDNA synthesis was performed using the SuperScript III first-strand synthesis SuperMix assay (Invitrogen) according to the manufacturer's protocol. As a template for the reaction, 1 μg of total RNA was used. Samples were incubated at 50°C for 50 min. Real-time PCR was performed on a Bio-Rad iCycler system using the Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) according to the manufacturer's protocol. Cycling conditions were as follows: 95°C for 2 min, 45 cycles of 30 s at 60°C, and 30 s at 72°C. After each PCR, the specificity of the amplifications was verified, and the threshold cycle above background was calculated using Bio-Rad iCycler software. Forward (F) and reverse (R) primers used for detection were as follows (in 5′–3′ orientation): for clp1, clp1-F 5′-GTCAGTTCGTTTGCGCCTAC-3′ and clp1-R 5′-GCATCGTCTCGTGCAACTTC-3′; for bW, bW-F 5′-GATCTCACCCAGCCAATCAC-3′ and bW-R 5′-GAGTTGATCGAGGCCGAATG-3′; for bE, bE-F 5′-GCACAACACCTTCCATTGAC-3′ and bE-R 5′-ACTGCTCCCGAATGTACT-3′; for ppi, ppi-F 5′-ACGCCGATTCACTTCGTC-3′ and ppi-R 5′-TCTTGGCAGTCTTGGGAAC-3′; for rbf1, RT_rbf1_F 5′-AGTACGAGCTACGACGGATTC-3′ and RT_rbf1_R 5′-GGGTAGGTGTTGGACACATTC-3′; for egl1, um06332RT_1 5′-CATCGCCAGATCCGTCAACACC-3′ and um06332RT_2 5′-AAGGTCGACAGCGCCAACACTG-3′; for dik6, um04130RT_1 5′-TTGTTCCACCCATCCTTCACGC-3′ and um04130RT_2 5′-GGATCGAGCGTCGAAACACAGC-3′; for polX, um01262RT_1 5′-GGATCGAGCGTCGAAACACAGC-3′ and um01262RT_2 5′-GGATCGAGCGTCGAAACACAGC-3′; for kpp6, um02331RT_1 5′-CTCCCATTCTGCACTCCGCAAC-3′ and um02331RT_2 5′-GACAACACCATAGGCGCCTTCG-3′; for rep1, um03924RT_1 5′-CCACTCCTTCGCCCAAGCCTAC-3′ and um03924RT_2 5′-ACCGTCCAGGATGCCAATGTTG-3′; for frb172, um04771RT_1 5′-AACACTCTGGCGCTCGTTCAGG-3′ and um04771RT_2 5′-GACCACTGAATCGATCCGCTGC-3′; and for lga2, lga2RT_1 5′-TCTATCACGGGCAAGAGGCTG-3′ and lga2RT_2 5′-ACTTTGCCGGTGACGACGTGAT-3′.
cRT-PCR
The procedure used was modified from Couttet et al. (1997). Total RNA (20 μg) was decapped with 2.5 units of tobacco acid phosphatase (Invitrogen) for 2 h at 37°C. Decapped RNA was purified (RNeasy) and on-column DNase digested (DNase set; Qiagen). RNA (1 μg) was circularized with 10 units of T4 RNA ligase (New England Biolabs) in the supplied buffer supplemented with 40 units of RNaseOUT ribonuclease inhibitor (Invitrogen) for 16 h at 16°C. After phenol/chloroform extraction, RNA was precipitated with ethanol. First-strand cDNA synthesis was performed using Superscript II RnaseH− reverse transcriptase (Invitrogen) with the clp1 gene-specific primer 5′-CGGATGTTGACATTGACGGGTGCG-3′ for 1 h at 50°C. For PCR amplification, the nested primers 5′-TTGACGGGTGCGAGAGGACGG-3′ and 5′-TGTCAACATCCGCAACTCTGCTAACCG-3′ were used. cRT-PCR products were cloned using the TOPO cloning kit (Invitrogen).
DNA Microarray Analysis
For DNA array analysis, custom-designed Affymetrix chips were used. Probe sets were designed based on the map-based sequencing assembly; for each predicted gene, 33 perfect match and the respective mismatch probes were designed, covering a region of 800 bp at the 3′-ends. The U. maydis DNA arrays address >6200 genes. Probe sets for the individual genes are available at http://mips.gsf.de/genre/proj/ustilago/. For DNA array analysis, strains were grown to an OD600 of 0.5 at 28°C in liquid array medium: 6.25% (w/v) salt solution, 30 mM l-Gln, and 1% (w/v) glucose, pH 7.0 (filter-sterilized). For induction of the crg1 promoter, cells were inoculated in liquid array medium containing 1% (w/v) arabinose instead of glucose as a carbon source. Target preparation and hybridization were performed according to the standard Affymetrix protocol using the Enzo BioArray RNA transcript labeling kit, except for a reaction temperature of 50°C for first-strand cDNA synthesis. Experiments were done in two biological replicates. Hybridization followed the Affymetrix protocol, and arrays were processed following the EukGE-WS2 protocol on a GeneChip Fluidics Station 400 and scanned on an Affymetrix GSC2500. Data analysis was performed using the Affymetrix Micro Array Suite 5.0 software and the dChip1.3 program (http://biosun1.harvard.edu/complab/dchip/).
Tagging of Nuclei with Triple eGFP
To create plasmid pMS76, oligonucleotides NLS_anti (5′-CATGCCGAATTCGACCTTTCTCTTCTTTTTTGGAGG-3′) and NLS_sense (5′-CATGGTCGAGCCTCCAAAAAAGAAGAGAAAGGTCGA-3′) were hybridized and ligated to an NcoI-NotI 3xeGFP HygR fragment of pUMA647 (K. Zarnack and M. Feldbrügge, unpublished data) and an 869-bp XbaI-NcoI mig2-5 promoter fragment of pJF (Farfsing et al., 2005) and integrated into pRU12 (Brachmann et al., 2001), cut with XbaI-NotI. Correct orientation of the NLS was verified by PCR. pMS76 was linearized with SspI for integration into the ip locus of FB1, FB2, SG200, and UVO159 (Brachmann et al., 2001).
Deletion of clp1
The deletion of clp1 was performed by a PCR-based approach (Kämper, 2004). The entire clp1 ORF was replaced by a hygromycin resistance cassette in the strains FB1, FB2, and SG200.
Replacement of clp1 by eGFP
pMS71 contains the eGFP gene fused to a 1-kb promoter region of clp1. The promoter region was amplified by PCR using primers 5′-GGTGGCCGCGTTGGCCGACATTATCATGATTGGGAATGGG-3′ and 5′-AGATCGCGCCTGGAGCGC-3′, generating an SfiI site at the 5′-end of clp1. The terminator region of clp1 was amplified using primers 5′-GTGGGCCTGAGTGGCCTGAATCCAACCATTCAACGCCTC-3′ and 5′-TTACGGAGCTCAAAGACACGC-3′, generating an SfiI site at the 3′-end of clp1. Both fragments were ligated to an SfiI eGFP HygR fragment of pUMA317 in a pCR2.1 (Invitrogen) backbone. The eGFP HygR fragment in pUMA317 is similar to that of pMF3H (Brachmann et al., 2004) but contains an altered SfiI site (5′-GGCCAACGCGGCC-3′) 5′ to the eGFP ORF that allows translational in-frame fusions (P. Becht, J. König, and M. Feldbrügge, unpublished data). The fusion product was excised with EcoRI and integrated into the respective site of pUC18. Subsequently, the hygromycin resistance cassette was replaced by a nourseothricin resistance cassette as a NotI fragment from pSL-nat. pSL-nat harbors a 1.4-kb NotI fragment with the bacterial nourseothricin resistance gene, driven by the U. maydis GAPDH promoter and terminated by the Saccharomyces cerevisiae cyc terminator from plasmid pBR322-Nat1 (Kojic and Holloman, 2000; G. Bakkeren, personal communication). pMS71 was linearized with SspI for U. maydis transformation.
Clp1:eGFP Fusion
The 3′-region of the clp1 ORF was amplified by PCR using primers 5′-GGTGGCCGCGTTGGCCGCCTCGAGTTTGGTGGATTG-3′ and 5′-TCCCAGGTCAGCCTTTGCC-3′, creating an SfiI site at the 3′-end. The 3′ UTR sequence was amplified using primers 5′-GTGGGCCTGAGTGGCCTGAATCCAACCATTCAACGC-3′ and 5′-TTACGGAGCTCAAAGACACGC-3′, creating an SfiI site at the 5′-end. Both fragments were ligated to an SfiI 3xeGFP HygR fragment of pUMA647 (K. Zarnack and M. Feldbrügge, unpublished data) in a pCR2.1 vector (Invitrogen) to create pMS74.
Induced Expression of clp1
The clp1 ORF was amplified by PCR from genomic DNA with the primers 5′-CATATGTCACCCCGTCACCAGC-3′ and 5′-GCGGCCGCTCACTCGAGTTTGGTGGATTG-3′, creating an NdeI site at the start and an NotI site following the stop codon. PCR products were cloned using the TOPO cloning kit (Invitrogen). After sequence verification, the clp1 ORF was cloned as an NdeI-NotI fragment into the respective sites of pRU12 (Brachmann et al., 2001). The resulting plasmid pRU-ATG1 was linearized with SspI and transformed into strain AB33 (Brachmann et al., 2001) to yield strain UMS84 (a2 Pnar:bW2,bE1 ipr[Pcrg:clp1]ips) or into strain JB1 (a1Δb) to yield strain UVO151 (a1Δb ipr[Pcrg:clp1]ips). JB1 (a1Δb) is a derivative of FB1 (Banuett and Herskowitz, 1989) in which the b2-locus was substituted with a 2.9-kb PvuII fragment harboring the hygromycin resistance cassette from plasmid pCM54 (Tsukuda et al., 1988), removing the entire bE2 ORF and the bW2 ORF to amino acid 624.
Microscopy
Microscopy analysis was performed using a Zeiss Axioplan 2 microscope. Photomicrographs were obtained with an Axiocam HrM camera, and the images were processed with Axiovision (Zeiss) and Photoshop (Adobe). For staining of nuclei with DAPI, Vectashield H-1200 (Vector Laboratories) was used. Cell wall components were stained with 2 μg/mL of Calcofluor-white (Sigma-Aldrich) in PBS. Chlorazole Black E staining of fungal cells in planta was performed as described (Brachmann et al., 2003).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XP_758585.1 (U. maydis clp1, hypothetical protein UM02438.1 of the U. maydis 521 genome sequence) and AB034196 (C. cinerea clp1). DNA array data for the clp1 induction were submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE4689.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Alignment of Clp1 to Predicted Proteins from Basidiomycetes.
Supplementary Material
Acknowledgments
We thank M. Feldbrügge for various plasmid constructs, U. Kües, K. Snetselaar, and U. Kämper for stimulating discussions, J. Pérez-Martín for supplying unpublished data, and V. Vincon for his excellent technical support. This work was supported by grants from the German Ministry of Science and Education for the U. maydis DNA array setup, from the Max-Planck-Society for U. maydis genome annotation, and from the International Research Training Group Transcriptional Control of Developmental Processes of the German Research Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jörg Kämper (kaemper@mpi-marburg.mpg.de).
Online version contains Web-only data.
References
- Badalyan, S.M., Polak, E., Hermann, R., Aebi, M., and Kues, U. (2004). Role of peg formation in clamp cell fusion of homobasidiomycete fungi. J. Basic Microbiol. 44 167–177. [DOI] [PubMed] [Google Scholar]
- Banuett, F. (1995). Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet. 29 179–208. [DOI] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1989). Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122 2965–2976. [DOI] [PubMed] [Google Scholar]
- Basse, C.W., and Farfsing, J.W. (2006). Promoters and their regulation in Ustilago maydis and other phytopathogenic fungi. FEMS Microbiol. Lett. 254 208–216. [DOI] [PubMed] [Google Scholar]
- Bauer, R., Oberwinkler, F., and Vanky, K. (1997). Ultrastructural markers and systematics in smut fungi and allied taxa. Can. J. Bot. 75 1273–1314. [Google Scholar]
- Bohlmann, R., Schauwecker, F., Basse, C., and Kahmann, R. (1994). Genetic regulation of mating and dimorphism in Ustilago maydis. In Advances in Molecular Genetics of Plant-Microbe Interactions, M.J. Daniels, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 239–245.
- Bortfeld, M., Auffarth, K., Kahmann, R., and Basse, C.W. (2004). The Ustilago maydis a2 mating-type locus genes lga2 and rga2 compromise pathogenicity in the absence of the mitochondrial p32 family protein Mrb1. Plant Cell 16 2233–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, A., Konig, J., Julius, C., and Feldbrugge, M. (2004). A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272 216–226. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Schirawski, J., Müller, P., and Kahmann, R. (2003). An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, A., Weinzierl, G., Kämper, J., and Kahmann, R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42 1047–1063. [DOI] [PubMed] [Google Scholar]
- Brown, A.J., and Casselton, L.A. (2001). Mating in mushrooms: Increasing the chances but prolonging the affair. Trends Genet. 17 393–400. [DOI] [PubMed] [Google Scholar]
- Bölker, M., Böhnert, H.U., Braun, K.H., Görl, J., and Kahmann, R. (1995). Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 248 547–552. [DOI] [PubMed] [Google Scholar]
- Bölker, M., Urban, M., and Kahmann, R. (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell 68 441–450. [DOI] [PubMed] [Google Scholar]
- Casselton, L.A. (2002). Mate recognition in fungi. Heredity 88 142–147. [DOI] [PubMed] [Google Scholar]
- Casselton, L.A., and Olesnicky, N.S. (1998). Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet, P., Fromont-Racine, M., Steel, D., Pictet, R., and Grange, T. (1997). Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. USA 94 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, P.R., Anagnostakis, S.L., and Puhalla, J.E. (1971). Pathogenicity resulting from mutation at the b locus of Ustilago maydis. Proc. Natl. Acad. Sci. USA 68 533–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfsing, J.W., Auffarth, K., and Basse, C.W. (2005). Identification of cis-active elements in Ustilago maydis mig2 promoters conferring high-level activity during pathogenic growth in maize. Mol. Plant Microbe Interact. 18 75–87. [DOI] [PubMed] [Google Scholar]
- Frieders, E.M., and McLaughlin, D.J. (2001). The heterobasidiomycete moss parasites Jola and Eocronartium in culture: Cytology, ultrastructure and anamorph. Mycol. Res. 105 734–744. [Google Scholar]
- Fuchs, U., Manns, I., and Steinberg, G. (2005). Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol. Biol. Cell 16 2746–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesy, R.M., and Day, P.R. (1965). The septal pores of Coprinus lagopus in relation to nuclear migration. Am. J. Bot. 52 287–293. [Google Scholar]
- Gillissen, B., Bergemann, J., Sandmann, C., Schröer, B., Bölker, M., and Kahmann, R. (1992). A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68 647–657. [DOI] [PubMed] [Google Scholar]
- Hiscock, S.J., and Kües, U. (1999). Cellular and molecular mechanisms of sexual incompatibility in plants and fungi. Int. Rev. Cytol. 193 165–295. [DOI] [PubMed] [Google Scholar]
- Holliday, R. (1961). The genetics of Ustilago maydis. Genet. Res. Camb. 2 204–230. [Google Scholar]
- Holliday, R. (1974). Ustilago maydis. In Handbook of Genetics, R.C. King, ed (New York: Plenum Press), pp. 575–595.
- Ikeda, K.-I., Nakamura, H., and Matsumoto, N. (2003). Mycelial incompatibility operative in pairings between single basidiospore isolates of Helicobasidium mompa. Mycol. Res. 107 847–853. [DOI] [PubMed] [Google Scholar]
- Inada, K., Morimoto, Y., Arima, T., Murata, Y., and Kamada, T. (2001). The clp1 gene of the mushroom Coprinus cinereus is essential for A-regulated sexual development. Genetics 157 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann, R., and Kämper, J. (2004). Ustilago maydis: How its biology relates to pathogenic development. New Phytol. 164 31–42. [DOI] [PubMed] [Google Scholar]
- Kamada, T. (2002). Molecular genetics of sexual development in the mushroom Coprinus cinereus. Bioessays 24 449–459. [DOI] [PubMed] [Google Scholar]
- Kämper, J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271 103–110. [DOI] [PubMed] [Google Scholar]
- Kämper, J., Reichmann, M., Romeis, T., Bölker, M., and Kahmann, R. (1995). Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81 73–83. [DOI] [PubMed] [Google Scholar]
- Kemp, R.F.O. (1980). A heterothallic species of Coprinus related to C. serquilinus with clamp connections on homokaryotic and dikaryotic mycelia. Trans. Br. Mycol. Soc. 74 411–418. [Google Scholar]
- Kojic, M., and Holloman, W.K. (2000). Shuttle vectors for genetic manipulations in Ustilago maydis. Can. J. Microbiol. 46 333–338. [DOI] [PubMed] [Google Scholar]
- Kothe, E. (2001). Mating-type genes for basidiomycete strain improvement in mushroom farming. Appl. Microbiol. Biotechnol. 56 602–612. [DOI] [PubMed] [Google Scholar]
- Kües, U. (2000). Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64 316–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr, D., Venkov, P., and Zlatanova, J. (1995). Transcriptional regulation in the yeast GAL gene family: A complex genetic network. FASEB J. 9 777–787. [DOI] [PubMed] [Google Scholar]
- Murata, Y., Fujii, M., Zolan, M.E., and Kamada, T. (1998). Molecular analysis of pcc1, a gene that leads to A-regulated sexual morphogenesis in Coprinus cinereus. Genetics 149 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhalla, J.E. (1968). Compatibility reactions on solid medium and interstrain inhibition in Ustilago maydis. Genetics 60 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhalla, J.E. (1970). Genetic studies on the b incompatibility locus of Ustilago maydis. Genet. Res. Camb. 16 229–232. [Google Scholar]
- Quadbeck-Seeger, C., Wanner, G., Huber, S., Kahmann, R., and Kämper, J. (2000). A protein with similarity to the human retinoblastoma binding protein 2 acts specifically as a repressor for genes regulated by the b mating type locus in Ustilago maydis. Mol. Microbiol. 38 154–166. [DOI] [PubMed] [Google Scholar]
- Raper, J.R. (1966). Genetics of Sexuality in Higher Fungi. (New York: The Ronald Press).
- Romeis, T., Brachmann, A., Kahmann, R., and Kämper, J. (2000). Identification of a target gene for the bE/bW homeodomain protein complex in Ustilago maydis. Mol. Microbiol. 37 54–66. [DOI] [PubMed] [Google Scholar]
- Rowell, J.B. (1955). Functional role of compatibility factors and an in vitro test for sexual incompatibility with haploid lines of Ustilago zea. Phytopathology 45 370–374. [Google Scholar]
- Rowell, J.B., and DeVay, J.E. (1954). Genetics of Ustilago zea in relation to basic problems of its pathogenicity. Phytopathology 44 356–362. [Google Scholar]
- Salo, V., Niini, S.S., Virtanen, I., and Raudaskoski, M. (1989). Comparative immunocytochemistry of the cytoskeleton in filamentous fungi with dikaryotic und multinuclieate hyphae. J. Cell Sci. 94 11–24. [Google Scholar]
- Sambrook, J., Frisch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schauwecker, F., Wanner, G., and Kahmann, R. (1995). Filament-specific expression of a cellulase gene in the dimorphic fungus Ustilago maydis. Biol. Chem. Hoppe Seyler 376 617–625. [DOI] [PubMed] [Google Scholar]
- Schuchardt, I., Assmann, D., Thines, E., Schuberth, C., and Steinberg, G. (2005). Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell 16 5191–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, B., Banuett, F., Dahl, M., Schlesinger, R., Schäfer, W., Martin, T., Herskowitz, I., and Kahmann, R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60 295–306. [DOI] [PubMed] [Google Scholar]
- Sleumer, O. (1931). Über sexualität und zytologie von Ustilago zeae (Beckm.) Unger. Zeitschr. Bot. 25 209–262 [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1992). Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84 193–203. [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1994). Light and electron microscopy of Ustilago maydis hyphae in maize. Mycol. Res. 98 347–355. [Google Scholar]
- Spellig, T., Bölker, M., Lottspeich, F., Frank, R.W., and Kahmann, R. (1994). Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 13 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, G., Schliwa, M., Lehmler, C., Bolker, M., Kahmann, R., and McIntosh, J.R. (1998). Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J. Cell Sci. 111 2235–2246. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., Wedlich-Soldner, R., Brill, M., and Schulz, I. (2001). Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 114 609–622. [DOI] [PubMed] [Google Scholar]
- Swamy, S., Uno, I., and Ishikawa, T. (1984). Morphogenic effects of mutations in the A and B incompatibility factors of Coprinus cinereus. J. Gen. Microbiol. 130 3219–3224. [Google Scholar]
- Swiezynski, K.M., and Day, P.R. (1960. a). Heterokaryon formation in Coprinus lagopus. Genet. Res. 1 114–128. [Google Scholar]
- Swiezynski, K.M., and Day, P.R. (1960. b). Migration of nuclei in Coprinus lagopus. Genet. Res. 1 129–139. [Google Scholar]
- Tsukuda, T., Carleton, S., Fotheringham, S., and Holloman, W.K. (1988). Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8 3703–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, M., Kahmann, R., and Bölker, M. (1996). Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251 31–37. [DOI] [PubMed] [Google Scholar]
- Weber, I., Assmann, D., Thines, E., and Steinberg, G. (2006). Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, I., Gruber, C., and Steinberg, G. (2003). A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 15 2826–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten, H.A., Bohlmann, R., Eckerskorn, C., Lottspeich, F., Bölker, M., and Kahmann, R. (1996). A novel class of small amphipathic peptides affect aerial hyphal growth and surface hydrophobicity in Ustilago maydis. EMBO J. 15 4274–4281. [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Lin, D.M., Peng, V., Ngai, J., and Speed, T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.