Abstract
Previously, we used the ability of the higher eukaryotic positive-strand RNA virus brome mosaic virus (BMV) to replicate in yeast to show that the yeast LSM1 gene is required for recruiting BMV RNA from translation to replication. Here we extend this observation to show that Lsm1p and other components of the Lsm1p-Lsm7p/Pat1p deadenylation-dependent mRNA decapping complex were also required for translating BMV RNAs. Inhibition of BMV RNA translation was selective, with no effect on general cellular translation. We show that viral genomic RNAs suitable for RNA replication were already distinguished from nonreplication templates at translation, well before RNA recruitment to replication. Among mRNA turnover pathways, only factors specific for deadenylated mRNA decapping were required for BMV RNA translation. Dependence on these factors was not only a consequence of the nonpolyadenylated nature of BMV RNAs but also involved the combined effects of the viral 5′ and 3′ noncoding regions and 2a polymerase open reading frame. High-resolution sucrose density gradient analysis showed that, while mutating factors in the Lsm1p-7p/Pat1p complex completely inhibited viral RNA translation, the levels of viral RNA associated with ribosomes were only slightly reduced in mutant yeast. This polysome association was further verified by using a conditional allele of essential translation initiation factor PRT1, which markedly decreased polysome association of viral genomic RNA in the presence or absence of an LSM7 mutation. Together, these results show that a defective Lsm1p-7p/Pat1p complex inhibits BMV RNA translation primarily by stalling or slowing the elongation of ribosomes along the viral open reading frame. Thus, factors in the Lsm1p-7p/Pat1p complex function not only in mRNA decapping but also in translation, and both translation and recruitment of BMV RNAs to viral RNA replication are regulated by a cell pathway that transfers mRNAs from translation to degradation.
Translation and turnover of mRNAs are intimately linked. Although aberrant control of mRNA stability and translation has been linked to serious diseases including cancer, the interplay between mRNA stability and translation are still poorly understood (for reviews, see references 61 and 66). One major pathway of mRNA turnover, conserved in all eukaryotes, is deadenylation-dependent mRNA decay (52, 65, 66). In this pathway, deadenylation of the 3′-terminal poly(A) by a cytoplasmic complex (62) triggers removal of the protective 5′ cap structure (decapping), allowing 5′ to 3′ exonucleolytic digestion. The yeast Saccharomyces cerevisiae has been a valuable model for identifying the factors and mechanisms of deadenylation-dependent mRNA decay and analyzing its interaction with translation (26, 29, 65, 66). Recent studies on the yeast Dhh1p decapping factor link deadenylation-dependent mRNA decapping and metazoan maternal mRNA translation repression during development and suggest that mRNA decapping and maternal mRNA storage may be alternate branches of a common pathway (13).
One set of yeast factors facilitating deadenylation-dependent mRNA turnover is Lsm1p-Lsm7p (25); these yeast factors belong to a family of small proteins that contain the conserved Sm motif. Sm and Lsm (Like Sm) proteins have been identified in all eukaryotes tested (52), in Archaea (1, 48), and in bacteria (40, 69). These proteins associate in heptameric or hexameric, doughnut-shaped complexes with RNA-binding capabilities (references 14 and 33 and references within). Heteroheptameric Sm and Lsm protein complexes are required for an expanding list of functions in eukaryotic RNA metabolism, including mRNA turnover (7, 57), tRNA processing (36), ribosome biogenesis (60), and telomere maintenance (53). In bacteria, the homohexameric Sm-like Hfq protein complex has been proposed to act as a chaperone facilitating RNA-RNA interactions regulating turnover and translation of a subset of bacterial mRNAs (40, 69). In yeast, Lsm1p-Lsm7p, encoded by the LSM1 to LSM7 genes, form a cytoplasmic heteroheptameric decapping activator complex. The Lsm1p-Lsm7p complex can interact with Pat1p, another deadenylation-dependent decapping factor, to form the Lsm1p-7p/Pat1p complex (6).
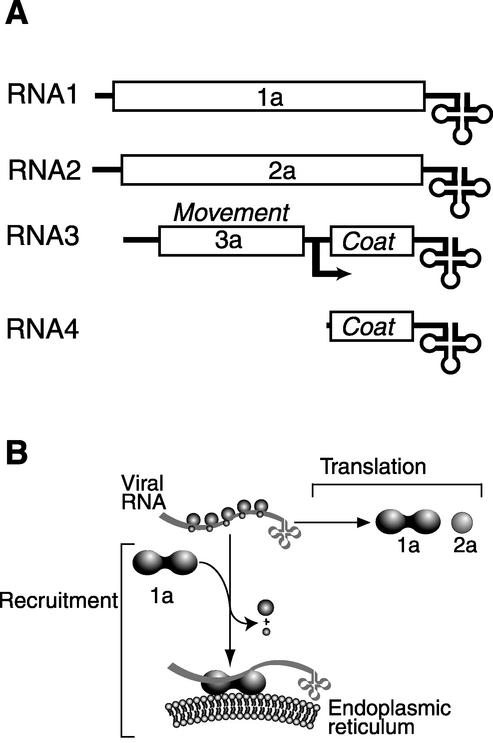
Viruses are obligate intracellular parasites that exploit existing host machineries in replicating and spreading their genomes. Positive-strand RNA viruses are a large group of viruses that cause serious acute and chronic diseases (64). One model for studying positive-strand RNA virus gene expression and RNA replication is brome mosaic virus (BMV), a member of the alphavirus-like superfamily of human, animal, and plant viruses. The BMV genome consists of three 5′-capped, messenger-sense genomic RNAs (2, 56) (Fig. 1A). Instead of the 3′ poly(A) of cellular mRNAs, BMV RNAs have a 3′ tRNA-like structure (3, 19, 46) that is aminoacylated in vivo by host enzymes (34). RNA1 and RNA2 encode essential viral RNA replication factors 1a and 2a. RNA3 is dispensable for RNA replication but encodes cell-to-cell movement and coat proteins required for systemic infection in BMV's natural plant hosts. Coat protein is translated only from a subgenomic mRNA, RNA4, initiated internally on negative-strand RNA3.
FIG. 1.
(A) Schematic diagram of the BMV genome (RNA1 to RNA3) showing ORFs (open boxes), NCRs (single lines), tRNA-like 3′ ends (cloverleaf), the subgenomic mRNA start site (bent arrow), and the subgenomic mRNA (RNA4). (B) Schematic diagram of BMV RNA translation and 1a-dependent recruitment from translation to replication on the ER.
BMV has recently been shown to share fundamental similarities in nucleic acid replication with retroviruses and double-stranded RNA viruses (50). BMV also has the nearly unique ability to direct viral RNA replication, subgenomic mRNA synthesis (31), and encapsidation (35) in S. cerevisiae, recapitulating all known features of BMV replication in plant cells. As in plant cells, BMV RNA replication in yeast is localized to the endoplasmic reticulum (ER) (43, 44), depends on the viral 1a protein and 2a polymerase, requires specific cis-acting RNA signals (55), and generates excess positive- to negative-strand RNA (31). Accordingly, yeast genetics has been used to identify a number of host factors required for BMV gene expression and RNA replication (17, 27, 38, 41).
Upon entering the cell, the genomes of all positive-strand RNA viruses must be translated to produce viral replication factors. These replication factors then recognize the viral RNA and recruit it out of translation and into a membrane-bound complex for replication (Fig. 1B). The transition between these alternate viral RNA states must be tightly regulated to allow sufficient translation of viral replication proteins but also efficient subsequent recruitment to replication. In addition, from the large pool of RNAs present in the cell, viruses must ensure that only viral RNAs are selected for replication. BMV RNAs are recruited from translation to replication by 1a, a multifunctional viral protein with key roles in assembly and function of the RNA replication complex (10, 30, 50). The yeast LSM1 gene is also required for efficient BMV RNA recruitment (17), suggesting a link between viral RNA recruitment and deadenylation-dependent mRNA turnover. In the present study we extend this observation to show that Lsm1p and other components of the Lsm1p-7p/Pat1p deadenylation-dependent decapping complex also are required for BMV RNA translation. We show that BMV uses the host deadenylation-dependent mRNA decapping and turnover pathway to distinguish viral replication from nonreplication templates at the stage of translation, well before RNA recruitment to replication. We also show that BMV RNA translation is blocked with a similar complement of ribosomes still on the RNA in yeast expressing a deficient Lsm1p-7p/Pat1p complex. In these yeast, ribosomes appear to be greatly slowed during translation elongation on the BMV RNA open reading frame (ORF), suggesting a similar mechanism of translation regulation as the subcellularly localized yeast ASH1 mRNA (9). Our results link BMV RNA translation and subsequent recruitment to viral RNA replication and show that factors of the Lsm1p-7p/Pat1p complex function not only in mRNA decapping but also in translation.
MATERIALS AND METHODS
Yeast and plasmids.
Standard yeast genetic techniques and media were used (5, 24). All yeast strains were from the S. cerevisiae genome deletion project (67) and were purchased from Research Genetics, Inc. (Invitrogen Corp., Carlsbad, Calif.). All mutant yeast strains used, including lsm1Δ, lsm6Δ, lsm7Δ, pat1Δ, upf1Δ, upf2Δ, upf3Δ, vps16Δ, edc1Δ, and edc2Δ, were derived from the parent wt strain BY4742 yeast (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) as described previously (67). The temperature-sensitive prt1-1 and lsm7Δ/prt1-1 yeast were constructed by replacing the genomic PRT1 ORF with the HIS3 ORF in BY4742 and lsm7Δ yeast, respectively, and providing the essential functions of Prt1p from YEpDETP-11, a plasmid expressing the temperature-sensitive prt1-1 allele (18).
Wild-type (wt) BMV RNA1, RNA2, and RNA3 were expressed from pB1AON1 (41), pB2NR3 (11), and pB3RQ39 (28), respectively. wt RNA4 was expressed from pB4MK1, constructed and kindly provided by Michael Krol. In short, the oligonucleotide linkers B3.RNA4A (5′-TCGACATTATTAATACGTA-3′) and B3.RNA4B (5′-TCGATACGTATTAATAATG-3′) were annealed and inserted into the SalI site of pB3TP8, creating a SnaBI site at the 5′ end of RNA4. The SnaBI/KpnI fragment from the resulting plasmid was exchanged for the SnaBI/KpnI fragment from pB3RQ39 to create pB4MK1. BMV/BMV/PolyA and GAL1/BMV/BMV mRNAs were expressed as described previously (41), and GAL1/BMV/PolyA mRNA was expressed from pB2YT5 (17). BMV/GFP/BMV and GAL1/GFP/PolyA mRNAs were expressed from pAON60 and pAON64. A green fluorescent protein (GFP) ORF with upstream BstBI and downstream StuI restriction sites was generated by PCR with the primers 1849 (5′-CACCAAGATGTCTTCGAAAGGTGAAGAATTATTCACTGGTGTTGTCCC-3′) and 1850 (5′-TCAACTGACGTTAGAGGCCTTTTGTACAATTCATCCATACCATGG3′) and a version of GFP optimized for yeast codon usage from yEGFP (15). The resulting PCR fragment was digested with BstBI and StuI and used to replace in frame the BstBI/StuI-flanked 2a ORF in pB2NR3 and pB2YT5, yielding pAON60 and pAON64, respectively.
RNA, protein, and polyribosome analysis.
Total yeast RNA isolation, Northern blot analysis, and total protein extractions and Western blot analysis with anti-1a, anti-2a, anti-CP, and anti-GFP were as described previously (16, 27, 31, 35, 55). Polyclonal anti-3a sera (39) were diluted 1:1,000.
Isolation of polyribosomes was as described previously (47) with some adjustments. In short, 0.1 mg of cycloheximide/ml was added to 300-ml yeast cultures at an optical density at 600 nm (OD600) of 0.6 to 1. The cultures were immediately chilled on ice, and all subsequent steps were carried out at 0 to 4°C with ice-cold buffer. Cells were washed twice with 24 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM KCl, 30 mM MgCl2, 6 mM dithiothreitol, 0.1 mg of cycloheximide/ml) and resuspended in 1 ml of final volume lysis buffer. Then, 200 U of RNasin (Promega), 200 U of Superase (Ambion), and 1 mg of heparin were added, and cells were disrupted by vortexing six times for 45 s with 500 μl of glass beads. The extracts were clarified by centrifuging the lysate and the resulting supernatant at 5,200 × g, then at 10,600 × g, and finally at 20,800 × g for 5 min each time. Next, 100 μl of extract was layered on top of a 12 ml of a 10 to 50% (wt/vol) sucrose gradient in lysis buffer with 0.5 mg of heparin/ml, 160 U of RNasin/ml, and 80 U of Superase/ml. Tubes were spun in a Beckman SW41 rotor at 39,000 rpm for 160 min at 4°C. An ISCO density gradient fractionator was used to collect 400-μl fractions for Fig. 7 and 8A and 100-μl fractions for Fig. 8B. RNA was isolated from 100-μl fractions by phenol-chloroform extraction, followed by precipitation with ethanol, and was analyzed by Northern blotting. Where indicated, EDTA was added to the sucrose gradient to a final concentration of 10 mM. For Fig. 9, yeast were grown at 26°C to an OD600 of 0.6 to 1, shifted to 37°C by the addition of an equal volume of medium prewarmed to 54°C, and incubated at 37°C for 30 min.
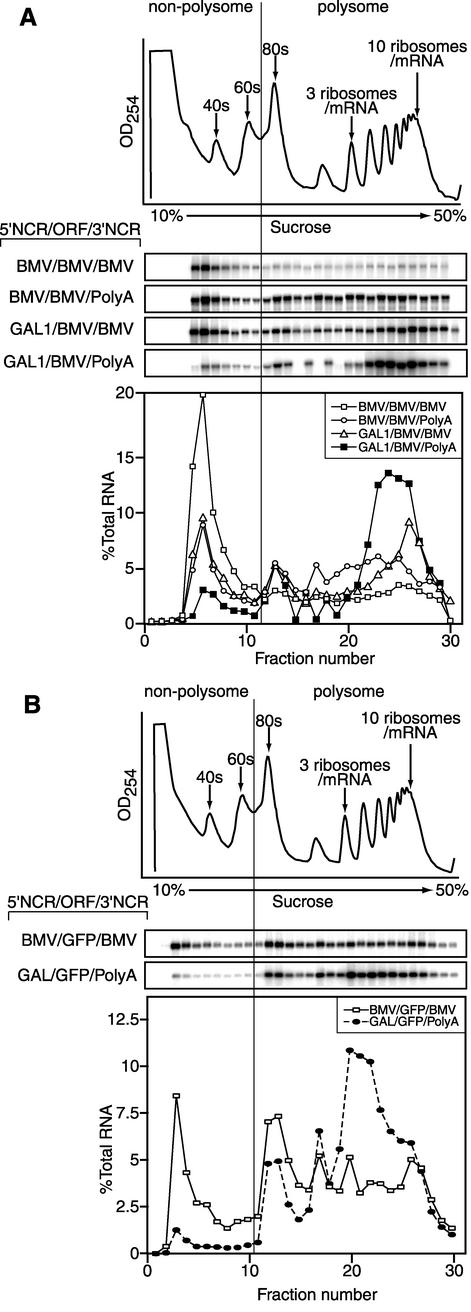
FIG. 7.
Sucrose density gradient sedimentation analysis of the ribosome and polysome association of RNA2 derivatives in wt yeast. (A) The top trace shows the UV absorbance profile at 254 nm of a cytoplasmic extract of wt yeast after sedimentation on a 10 to 50% (wt/vol) sucrose gradient. Aligned below this are Northern blots, probed for the 2a ORF, of fractions from identical gradients, centrifuged in parallel, of extracts from yeast expressing the indicated RNA2 derivatives from Fig. 6A. The bottom curves plot the amount of RNA2-derived mRNA in each fraction, expressed as a percentage of the total amount of RNA2-derived mRNA recovered over the gradient. (B) Same as in panel A but for mRNAs with the 2a ORF replaced by the GFP ORF. Each experiment was repeated three or more times, and representative data are shown. The vertical line through the UV absorbance profiles, Northern blots, and curves demarcates the point between ribosome-associated and non-ribosome-associated RNA.
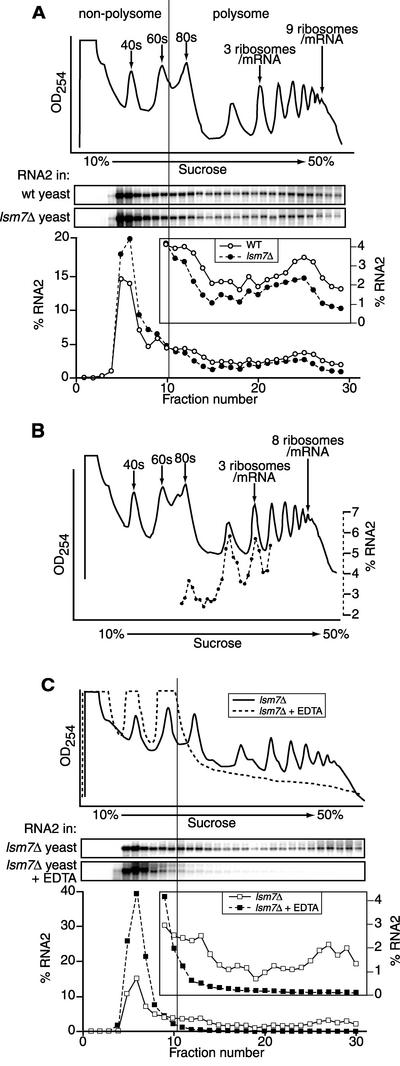
FIG. 8.
Distribution of RNA2 in polysome and nonpolysome fractions in wt and lsm7Δ yeast. (A) The top trace shows the UV absorbance profile at 254 nm of a cytoplasmic extract from wt yeast expressing wt RNA2, after sedimentation on a 10 to 50% sucrose gradient. Aligned below this are Northern blots of RNA2 in fractions from identical gradients, centrifuged in parallel, of extracts of wt and lsm7Δ yeast expressing RNA2. The bottom curves plot the amount of RNA2 in each fraction, expressed as a percentage of the total amount of RNA2 recovered over the gradient. The inset in the lower panel plots samples 10 to 29 on an expanded scale of 0 to 4% total RNA2. The vertical line through the UV absorbance profiles, Northern blots, and curves demarcates the point between ribosome-associated and non-ribosome-associated RNA. (B) Coincidence of RNA2 with ribosome peaks in polysome fractions. UV absorbance profile at 254 nm of cellular extracts from lsm7Δ yeast expressing RNA2 (solid line) and quantitation of Northern blot analysis of small fractions in the middle of the gradient (dotted line). The amount of RNA2 in each fraction is expressed as a percentage of the total amount of RNA2 recovered in these fractions. Each experiment was repeated three or more times, and representative data are shown. (C) The traces in the top panel show the UV absorbance profile at 254 nm of a cytoplasmic extract from lsm7Δ yeast expressing wt RNA2, after sedimentation on a 10 to 50% sucrose gradient without EDTA (solid line) and with EDTA (dashed line). Aligned below this are Northern blots of RNA2 in fractions from these gradients, centrifuged in parallel. The bottom curves plot the amount of RNA2 in each fraction, expressed as a percentage of the total amount of RNA2 recovered over the gradient. The inset in the lower panel plots samples 9 to 30 on an expanded scale of 0 to 4% total RNA2. The vertical lines through the UV absorbance profiles, Northern blots, and curves demarcate the point between ribosome-associated and non-ribosome-associated RNA.
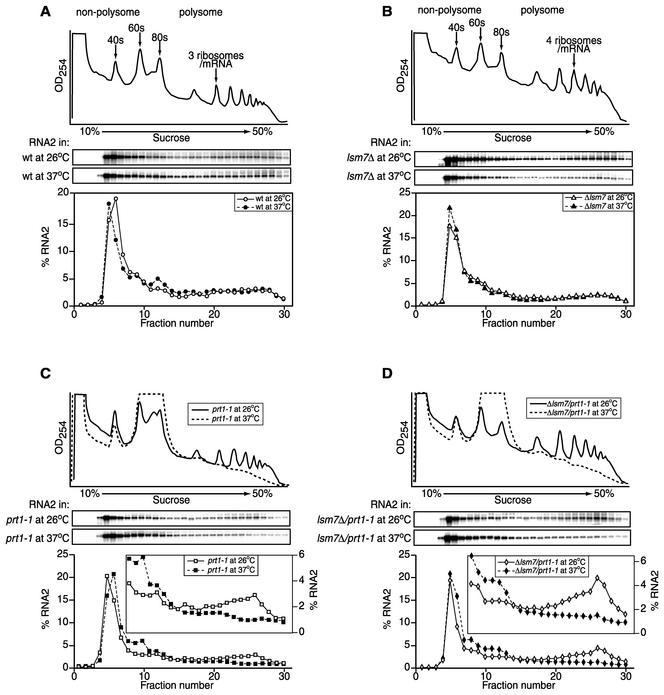
FIG. 9.
Distribution of RNA2 in polysome and nonpolysome fractions in wt (A), lsm7Δ (B), prt1-1 (C), and lsm7Δ/prt1-1 (D) yeast at 26 and 37°C. The traces in the top panels show the UV absorbance profiles at 254 nm of cytoplasmic extracts from the relevant yeast strains expressing wt RNA2 grown at 26°C (solid lines) and grown at 26°C, followed by incubation at 37°C for 30 min (dotted lines), after sedimentation on a 10 to 50% sucrose gradient. Aligned below this are Northern blots of RNA2 in fractions from these gradients, centrifuged in parallel. The bottom curves plot the amount of RNA2 in each fraction, expressed as a percentage of the total amount of RNA2 recovered over the gradient. The insets in lower panels of panels C and D plot samples 8 to 30 on an expanded scale of 0 to 6% total RNA2.
RESULTS
Components of the Lsm1p-7p/Pat1p complex are required for BMV RNA2 translation.
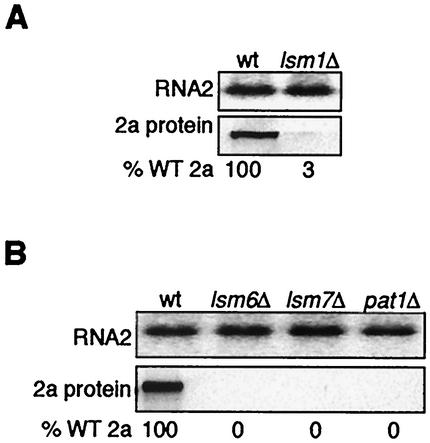
Since the LSM1 gene facilitates recruitment of BMV RNA to the replication complex and since BMV RNA translation and recruitment to the replication complex are linked, we assayed the effect of an LSM1 gene deletion on viral RNA translation. A plasmid expressing wt BMV RNA2 under the control of the yeast GAL1 promoter was introduced into wt yeast and an isogenic LSM1 deletion strain, lsm1Δ. After GAL1 promoter induction in galactose medium, RNA2 and 2a protein levels were compared in extracts from wt and lsm1Δ yeast (Fig. 2A). RNA2 levels were unchanged in lsm1Δ yeast. However, 2a protein accumulation in lsm1Δ yeast was decreased by 33-fold, suggesting inhibition of RNA2 translation.
FIG. 2.
Inhibition of RNA2 translation in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ mutant yeast. (A and B) Northern and Western blot analysis of RNA2 and 2a protein in wt and lsm1Δ yeast (A) and in wt and lsm6Δ, lsm7Δ, and pat1Δ yeast (B). Northern blots were hybridized with positive-strand-specific RNA probes from the 2a gene. The percent wt (% WT) 2a protein values are averages of three or more experiments.
In an independent genetic screen of ethyl methanesulfonate-mutagenized yeast for inhibition of BMV-directed marker gene expression (41), we found the yeast LSM6 gene also to be required for BMV RNA replication (A. O. Noueiry and P. Ahlquist, unpublished results). Subsequently, it was shown that yeast LSM1 and LSM6 plus LSM2, LSM3, LSM4, LSM5, LSM7, and PAT1 encode a cytoplasmic Lsm1p-7p/Pat1p complex that facilitates decapping of deadenylated mRNAs, leading to degradation by 5′ exonuclease Xrn1p (7, 57). These data suggested possible functional associations of viral RNA translation and replication with this larger host complex required for deadenylation-dependent mRNA turnover. To test whether Lsm1p-7p/Pat1p might be required for BMV RNA translation, RNA2 and 2a protein levels were compared in extracts from wt yeast and the lsm6Δ, lsm7Δ, and pat1Δ deletion strains (Fig. 2B). Together with the LSM1 gene tested above, these genes represent the full complement of nonessential genes encoding components of the Lsm1p-7p/Pat1 complex in yeast (67). As in lsm1Δ yeast, BMV RNA2 levels were unchanged in lsm6Δ, lsm7Δ, and pat1Δ yeast, whereas 2a protein was undetectable. These results implied that the deadenylation-dependent Lsm1p-7p/Pat1 decapping complex was required for translating RNA2.
mRNA decay genes not specific for deadenylated mRNA turnover are not required for RNA2 translation.
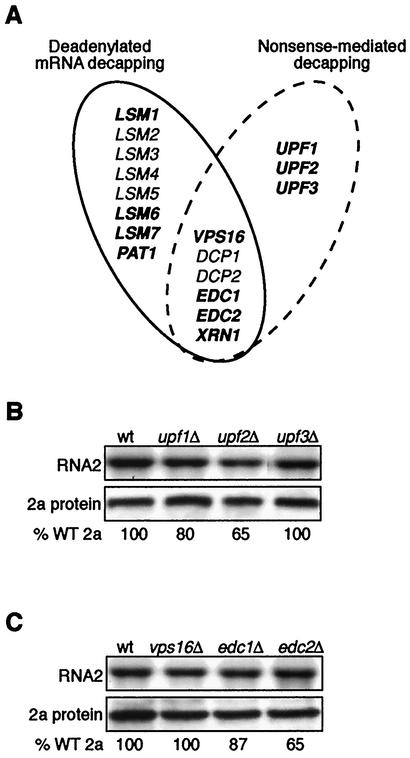
Nonsense-mediated mRNA decay (NMD) is another pathway of 5′ to 3′ mRNA turnover related to deadenylated mRNA turnover. In NMD, aberrant polyadenylated messages containing premature nonsense codons are decapped and degraded 5′ to 3′ by Xrn1p. The deadenylation-dependent and NMD pathways each require shared and pathway-specific genes (Fig. 3A). Deadenylation-dependent mRNA turnover specifically requires the genes that encode the Lsm1p-7p/Pat1p complex, whereas NMD specifically requires the UPF1 to -3 genes. Both pathways also require Vps16p, the decapping enzyme Dcp1p, its associated proteins Dcp2p and Edc1p-2p, and 5′ to 3′ exonuclease Xrn1p. To investigate the range of genes in these pathways that were also required for RNA2 translation, we tested RNA2 translation in isogenic strains with deletions of individual genes specific for the NMD pathway or common to the deadenylation-dependent and NMD mRNA turnover pathways.
FIG. 3.
(A) Venn diagram representing relationships between yeast genes required for the deadenylation-dependent and NMD mRNA decapping and turnover pathways. The solid and dashed ellipses surround genes for deadenylation-dependent and NMD mRNA turnover pathways, respectively. Genes in the intersection are involved in both pathways. Genes not essential for survival in the yeast strain BY4742 are in boldface. (B) Northern and Western blot analysis of RNA2 and 2a protein accumulation in wt and upf1Δ, upf2Δ, and upf3Δ yeast. (C) Same as in panel B for wt and vps16Δ, edc1Δ, and edc2Δ yeast. Northern blots were hybridized with positive-strand specific RNA probes from the 2a gene. The percent wt (% WT) 2a protein values are averages of three or more experiments.
First, we tested RNA2 and 2a protein levels in extracts from wt yeast and the isogenic, NMD-specific deletion strains upf1Δ, upf2Δ, and upf3Δ (Fig. 3B). As with gene deletions in the LSM1-7/PAT1 complex, RNA2 levels were unchanged in upf1Δ, upf2Δ, and upf3Δ yeast. However, unlike the dramatic inhibition of 2a protein accumulation seen with deletion of genes specific to deadenylation-dependent mRNA decay (Fig. 2), 2a protein accumulation was only slightly reduced to 80 and 65% of the wt in upf1Δ and upf2Δ yeast, respectively, and was unaltered in upf3Δ yeast.
Second, we tested BMV RNA2 and 2a protein levels in extracts from wt yeast and the isogenic vps16Δ, edc1Δ, and edc2Δ strains, representing nonessential genes required for both deadenylation-dependent and NMD mRNA turnover pathways (Fig. 3C). BMV RNA2 levels were unchanged in vps16Δ, edc1Δ, and edc2Δ yeast. As with the UPF gene deletions, 2a protein accumulation was unaltered in vps16Δ yeast and was only partially reduced to 87 and 65% of wt in edc1Δ and edc2Δ yeast, respectively. Thus, while genes specifically required for deadenylation-dependent turnover of mRNAs were essential for RNA2 translation (Fig. 2), genes specific for the NMD pathway or common to both the NMD and the deadenylation-dependent mRNA turnover pathways were not required.
Lsm1p-7p/Pat1p complex factors are selectively required for translating all BMV RNAs destined for replication.
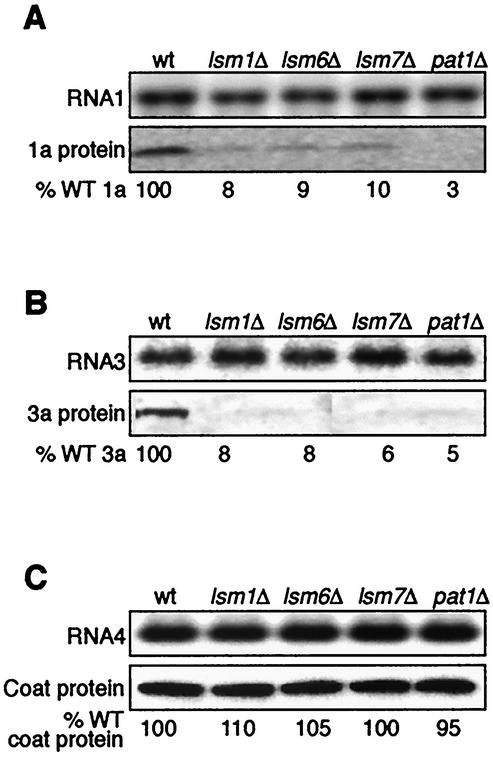
We previously reported that a mutant allele of the essential yeast translation initiation gene DED1 specifically inhibited translation of BMV RNA2 but not the related BMV RNA1, showing that one or more aspects of translation initiation are regulated differently for RNA2 than RNA1. For comparison, we tested the dependence of RNA1 translation on the dispensable factors of the Lsm1p-7p/Pat1p complex. A plasmid expressing wt RNA1 under the control of the yeast GAL1 promoter was introduced into wt and isogenic lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast, representing the Lsm1p-7p/Pat1p complex. BMV RNA1 levels were unchanged in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast (Fig. 4A). However, in contrast to unimpaired RNA1 translation in mutant DED1 yeast (41), 1a protein accumulations in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast strains were reduced to 8, 9, 10, and 3% of the wt yeast level, respectively.
FIG. 4.
Inhibition of genomic RNA1 and RNA3 but not subgenomic RNA4 translation in LSM1/PAT1 mutant yeast. (A to C) Northern and Western blot analysis of the accumulation of RNA1 and 1a protein (A), RNA3 and 3a protein (B), and RNA4 and coat protein (C) in wt and lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast. Northern blots were hybridized with positive-strand specific RNA probes for the 3′ tRNA-like ends conserved on all BMV RNAs. The percent wt (% WT) 1a, 3a, or coat protein values are averages of three or more experiments.
To determine whether all BMV genomic RNAs required the Lsm1p-7p/Pat1p complex for translation, a plasmid expressing full-length wt BMV RNA3 under the control of the yeast GAL1 promoter was introduced into wt and isogenic lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast. BMV RNA3 is bicistronic (Fig. 1), but only the 5′ terminal 3a ORF is translated directly from RNA3. Similar to results with RNA1 and RNA2, BMV RNA3 levels were unchanged in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast, but 3a protein levels were reduced to 8, 8, 6, and 5% of wt yeast, respectively (Fig. 4B). These results show that all BMV genomic RNAs are dependent on the LSM1/PAT1 complex for translation and that this viral RNA translation defect is distinct from the DED1 translation initiation defect observed specifically for BMV RNA2.
During viral replication, the 3′-terminal coat protein ORF of RNA3 is expressed by synthesis of RNA4, a subgenomic viral mRNA transcribed from negative-strand RNA3 (Fig. 1A). To find out whether the nonreplicating RNA4 depends on the Lsm1p-7p/Pat1p complex for translation, a plasmid expressing wt BMV RNA4 under the control of the GAL1 promoter was introduced into wt and isogenic lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast representing the Lsm1p-7p/Pat1p complex. BMV RNA4 levels were unchanged in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast. However, in contrast to RNA1, RNA2, and RNA3 translation, coat protein accumulation levels were the same for wt, lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast. These results show that only genomic BMV RNAs destined for replication are dependent on the Lsm1p-7p/Pat1p complex for translation.
Normal cell growth and translation in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast.
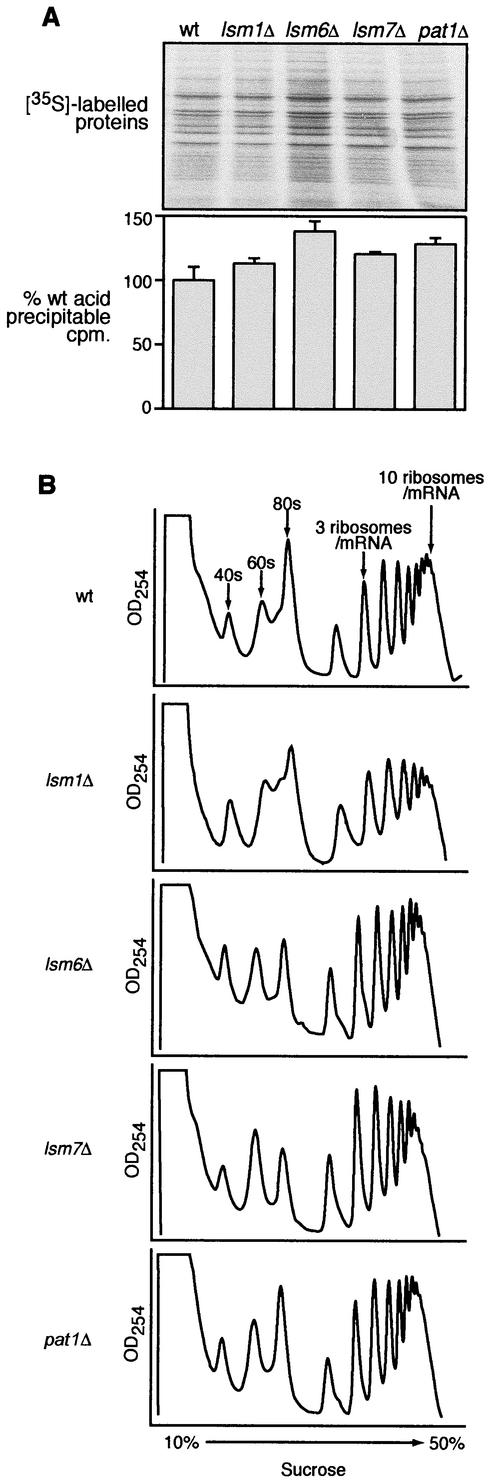
lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ deletion strains grew robustly, with a cell doubling rate indistinguishable from that of isogenic wt yeast, suggesting normal general cellular functions, including translation. To determine whether the effects on translation were general to most yeast mRNAs or specific to BMV RNAs, we tested the incorporation of [35S]methionine-cysteine into peptides in wt and lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast (Fig. 5A). Incorporation of [35S]methionine-cysteine into acid-precipitable peptides was the same as for the wt or slightly higher than for the wt in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast (Fig. 5A, lower panel). Similarly, levels of 35S-labeled proteins observed on a sodium dodecyl sulfate-polyacrylamide gel were at the wt level or slightly increased in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast, and the pattern of protein bands was unchanged (Fig. 5A, upper panel).
FIG. 5.
General cellular translation is unaffected in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ mutant yeast. (A) Polyacrylamide gel autoradiograph (upper panel) and acid-precipitable counts (lower panel) from 35S-labeled protein extracts from wt and lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast. The percent wt (% WT) acid-precipitable counts are averages of three or more experiments. (B) UV absorbance profile at 254 nm of cellular extracts from wt and lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast fractionated on 10 to 50% (wt/vol) sucrose gradients. Each experiment was repeated three or more times, and representative data are shown.
In some but not all (13, 68) genetic backgrounds, deleting the PAT1 gene results in slow growth and a decrease in the amount of polysomes, as observed by fractionation on sucrose density gradients. To test this in our yeast deletions, cellular extracts from wt and isogenic lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast were prepared and fractionated on 10 to 50% sucrose gradients (Fig. 5B). The distribution of polysomes showed no significant differences between the wt and the isogenic lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast (Fig. 5B). These results indicate that overall cellular translation levels and ribosome loading in lsm1Δ, lsm6Δ, lsm7Δ, and pat1Δ yeast are the same as for the wt yeast.
Viral 5′ NCR, 3′ NCR, and 2a ORF contribute to LSM1, LSM6, and PAT1 dependence of viral RNA translation.
Secondary structure analysis, sequence comparison, and mapping of the 5′ noncoding region (NCR) of RNA2 has identified subdomains involved in attenuating translation initiation of RNA2 and in making this RNA especially dependent on the yeast essential translation initiation factor DED1 (41). To identify the sequences responsible in cis for the selective inhibition of BMV RNA2 translation in yeast lacking components of the Lsm1p-7p/Pat1p complex, the 5′ and 3′ NCRs and the 2a ORF of BMV RNA2 were separately or simultaneously replaced with nonviral sequences, and the resulting RNAs were assayed for translation in wt yeast and the relevant isogenic deletion strains. Similar results were obtained in lsm1Δ, lsm6Δ, and pat1Δ yeast in these and later experiments. Moreover, similar results in the same strains were also obtained by using analogous replacements of the 5′ and 3′ NCRs and ORFs of BMV genomic RNA3. For simplicity, 2a mRNA translation results from wt and lsm7Δ yeast will be presented here and in subsequent sections to illustrate representative results.
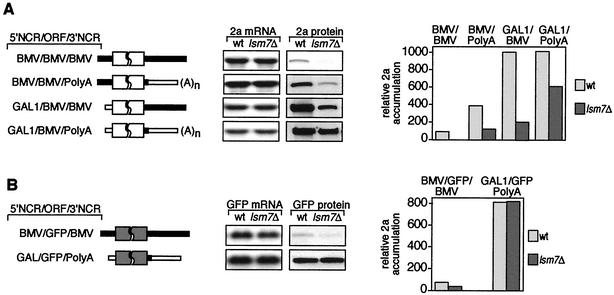
To address the role of the RNA2 NCRs, the viral 5′ and 3′ NCRs were replaced with the yeast GAL1 5′ leader and ADH1 terminator-derived polyadenylation signals, respectively, to produce three chimeric 2a mRNAs (41). BMV/BMV/PolyA contains the wt RNA2 5′ NCR and the polyadenylated ADH1 3′ NCR, GAL1/BMV/BMV contains the GAL1 5′ NCR and wt RNA2 3′ NCR, and GAL1/BMV/PolyA contains the GAL1 5′ NCR and the polyadenylated ADH1 3′ NCR (Fig. 6A). To provide a measure of the translational activity of each 2a mRNA in wt and lsm7Δ yeast, relative 2a accumulation was calculated as the ratio between the 2a protein level and the 2a mRNA level. In wt yeast (Fig. 6A), replacing the RNA2 tRNA-like 3′ NCR with the polyadenylated ADH1 3′ NCR increased 2a translation nearly fourfold. As noted previously (41), replacing the RNA2 5′ NCR had an even greater effect, increasing 2a translation 10-fold either alone or in combination with the ADH1 3′ NCR substitution.
FIG. 6.
Mapping of RNA2 determinants contributing to dependence of translation on LSM7. (A) Schematic diagrams of 2a mRNAs showing 2a ORF (white box), wt RNA2 5′ and 3′ NCRs (solid bars), and GAL1 mRNA 5′ and ADH1 3′ NCRs (open bars). Northern and Western blot analyses of 2a mRNA and protein accumulation in wt and lsm7Δ yeast containing these mRNAs are shown in the center. The histogram compares relative 2a protein accumulation for all combinations. Relative 2a accumulation was defined as the ratio between 2a protein and mRNA levels and was normalized to relative 2a accumulation for wt RNA2 (BMV/BMV/BMV) in wt yeast (100%). (B) Same as in panel A but for mRNAs with the 2a ORF replaced by the GFP ORF (gray box). The percent wt 2a protein values are averages of three or more experiments.
When lsm7Δ yeast was compared to wt yeast (Fig. 6A), the levels of RNA2 and its chimeric 2a mRNA derivatives were unaltered, but the relative 2a protein accumulation was inhibited to various degrees. The LSM7 dependence of 2a accumulation was greatest for wt RNA2, for which 2a accumulation in lsm7Δ yeast remained undetectable. Replacing the BMV 5′ or 3′ NCRs with the yeast GAL1 leader or ADH1 polyadenylation signal enhanced 2a accumulation in lsm7Δ yeast from 3% to 20 or 30%, respectively, of that in wt yeast. Simultaneously replacing both the BMV 5′ and 3′ NCRs enhanced 2a accumulation in lsm7Δ yeast additively. However, even in this case, 2a accumulation in lsm7Δ yeast remained only 60% of that in wt yeast. This implied that the LSM7 dependence of 2a translation was not only due to the RNA2 5′ and 3′ NCRs but also in part to contributions of the 2a ORF itself.
To further test whether the 2a ORF contributed to the LSM7 dependence of RNA2 translation, the 2a ORF of wt RNA2 and GAL1/BMV/PolyA was replaced with the GFP ORF to produce BMV/GFP/BMV and GAL1/GFP/PolyA (Fig. 6B). In wt yeast, the relative accumulation of GFP protein was 10-fold higher for GAL1/GFP/PolyA mRNA than for BMV/GFP/BMV mRNA, showing that the RNA2 5′ and 3′ NCRs attenuate the translation of a reporter ORF to the same degree as the 2a ORF. In lsm7Δ yeast, BMV/GFP/BMV and GAL1/GFP/PolyA transcript levels were unchanged compared to wt yeast. GFP protein accumulation from BMV/GFP/BMV mRNA was partially inhibited to 60% of that in wt yeast, whereas GFP accumulation from GAL1/GFP/PolyA mRNA, containing no viral RNA sequences, was equal to that in wt yeast. Thus, the RNA2 5′ and 3′ NCRs can confer LSM7 dependence on the translation of a reporter ORF, but the 2a ORF also contributes to LSM7 dependence. Together, the results presented in Fig. 6 show that, in contrast to the 5′ NCR-linked dependence of RNA2 translation on DED1, the dependence of RNA2 translation on LSM7 was due to combined effects of the RNA2 5′ NCR, 3′ NCR, and 2a ORF.
Dependence of viral RNA translation on Lsm1p-7p/Pat1p complex factors correlates with an untranslated cytoplasmic viral mRNP peak.
To further understand viral RNA dynamics and the contribution of Lsm1p-7p/Pat1p complex factors to BMV RNA translation, we used sucrose density gradient centrifugation to determine the degree of ribosome association for various BMV RNA2 derivatives (Fig. 7A). Appropriate extracts from wt yeast expressing wt RNA2 or the 5′ NCR, 3′ NCR, or 2a ORF replacement derivatives described above were fractionated on 10 to 50% sucrose gradients. Total RNA was isolated from fractions across the gradient and analyzed by Northern blotting (Fig. 7A).
wt RNA2 fractionated into two pools in the sucrose gradient. A total of 60% of the RNA was recovered as a single peak in the top fractions devoid of ribosomes, indicating that this RNA was not translated. Although not associated with ribosomes, this RNA2 peak sedimented just above the 40s peak. Since this implies an association with proteins in one or more ribonucleoprotein (mRNP) complexes, this peak is referred to hereafter as the mRNP peak. The remaining 40% of RNA2 was evenly distributed throughout the fractions containing polysomes. Replacing the wt viral 5′ or 3′ NCRs with yeast sequences had distinct effects on the distribution of RNA derivatives between the viral mRNP peak and ribosomal fractions and within the ribosomal fractions. Replacing either the 5′ or 3′ NCRs decreased the relative amount of RNA2 in the mRNP peak similarly from 60 to 30% of total RNA. Moreover, replacing the wt RNA2 5′ NCR but not the 3′ NCR shifted the distribution of RNA2 in the fractions containing ribosomes toward the bottom of the gradient. Since the efficiency of translation initiation on a mRNA correlates with the average number of ribosomes the mRNA bears, the increased ribosome occupancy observed when the viral 5′ NCR was replaced further confirms the conclusions described above that the viral 5′ NCR attenuates wt RNA2 translation initiation (Fig. 6A). Replacing both wt RNA2 5′ and 3′ NCRs further decreased the mRNP peak to 10% of total mRNA and further shifted the mRNA distribution toward the bottom of the gradient. Thus, the viral 5′ and 3′ NCRs contribute additively to the mRNP peak.
Similarly, we determined the distribution of BMV/GFP/BMV and GAL1/GFP/PolyA RNAs in cell lysates fractionated on 10 to 50% sucrose gradients (Fig. 7B). Like wt RNA2, the BMV/GFP/BMV mRNA containing viral NCRs exhibited a prominent mRNP peak in the fractions devoid of ribosomes, with the remaining mRNA distributed in the polysome-containing fractions. However, the relative distribution of mRNA in the mRNP peak and polysome fractions differed from the wt RNA2 distribution, with only 25% of the RNA recovered in the mRNP peak, and the remaining 75% in the polysome fractions. For the GAL1/GFP/PolyA mRNA containing no viral sequences, only 4% of the total mRNA was found in an mRNP peak. The remainder of the mRNA was in the fractions containing ribosomes, primarily in the bottom of the gradient, suggesting efficient translation initiation. Therefore, the 2a ORF contributed to the accumulation of 2a mRNA in the mRNP peak. These results corroborated the role of the viral 5′ NCR in attenuating translation initiation and showed that the viral 5′ and 3′ NCRs were sufficient to induce mRNA accumulation in the mRNP peak. Comparison of these results with the mapping results in Fig. 6 shows that the presence and intensity of the viral mRNP peak correlates with the dependence of RNA2 translation on genes of the Lsm1p-7p/Pat1p complex.
Untranslated BMV RNA2 in lsm7Δ yeast is associated with ribosomes.
To test whether the block of BMV RNA translation in lsm7Δ yeast is at translation initiation, elongation, or both, the distribution of viral RNA2 on 10 to 50% sucrose gradients of cell lysates from wt and lsm7Δ yeast was determined. Despite the dramatic inhibition of RNA2 translation in lsm7Δ yeast (Fig. 2B), viral RNA2 distribution in the sucrose gradient in lsm7Δ yeast was similar to the distribution of RNA2 in wt yeast (Fig. 8A; see also Fig. 9 below). A fraction of the viral RNA was present in a mRNP complex coincident with the RNA2 mRNP complex in wt yeast, and the rest of the RNA was evenly distributed in the higher-molecular-weight polysome-containing fractions of the gradient. A consistent but relatively minor increase in the mRNP peak in lsm7Δ yeast was seen, with 70% of total RNA2 present in the mRNP peak in lsm7Δ yeast, compared to 60% in wt yeast. However, this difference was insufficient to account for the complete loss of 2a protein accumulation observed in lsm7Δ yeast. Thus, although RNA2 was not detectably translated in lsm7Δ yeast, the fraction of RNA2 sedimenting in the ribosome-containing lower portion of the gradient was similar to that of wt yeast.
To determine whether the portion of RNA2 sedimenting as high-molecular-mass complexes in lsm7Δ yeast was ribosome associated, we used higher-resolution analysis (47) to determine whether the fine structure distribution of RNA2 in the gradient coincided with the peaks of one, two, or more ribosomes as measured by UV absorbance. To that end, an extract from lsm7Δ yeast was fractionated on a 10 to 50% sucrose gradient, and small, high-resolution fractions were collected from the middle of the gradient. The level of RNA2 in each fraction was determined by Northern blot analysis. As shown in Fig. 8B, the distribution of RNA2 was highly discontinuous, with peaks and valleys precisely following the position of the 80s and multiple ribosome positions in the polysome profile. Thus, these results indicate that, despite the block to translation, BMV RNA2 was lsm7Δ yeast that is associated with ribosomes in lsm7Δ yeast.
As a further test of RNA2 association with ribosomes in lsm7Δ yeast, an extract from lsm7Δ yeast was fractionated on a sucrose gradient containing 10 mM EDTA. As shown in Fig. 8C (upper panel), the presence of EDTA dissociates monosomes and polysomes into 40s and 60s subunits, releasing any mRNA associated with ribosomes (37). As expected for an mRNA associated with ribosomes, the addition of EDTA shifted RNA2 almost completely out of the lower portion of the gradient and into the non-ribosome-associated mRNP peak at the top (Fig. 8C).
Inhibiting translation initiation reduces ribosome-association of RNA2 in wt and lsm7Δ yeast.
The results presented above suggested that inhibition of RNA2 translation in lsm7Δ yeast was primarily at the level of translation elongation. However, since the fraction of RNA2 associated with ribosomes was low even in wt yeast (Fig. 7), it was possible that even substantial further impairment of initiation, if it occurred in lsm7Δ yeast, might not produce a substantial change in RNA2 distribution in the polysome gradients. To directly address this concern, we determined the distribution of RNA2 in polysomes from strains harboring a recognized general translation initiation defect. PRT1 encodes an essential component of the translation initiation factor eIF3 complex, which brings the eIF2-GTP-Met-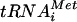 ternary complex to the 40s ribosomal subunit during initiation (reference 63 and references therein). For yeast with the temperature-sensitive allele prt1-1, cell growth and translation initiation are normal at 26°C but are rapidly inhibited at 37°C. In sucrose gradients, this inhibition of translation initiation upon switching to the nonpermissive temperature can be easily observed as a drastic reduction in polysomes and a corresponding increase in monosomes (Fig. 9C and D, upper panels). Accordingly, the prt1-1 allele was introduced into wt and lsm7Δ strains to produce isogenic prt1-1 and lsm7Δ/prt1-1 strains. wt, lsm7Δ, prt1-1, and lsm7Δ/prt1-1 yeast expressing RNA2 were grown at 26 or at 26°C, followed by growth at 37°C for 30 min, and cell lysates were fractionated on sucrose gradients to determine the distribution of RNA2. The polysome profile of extracts from wt or lsm7Δ yeast at 37°C was similar to that of yeast grown at 26°C (Fig. 9A and B). In all of these cell types, the effects of 37°C on total polysome profiles were mirrored by parallel changes in the gradient distribution of RNA2. Thus, in wt and lsm7Δ yeast, there was no difference between yeast grown at 26°C and yeast incubated at 37°C for 30 min (Fig. 9A and B). However, in prt1-1 and lsm7Δ/prt1-1 yeast, 30 min at 37°C resulted in a clear redistribution of RNA2 from the polysomal fractions to the monosomes, a finding consistent with inhibition of translation initiation (Fig. 9C and D). As a further test, LSM7 was also deleted from TP11B-2-2 (8), another prt1-1-containing yeast strain, to produce TP11B-2-2/lsm7Δ yeast. As in prt1-1 and lsm7Δ/prt1-1 yeast, TP11B-2-2 and TP11B-2-2/lsm7Δ yeast incubated 30 min at 37°C resulted in a clear redistribution of RNA2 from the polysomal fractions to the monosomes, a finding consistent with an inhibition of translation initiation irrespective of strain background (results not shown). Thus, blocking translation initiation produced clearly visible, parallel reductions in ribosome-associated RNA2 in both wt and lsm7Δ yeast. Accordingly, the lsm7Δ mutation still allows significant translation initiation but blocks RNA2 translation while ribosomes are still associated with the RNA.
ternary complex to the 40s ribosomal subunit during initiation (reference 63 and references therein). For yeast with the temperature-sensitive allele prt1-1, cell growth and translation initiation are normal at 26°C but are rapidly inhibited at 37°C. In sucrose gradients, this inhibition of translation initiation upon switching to the nonpermissive temperature can be easily observed as a drastic reduction in polysomes and a corresponding increase in monosomes (Fig. 9C and D, upper panels). Accordingly, the prt1-1 allele was introduced into wt and lsm7Δ strains to produce isogenic prt1-1 and lsm7Δ/prt1-1 strains. wt, lsm7Δ, prt1-1, and lsm7Δ/prt1-1 yeast expressing RNA2 were grown at 26 or at 26°C, followed by growth at 37°C for 30 min, and cell lysates were fractionated on sucrose gradients to determine the distribution of RNA2. The polysome profile of extracts from wt or lsm7Δ yeast at 37°C was similar to that of yeast grown at 26°C (Fig. 9A and B). In all of these cell types, the effects of 37°C on total polysome profiles were mirrored by parallel changes in the gradient distribution of RNA2. Thus, in wt and lsm7Δ yeast, there was no difference between yeast grown at 26°C and yeast incubated at 37°C for 30 min (Fig. 9A and B). However, in prt1-1 and lsm7Δ/prt1-1 yeast, 30 min at 37°C resulted in a clear redistribution of RNA2 from the polysomal fractions to the monosomes, a finding consistent with inhibition of translation initiation (Fig. 9C and D). As a further test, LSM7 was also deleted from TP11B-2-2 (8), another prt1-1-containing yeast strain, to produce TP11B-2-2/lsm7Δ yeast. As in prt1-1 and lsm7Δ/prt1-1 yeast, TP11B-2-2 and TP11B-2-2/lsm7Δ yeast incubated 30 min at 37°C resulted in a clear redistribution of RNA2 from the polysomal fractions to the monosomes, a finding consistent with an inhibition of translation initiation irrespective of strain background (results not shown). Thus, blocking translation initiation produced clearly visible, parallel reductions in ribosome-associated RNA2 in both wt and lsm7Δ yeast. Accordingly, the lsm7Δ mutation still allows significant translation initiation but blocks RNA2 translation while ribosomes are still associated with the RNA.
DISCUSSION
Genetic and biochemical results show that RNA virus replication requires host factors (4, 20, 27, 38, 41, 54). Prior studies in this area most often have characterized single, seemingly unrelated host gene products used by RNA viruses at different steps of infection. Here we show that LSM1, LSM6, LSM7, and PAT1, representing the full complement of nonessential genes in the Lsm1p-7p/Pat1p complex (67) that facilitates deadenylation-dependent mRNA decapping (61, 66), all were required for translation of BMV genomic RNAs. In addition, the dependence of BMV translation on mRNA turnover factors was specific to the deadenylation-dependent pathway. Genes required for the related NMD pathway, or genes common to both mRNA turnover pathways, were not required for BMV translation (Fig. 4). These parallels suggest that the functions of LSM1, LSM6, LSM7, and PAT1 in BMV translation are related to their functions in deadenylation-dependent mRNA decay and to the known Lsm1p-7p/Pat1p complex. Since LSM1 is also required for efficient BMV RNA recruitment to the replication complex (17), our results link BMV RNA translation and recruitment from translation to RNA replication to a pathway that facilitates cellular mRNA turnover.
Role of Lsm1p-7p/Pat1p complex genes in BMV RNA expression and replication.
During infection, positive-strand RNA viruses must ensure that only viral genomic RNAs destined for replication are selected from the large pool of RNAs present in the cell for recruitment to the replication complex. For BMV, such recruitment of genomic RNA1, RNA2, and RNA3, but not subgenomic RNA4 or cellular RNA, from translation to replication is mediated by the viral 1a protein (11, 30, 50). In the present study we found that LSM1, LSM6, LSM7, and PAT1 were required for translating BMV genomic RNA1, RNA2, and RNA3, but not subgenomic RNA4 (Fig. 4) or the vast majority of cell mRNAs (Fig. 5). Therefore, BMV genomic RNAs suitable for replication already were distinguished from nonreplication templates at translation, well before 1a-mediated recruitment of viral RNAs to replication.
Translation of all BMV genomic RNAs depended on the Lsm1p-7p/Pat1p complex, suggesting the possible involvement of features shared by all genomic RNAs. To date, the only known features common to all BMV genomic RNAs are the short box B motifs conserved in replication enhancer elements required for recruitment of BMV RNAs from translation to replication (11, 30, 55), and the 3′-terminal tRNA-like structure (3, 19, 46). Although either or both of these elements may contribute, our results show that neither is essential. Rather, multiple RNA2 sequences including the 5′ and 3′ UTRs and ORF contribute additively to Lsm1p-7p/Pat1p dependence of translation (Fig. 6 and see also below). Recent results show that the nuclear Lsm2p-8p complex associates with pre-tRNA primary transcripts in tRNA processing (36), suggesting that interaction of the 3′ tRNA-like end could be involved in recruiting the Lsm1p-7p/Pat1p complex. Again, however, the 3′ tRNA-like end was neither essential nor sufficient for translational dependence on the Lsm1p-7p/Pat1p complex. Translation of an RNA2 derivative with the 3′ tRNA-like end replaced with poly(A) was still facilitated by LSM7 (Fig. 6A and related results). Moreover, the BMV RNA4 subgenomic mRNA, which bears the same 3′ tRNA-like end as RNA3 (Fig. 1A), showed no translational dependence on the Lsm1p-7p/Pat1p complex (Fig. 4).
In lsm7Δ yeast, translation of RNA2 was inhibited, whereas RNA2 remained associated with nearly wt levels of ribosomes (Fig. 8A and B and 9C and D, 26°C). In contrast, when translation initiation was inhibited by 37°C inactivation of essential translation initiation factor Prt1p, the level of RNA2 in polysome-associated fractions declined dramatically and equally in wt and lsm7Δ yeast (Fig. 9C). The rapidly sedimenting fraction of RNA2 in polysome gradients therefore was ribosome associated, as was also confirmed by the high-resolution analysis of Fig. 8B. Moreover, although the fraction of RNA2 associated with polysomes was low even in wt yeast (Fig. 7), the decrease in ribosome association caused by prt1-1 inhibition of translation initiation was clearly observable (Fig. 9C and D). Thus, the lsm7Δ mutation allowed significant translation initiation and ribosome loading on RNA2 but blocked translation with ribosomes still present on RNA2. Thus, in lsm7Δ yeast, ribosomes are stalled or greatly slowed on RNA2.
Several other classes of mRNAs are translationally repressed while still associated with ribosomes. Among these are the yeast ASH1 and HAC1 mRNAs (9, 47) and some developmentally regulated metazoan mRNAs (12, 42, 51; see also below). In particular, translational repression of BMV genomic RNAs shows several similarities to that of yeast ASH1 mRNA. Asymmetric segregation of Ash1p to the daughter cell nucleus is regulated by slowing translation elongation on ASH1 mRNA during subcellular localization of the ASH1 mRNA to the bud tip (9). Like ASH1 mRNA, BMV genomic RNA destined for replication must be localized subcellularly, in this case to the ER membrane (50). Inhibition of elongation on ASH1 mRNA involves the additive action of multiple cis elements in the ASH1 ORF and 3′ NCR, just as the LSM dependence of BMV RNA2 translation was due to the combined effects of sequences in the 5′ NCR, 3′ NCR, and 2a ORF (Fig. 6). Moreover, ribosomes on ASH1 mRNA are greatly slowed but not completely stopped. Similarly, in lsm7Δ yeast, ribosomes on BMV RNA2 must have been greatly slowed (since translation was inhibited) but not completely stopped since, when translation initiation was blocked in lsm7Δ/prt1-1 yeast at 37°C, most ribosomes were cleared from the RNA after 30 min (Fig. 9D). Given the size of RNA2 and the normal speed of ribosome translocation of 10 codons per s (9), such clearance within 30 min is consistent with a ≥20-fold slowing of elongation.
How does the Lsm1p-7p/Pat1p complex overcome ribosome stalling on BMV RNA2? Recent experiments suggest that the Lsm1p-7p/Pat1p complex facilitates mRNP rearrangement and displaces factors to promote yeast mRNA decapping (58). Since the cis elements in ASH1 mRNA, and potentially BMV RNA2, are thought to inhibit ribosome passage by their secondary structure and bound proteins (9), similar mRNP rearrangement and protein displacement may underlie the role of the Lsm1p-7p/Pat1p complex in facilitating BMV translation elongation. Another factor that interacts with the Lsm1p-7p/Pat1p complex to facilitate decapping, the helicase Dhh1p, might participate in overcoming secondary structure or protein blocks in RNA2. Consistent with this possibility, we recently found that Dhh1p was required for BMV RNA translation (Noueiry and Ahlquist, unpublished). In addition to mediating translation, Lsm1p-7p/Pat1p enhancement of ribosome translocation may be related to the LSM1 requirement for BMV RNA recruitment from translation into the replication complex, which depends on clearing ribosomes from the RNA (17, 30, 50).
Deadenylation-dependent mRNA decapping and regulation of mRNA translation and turnover.
Our findings show that factors in the Lsm1p-7p/Pat1p complex facilitate two alternate, successive fates of BMV genomic RNAs: translation and 1a-dependent translational repression and recruitment to ER-localized RNA replication complexes (17, 50). Metazoan maternal mRNAs are also shuttled between alternate fates, including translation, translationally repressed storage, subcellular localization, and turnover (32, 45) and, as noted below, the Lsm1p-7p/Pat1p complex has been linked to some of these transitions (13). Intriguingly, the regulation of maternal mRNAs and certain other developmentally regulated metazoan mRNAs collectively share several unusual features with the dependence of BMV RNA translation on deadenylation-dependent mRNA turnover factors (1). Like BMV RNA, some developmentally regulated mRNAs require sequences in both 5′ and 3′ NCRs (21, 23), and sometimes even in the ORF (32, 49, 59) for proper regulation of subcellular localization, translation, storage, and turnover (2). Like BMV RNA, some developmentally regulated mRNAs remain associated with stalled ribosomes when not translated (12, 42, 51) (3). Storage and translation of maternal mRNAs are regulated by 3′ deadenylation and readenylation by mechanisms related to the deadenylation-dependent mRNA turnover pathway. When not translated, these mRNAs are stored in the cytoplasm in mRNP complexes (22, 32, 45) reminiscent of the translationally inactive BMV RNA mRNP peak.
Because both maternal mRNA storage and mRNA decapping involve Lsm1p-associated helicase Dhh1p and its homologs and lead to a translationally repressed state, Coller et al. suggested that these processes may be alternate outcomes of a common deadenylation pathway, both modulated by the Lsm1p-7p/Pat1p complex and associated factors (13). Our results, showing that Lsm1p-7p and associated factors modulate BMV RNA translation and recruitment from translation to replication, complement and extend these observations and suggest that these conserved factors might similarly facilitate translation of some posttranscriptionally regulated cellular mRNAs, such as maternal mRNAs. Thus, dependence of BMV RNA translation and replication on factors of the Lsm1p-7p/Pat1p complex may be a useful model able to provide further insights into broader mechanisms of posttranscriptional mRNA regulation and further functions of the Lsm1p-7p/Pat1p complex in such regulatory pathways.
Acknowledgments
We thank Michael Krol for plasmid pB4MK1, Greg Gromowski for excellent technical assistance, Arlen W. Johnson and Christine A. Barnes for sharing yeast strains and clones, and members of our laboratory for helpful discussions.
This work was supported by National Institutes of Health grant GM35072. P.A. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Luhrmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 3.Ahlquist, P., R. Dasgupta, and P. Kaesberg. 1981. Near identity of 3- RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell 23:183-189. [DOI] [PubMed] [Google Scholar]
- 4.Andino, R., N. Boddeker, D. Silvera, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76-82. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 6.Bonnerot, C., R. Boeck, and B. Lapeyre. 2000. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 20:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouveret, E., G. Rigaut, A. Shevchenko, M. Wilm, and B. Seraphin. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19:1661-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, J. T., X. Yang, and A. W. Johnson. 2000. Inhibition of mRNA turnover in yeast by an xrn1 mutation enhances the requirement for eIF4E binding to eIF4G and for proper capping of transcripts by Ceg1p. Genetics 155:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chartrand, P., X. H. Meng, S. Huttelmaier, D. Donato, and R. H. Singer. 2002. Asymmetric sorting of Ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell 10:1319-1330. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, I. E., D. Wyckoff, and E. R. Gavis. 2000. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr. Biol. 10:1311-1314. [DOI] [PubMed] [Google Scholar]
- 13.Coller, J. M., M. Tucker, U. Sheth, M. A. Valencia-Sanchez, and R. Parker. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, B. M., S. J. Harrop, G. D. Kornfeld, I. W. Dawes, P. M. Curmi, and B. C. Mabbutt. 2001. Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. 309:915-923. [DOI] [PubMed] [Google Scholar]
- 15.Cormack, B., G. Bertram, M. Egerton, N. Gow, S. Falkow, and A. Brown. 1997. Yeast-enhanced green flourescent protein (yEGFP): a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 16.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans, D. R., C. Rasmussen, P. J. Hanic-Joyce, G. C. Johnston, R. A. Singer, and C. A. Barnes. 1995. Mutational analysis of the Prt1 protein subunit of yeast translation initiation factor 3. Mol. Cell. Biol. 15:4525-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felden, B., C. Florentz, R. Giege, and E. Westhof. 1994. Solution structure of the 3′-end of brome mosaic virus genomic RNAs: conformational mimicry with canonical tRNAs. J. Mol. Biol. 235:508-531. [DOI] [PubMed] [Google Scholar]
- 20.Gamarnik, A. V., and R. Andino. 1996. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 15:5988-5998. [PMC free article] [PubMed] [Google Scholar]
- 21.Gebauer, F., D. F. Corona, T. Preiss, P. B. Becker, and M. W. Hentze. 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 18:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray, N. K., and M. Wickens. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399-458. [DOI] [PubMed] [Google Scholar]
- 23.Gunkel, N., T. Yano, F. H. Markussen, L. C. Olsen, and A. Ephrussi. 1998. Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes Dev. 12:1652-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., San Diego, Calif.
- 25.He, W., and R. Parker. 2000. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 12:346-350. [DOI] [PubMed] [Google Scholar]
- 26.Hilleren, P., and R. Parker. 1999. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33:229-260. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa, M., J. Diez, M. Restrepo-Hartwig, and P. Ahlquist. 1997. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc. Natl. Acad. Sci. USA 94:13810-13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa, M., M. Janda, M. A. Krol, and P. Ahlquist. 1997. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J. Virol. 71:7781-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson, A., and S. W. Peltz. 1999. Tools for turnover: methods for analysis of mRNA stability in eukaryotic cells. Methods 17:1-2. [DOI] [PubMed] [Google Scholar]
- 30.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 32.Johnstone, O., and P. Lasko. 2001. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35:365-406. [DOI] [PubMed] [Google Scholar]
- 33.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Luhrmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96:375-387. [DOI] [PubMed] [Google Scholar]
- 34.Kohl, R. J., and T. C. Hall. 1974. Aminoacylation of RNA from several viruses: amino acid specificity and differential activity of plant, yeast and bacterial synthetases. J. Gen. Virol. 25:257-261. [DOI] [PubMed] [Google Scholar]
- 35.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96:13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kufel, J., C. Allmang, L. Verdone, J. D. Beggs, and D. Tollervey. 2002. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell. Biol. 22:5248-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhn, K. M., J. L. DeRisi, P. O. Brown, and P. Sarnow. 2001. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 21:916-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, W. M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miglietta, J. 1987. Ph.D. thesis. University of Wisconsin, Madison.
- 40.Moller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 41.Noueiry, A. O., J. Chen, and P. Ahlquist. 2000. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA 97:12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen, P. H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671-680. [DOI] [PubMed] [Google Scholar]
- 43.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richter, J. D. 1999. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 63:446-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rietveld, K., C. W. A. Pleij, and L. Bosch. 1983. Three dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 2:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruegsegger, U., J. H. Leber, and P. Walter. 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107:103-114. [DOI] [PubMed] [Google Scholar]
- 48.Salgado-Garrido, J., E. Bragado-Nilsson, S. Kandels-Lewis, and B. Seraphin. 1999. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 18:3451-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunders, C., and R. S. Cohen. 1999. The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol. Cell 3:43-54. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 51.Seggerson, K., L. Tang, and E. G. Moss. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243:215-225. [DOI] [PubMed] [Google Scholar]
- 52.Seraphin, B. 1995. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4, and U5 snRNPs. EMBO J. 14:2089-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seto, A. G., A. J. Zaug, S. G. Sobel, S. L. Wolin, and T. R. Cech. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401:177-180. [DOI] [PubMed] [Google Scholar]
- 54.Strauss, J. H., and E. G. Strauss. 1999. With a little help from the host. Science 283:802-804. [DOI] [PubMed] [Google Scholar]
- 55.Sullivan, M., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan, M., and P. Ahlquist. 1997. cis-Acting signals in bromovirus RNA replication and gene expression: networking with viral proteins and host factors. Semin. Virol. 8:221-230. [Google Scholar]
- 57.Tharun, S., W. He, A. E. Mayes, P. Lennertz, J. D. Beggs, and R. Parker. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404:515-518. [DOI] [PubMed] [Google Scholar]
- 58.Tharun, S., and R. Parker. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8:1075-1083. [DOI] [PubMed] [Google Scholar]
- 59.Thio, G. L., R. P. Ray, G. Barcelo, and T. Schupbach. 2000. Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 221:435-446. [DOI] [PubMed] [Google Scholar]
- 60.Tomasevic, N., and B. A. Peculis. 2002. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell. Biol. 22:4101-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69:571-595. [DOI] [PubMed] [Google Scholar]
- 62.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 63.Valasek, L., K. H. Nielsen, and A. G. Hinnebusch. 2002. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 21:5886-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Regenmortel, M. H. V. (ed.). 2000. Virus taxonomy. Academic Press, Inc., San Diego, Calif.
- 65.Vasudevan, S., and S. W. Peltz. 2001. Regulated ARE-mediated mRNA decay in Saccharomyces cerevisiae. Mol. Cell 7:1191-1200. [DOI] [PubMed] [Google Scholar]
- 66.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 67.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 68.Wyers, F., M. Minet, M. E. Dufour, L. T. Vo, and F. Lacroute. 2000. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol. Cell. Biol. 20:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]