Abstract
RhoE belongs to the Rho GTPase family, the members of which control actin cytoskeletal dynamics. RhoE induces stress fiber disassembly in a variety of cell types, whereas RhoA stimulates stress fiber assembly. The similarity of RhoE and RhoA sequences suggested that RhoE might compete with RhoA for interaction with its targets. Here, we show that RhoE binds ROCK I but none of the other RhoA targets tested. The interaction of RhoE with ROCK I was confirmed by coimmunoprecipitation of the endogenous proteins, and the two proteins colocalized on the trans-Golgi network in COS-7 cells. Although RhoE and RhoA were not able to bind ROCK I simultaneously, RhoE bound to the amino-terminal region of ROCK I encompassing the kinase domain, at a site distant from the carboxy-terminal RhoA-binding site. Overexpression of RhoE inhibited ROCK I-induced stress fiber formation and phosphorylation of the ROCK I target myosin light chain phosphatase. These data suggest that RhoE induces stress fiber disassembly by directly binding ROCK I and inhibiting it from phosphorylating downstream targets.
Rho family proteins are involved in regulating cytoskeletal organization and cell motility responses. Most Rho proteins are GTPases and cycle between an active, GTP-bound conformation that interacts with downstream targets and an inactive, GDP-bound conformation. The members of the Rnd subfamily (Rnd1, Rnd2, and RhoE/Rnd3) of Rho proteins are an exception to this, in that they bind only detectably to GTP, not GDP, and have very low if any GTPase activity (9, 15, 30). Similarly, RhoH/TTF is GTPase defective (25).
Different members of the Rho family induce distinct changes to the actin cytoskeleton, including stress fiber formation and extension of filopodia and lamellipodia (16). The sequences of three highly related Rho proteins, RhoA, RhoB, and RhoC, differ significantly only at their carboxy termini, and the proteins are generally referred to together as “Rho.” Rho stimulates actomyosin-based contractility through its downstream targets ROCK I/ROKβ and ROCK II/Rho-kinase/ROKα (referred to here as ROCKs), and this is required for stress fiber formation in cultured fibroblasts, epithelial cells, and endothelial cells. ROCKs control the formation of stress fibers by inactivating myosin light chain phosphatase (MLCP), thus maintaining myosin light chain in an active form (23), and by activating LIM kinase, which subsequently inhibits the actin cable-severing protein cofilin (28). Other Rho targets involved in Rho-induced actin reorganization include the Dia proteins, protein kinase C-related kinases (PRKs), and phosphatidylinositide 4-phosphate 5-kinases (33, 45, 47).
In contrast to Rho, RhoE/Rnd3 or Rnd1 overexpression in fibroblasts and epithelial cells induces a decrease in stress fibers (15, 30), and expression of RhoE stimulates cell migration (15). RhoE but not Rnd1 or Rnd2 expression can be induced by expressing activated Raf in MDCK cells and could thereby contribute to Raf-induced loss of stress fibers (17). The mechanisms that RhoE employs to induce loss of stress fibers are not known, although a Rnd1/RhoE-interacting protein, Socius, has recently been implicated in this response (21). One hypothesis is that RhoE competes with Rho for interaction with its targets. Analysis of the crystal structures of RhoA-GDP and RhoA-GTP revealed that the main changes in structure between the GDP- and GTP-bound forms occur in two surface loops, known as switch I (amino acids 28 to 44) and switch II (amino acids 61 to 76) (18). Downstream effector proteins need to recognize the conformational changes in these regions in order to differentiate between the GDP- and GTP-bound forms of Rho. Mutational analysis together with the crystal structure of RhoA bound to PRK1 has shown that switch I is involved in interaction with downstream targets (2, 18). The equivalent region of RhoE is highly homologous to RhoA, and comparison of RhoE and RhoA structures shows that the RhoE switch I region is most similar in conformation to RhoA bound to the nonhydrolyzable GTPγS and not RhoA-GDP (14). However, other regions of Rho in addition to switch I are required for interaction with various downstream targets (2, 18), and these are less conserved between RhoE and RhoA (14).
Here, we have investigated whether RhoE is able to interact with a panel of known RhoA targets. Interestingly, we observe that RhoE interacts with ROCK I but that this is not through the RhoA-binding site on ROCK I. In addition, we show that RhoE inhibits ROCK I-induced stress fiber formation and phosphorylation of its downstream target MLCP. These data suggest that RhoE acts to reduce stress fibers by directly interacting with ROCK I.
MATERIALS AND METHODS
Antibodies.
The rabbit antiserum against an N-terminal peptide of RhoE (SQKLSSKSIMDPNQNVK) was produced by Eurogentec, Seraing, Belgium, and the antibody was affinity purified with the peptide column. RhoE monoclonal antibodies were generated in mice against purified recombinant full-length RhoE according to standard procedures. Mouse monoclonal (9E10) and rabbit polyclonal anti-myc antibodies and polyclonal goat anti-ROCK I antibody (C-19) were purchased from Santa Cruz Biotechnology. Antipolyhistidine (clone HIS-1) and anti-FLAG (M2) antibodies were from Sigma. Monoclonal TGN38 and ROCK I antibodies were from BD Transduction Laboratories, anti-MLCP (MLCP/MYPT1) was from Covance, and anti-phospho-MYPT1 (Thr696) was from Upstate Biotechnology Inc.
Expression vectors.
The expression vector encoding FLAG-RhoE was generated by inserting the full-length mouse RhoE cDNA between the EcoRI and HindIII sites of pCMV5-FLAG. The coding regions for residues 375 to 727, 1 to 420, and 76 to 420 of ROCK I were cloned into KpnI and XhoI sites of pCAG expression vector with an N-terminal myc tag. pRK5-myc-RhoA wild type (wt) and pRK5-myc-RhoA-V14 were kindly provided by Alan Hall (MRC Laboratory of Molecular Cell Biology, London, United Kingdom). Expression vectors encoding FLAG-tagged wt mDia1 (pFL-C1-mDia1) (47), full-length citron kinase (pCAG-myc-citron kinase) and its C-terminally truncated mutant (pCAG-myc-citron kinase Δ1) (27), full-length ROCK I (pCAG-myc-p160 wt), and three C-terminally truncated mutants (pCAG-myc-p160Δ1, -Δ3, and -Δ5) (19) were kindly provided by Shuh Narumiya (Kyoto University, Kyoto, Japan). A truncation mutant of ROCK I from amino acids 906 to 1024 that covers the RhoA-binding domain (RBD) (pQE-11-His-M2-2; Shuh Narumiya) was digested with BamHI and NotI, blunted, and inserted into the EcoRI site of pCAG plasmid to express His-M2-2 in mammalian cells. pcDNA3-PRK1-myc (29) was kindly provided by Peter Parker (Cancer Research UK, London, United Kingdom).
Cell culture, transfection, and microinjection.
COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum and penicillin-streptomycin (Invitrogen). For transfection, cells were washed with 5 ml of cold electroporation buffer (120 mM KCl, 10 mM K2PO4 · KH2PO4 [pH 7.6], 25 mM HEPES [pH 7.6], 2 mM MgCl2, 0.5% Ficoll). The buffer was removed, and cells were resuspended in 250 μl of cold electroporation buffer and electroporated at 250 V and 960 μF with 10 μg of DNA. The cells were then plated on 10-cm-diameter dishes and incubated at 37°C for 24 h. Swiss 3T3 cells were grown in DMEM containing 10% fetal calf serum. For microinjection, cells were seeded on glass coverslips 2 days before being starved with DMEM without serum for 24 h. The starved cells were microinjected with expression construct DNA and incubated for 6 h. For platelet-derived growth factor (PDGF) treatment, the starved Swiss 3T3 cells were incubated with 25 ng of PDGF/ml in DMEM without serum.
GST pull-down, immunoprecipitation, and immunoblotting.
Recombinant RhoA-V14 (constitutively active RhoA) was expressed as glutathione S-transferase (GST) fusion protein and purified as previously described (35). Recombinant GST-RhoE was purified by the same procedure with slight modifications (15). For cell lysates and GST pull-down assays, transfected COS-7 cells were lysed in lysis buffer (1% NP-40, 20 mM Tris-HCl [pH 8], 130 mM NaCl, 10 mM NaF, 1% aprotinin, 10 μg of leupeptin/ml, 1 mM dithiothreitol, 0.1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride). After removal of insoluble material, the cell lysate was incubated for 2 h at 4°C with the recombinant proteins on glutathione beads (Amersham Pharmacia Biotech). Beads were washed three times with lysis buffer before the proteins were eluted in Laemmli sample buffer. For immunoprecipitations, clarified lysates were incubated with anti-FLAG M2 antibody (Sigma) or anti-myc antibody (Santa Cruz) covalently linked to agarose or with polyclonal RhoE antibody and protein A beads (Amersham Pharmacia Biotech) for 2 h at 4°C. The bound proteins were, after thorough washes of the matrix with lysis buffer, eluted in Laemmli sample buffer. To analyze direct interaction between RhoE and ROCK I, proteins binding immunoprecipitated myc-ROCKΔ1 were removed by washes with lysis buffer containing 0.5 M NaCl. myc-ROCKΔ1 was eluted with 20 μg of myc peptide (Sigma)/ml in lysis buffer and dialyzed against phosphate-buffered saline (PBS) before the GST pull-down assay. A sample was also resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gel was silver stained. For immunoblotting, proteins were resolved by SDS-PAGE and transferred to an Immobilon P membrane (Millipore). Nonspecific binding of antibodies was blocked with 5% nonfat dried milk-0.05% Tween 20 in TBS (25 mM Tris-HCl [pH 7.4], 137 mM NaCl, 3 mM KCl). Rabbit anti-myc antibody was used for detection of myc-tagged proteins. The bound antibodies were visualized with horseradish peroxidase-conjugated goat anti-immunoglobulin G antibodies and the enhanced chemiluminescence system (ECL; Amersham Pharmacia Biotech).
Yeast two-hybrid analysis.
The mouse RhoE cDNA lacking the C-terminal sequence encoding the CAAX box was inserted into the SmaI site of pGBT9. pGBT9-RhoA-V14/S190, pGAD-ROCK I (amino acids 349 to 1025), pGAD-kinectin (amino acids 1053 to 1327), pGAD-mNET (amino acids 1 to 595), pGAD-mDia2 (amino acids 47 to 800), pGAD424-PAK (amino acids 1 to 252), pGAD-PKN (amino acids 1 to 942), pVP16-citron (amino acids 674 to 870), and pVP16-rhophilin (amino acids 1 to 130) have been previously described (37) and were kindly provided by Richard Treisman (Cancer Research UK). Transformation of HF7C cells (Clontech) was carried out according to the supplier's protocol. The interaction of the two encoded proteins was evaluated as the ability of the transformed yeast cells to grow on plates lacking histidine with increasing concentrations of 3-aminotriazole.
Immunofluorescence microscopy.
Swiss 3T3 cells on coverslips were fixed for 20 min with 4% paraformaldehyde in PBS. The fixed cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min, and the cells were incubated with primary antibodies for 30 min. Actin filaments were localized by incubating the cells with 0.1 μg of tetramethyl rhodamine isothiocyanate (TRITC)-labeled phalloidin (Sigma)/ml. For secondary antibodies fluorescein isothiocyanate-conjugated donkey anti-mouse, indodicarbocyanine (Cy5)-conjugated donkey anti-goat, and TRITC-conjugated donkey anti-rabbit antibodies with minimal cross-reaction to different species (Jackson ImmunoResearch) were used for 30 min. The specimens were mounted in Mowiol (Calbiochem) containing p-phenylenediamine (Sigma), and images were generated with a Zeiss LSM 510 confocal microscope.
Cell fractionation.
Cell fractionation was performed essentially as previously described (36). In brief, Swiss 3T3 cells were washed with PBS and suspended in 10 mM Tris-HCl (pH 7.5)-10 mM KCl-250 mM sucrose-10 mM NaF-1% aprotinin-10 μg of leupeptin/ml-1 mM dithiothreitol-0.1 mM Na3VO4-1 mM phenylmethylsulfonyl fluoride. The cells were broken by repeated passages through a 21-gauge needle, and after removal of intact cells and nuclei (500 × g, 10 min), the postnuclear supernatant was centrifuged at 150,000 × g for 30 min. The pellet (particulate fraction) and supernatant (cytosol) were analyzed by immunoblotting.
RESULTS
RhoE interacts with ROCK I but not with other RhoA targets.
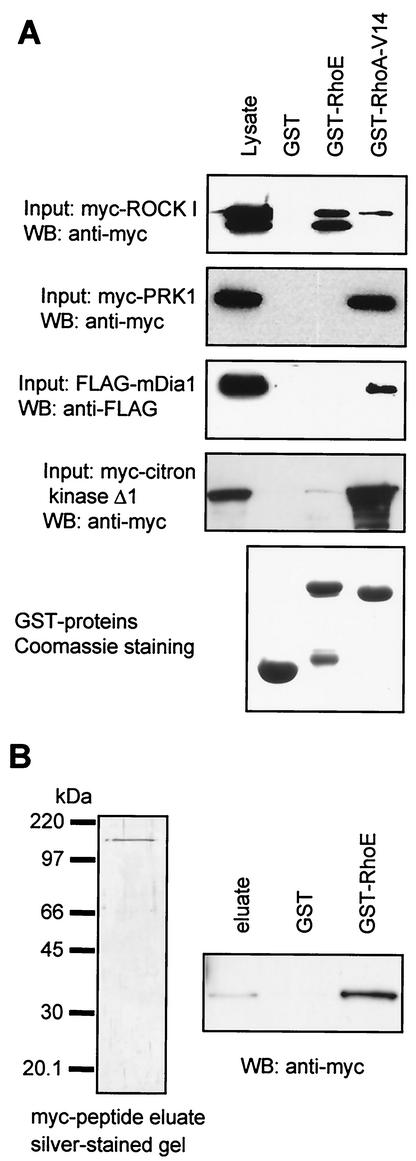
Exogenous expression of RhoE leads to loss of stress fibers in fibroblasts and epithelial cells (15, 30). RhoA-induced stress fiber formation involves its targets ROCKs and Dia (33), and possibly PRKs (45). We therefore investigated whether RhoE could interact with these RhoA targets. Expression vectors encoding myc-ROCK I, FLAG-mDia1, and myc-PRK1 were transfected into COS-7 cells. GST-RhoE pulled down ROCK I from lysates of these transfected cells (Fig. 1A). GST-RhoE interacted preferentially with a faster-migrating version of ROCK I rather than full-length ROCK I (Fig. 1A, lower band). This could be a cleaved version of ROCK I, as ROCK I can be cleaved by caspase 3 (7, 39). RhoE could also bind, however, to full-length ROCK I (Fig. 1A, upper band) and to the caspase 3-resistant ROCK I mutant (data not shown) (7). In contrast, GST-RhoE did not interact with mDia1 or PRK1 (Fig. 1A). The RhoA target most closely related to ROCKs is citron kinase. GST-RhoE interacted very weakly with myc-citron kinase (data not shown) and its activated, C-terminally truncated mutant Δ1 (Fig. 1A). As expected, the constitutively active RhoA mutant, GST-RhoA-V14, was able to bind to ROCK I, mDia1, citron kinase, and PRK1 (Fig. 1A).
FIG. 1.
RhoE interacts with ROCK I but not with PRK1 or mDia1 in vitro. (A) COS-7 cells were transiently transfected with expression vectors encoding myc-tagged ROCK I, myc-PRK1, FLAG-mDia1, or myc-citron kinase Δ1. Cells were lysed after 24 h, and cell lysates were incubated with GST, GST-RhoE, or GST-RhoA-V14 on glutathione beads. The precipitated complexes were analyzed by SDS-PAGE and immunoblotting. The amounts of GST fusion proteins were determined by Coomassie blue staining of the gel. (B) myc-ROCKΔ1 produced in COS-7 cells was immunoprecipitated with anti-myc antibody, washed with 0.5 M NaCl to remove ROCK-binding proteins, and eluted with myc-peptide. A sample of the eluate was resolved by SDS-PAGE and silver stained. The eluate was incubated with GST or GST-RhoE on glutathione beads, and the complexes were analyzed by SDS-PAGE and immunoblotting. WB, Western blot.
To test if the interaction between RhoE and ROCK I was direct, myc-ROCKΔ1, a constitutively active C-terminally truncated ROCK I (19), was expressed in COS-7 cells. After immunoprecipitation with an anti-myc antibody and thorough washes with high salt to remove all ROCKΔ1-binding proteins, myc-ROCKΔ1 was eluted by competition with a myc peptide. Only one protein was detected in this eluate (Fig. 1B, silver stain), which was myc-ROCKΔ1, as it was recognized by anti-myc antibody, and this bound specifically to GST-RhoE but not to GST (Fig. 1B). This indicates that RhoE and ROCK I interact directly.
Consistent with the in vitro binding results, RhoE was unable to interact with the RBDs of mDia1, PRK1, and a number of other RhoA targets in a yeast two-hybrid assay (Table 1). As a positive control, RhoA-V14 interacted with all the targets (Table 1). Although RhoE interacted with the C-terminal domain of citron kinase, this interaction was very weak in comparison to RhoA binding to the kinase. It was not possible to detect an interaction between RhoE and a region of ROCK I from amino acids 349 to 1025, which did not include the N terminus and kinase domain, although RhoA-V14 could interact with this region (Table 1). This suggests that the N terminus of ROCK I could be required for RhoE but not for RhoA interaction.
TABLE 1.
Yeast two-hybrid analysis of RhoA-V14 and RhoE interaction with Rho GTPase targets
| Target | Amino acids | Interactiona
|
|
|---|---|---|---|
| RhoA-V14 | RhoE | ||
| Citron kinase | 674-870 | +++ | + |
| Kinectin | 1053-1327 | +++ | − |
| mDia2 | 47-800 | +++ | − |
| mNET | 1-595 | ++ | +/− |
| PAK | 1-252 | − | − |
| PRK1/PKN | 1-942 | + | − |
| Rhophilin | 1-130 | +++ | − |
| ROCK I | 349-1025 | +++ | − |
Semiquantitative assay of HIS3 activity by growth on plates containing increasing amounts of aminotriazole: −, no growth on plates lacking histidine; +, ++, and +++, growth on plates containing 0, 5, or 10 mM 3-aminotriazole, respectively.
RhoE binds to the N-terminal region of ROCK I.
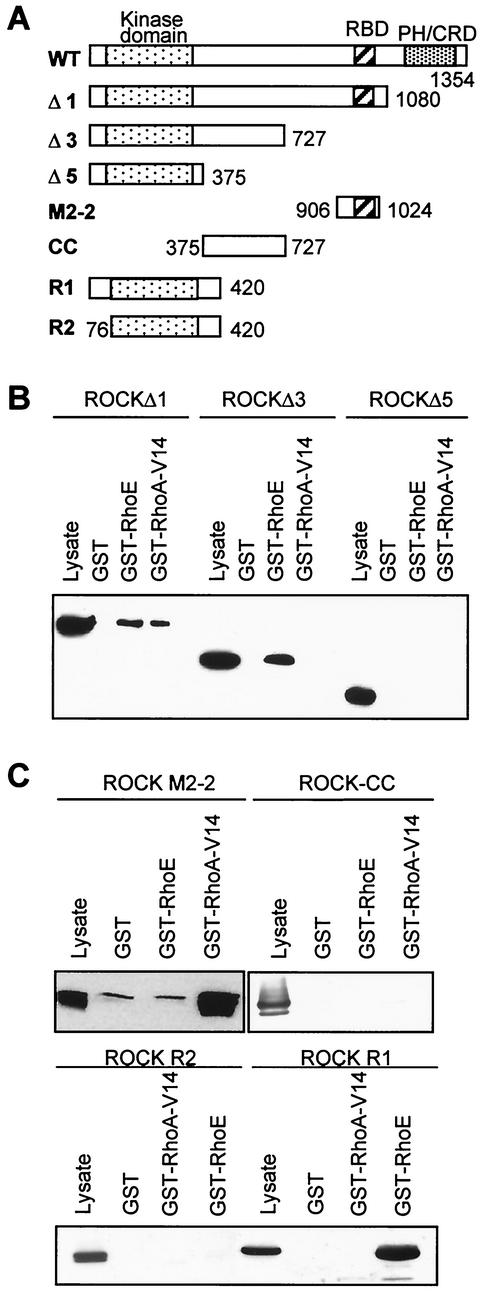
To determine the region of ROCK I which interacts with RhoE, a series of truncation mutants of myc-ROCK I was used (Fig. 2A). GST-RhoA-V14 interacted with ROCK I wt and ROCKΔ1 but not with ROCKΔ3 or ROCKΔ5, consistent with previous studies in which the major site of RhoA interaction with ROCK I was mapped to amino acids 934 to 1015 (11). In contrast, GST-RhoE interacted with ROCK I wt, ROCKΔ1, and ROCKΔ3 but not with ROCKΔ5 (Fig. 2B). In addition, RhoA-V14 but not RhoE bound to the RhoA-binding site of ROCK I (ROCK M2-2, amino acids 906 to 1024 [11]) (Fig. 2C). These results indicate that RhoE does not bind to the previously mapped RhoA-binding site and that the region of ROCK I between amino acids 375 and 727 is required for RhoE binding. To determine whether this region is sufficient for RhoE binding, we studied the interaction of myc-ROCK-CC (coiled-coil domain, amino acids 375 to 727 of ROCK I) with RhoE. As shown in Fig. 2C, ROCK-CC did not bind either RhoE or RhoA-V14, suggesting that amino acids within the N-terminal region of ROCK I (residues 1 to 375) are also needed for RhoE binding. This is consistent with the yeast two-hybrid analysis, which indicates that the region from amino acids 349 to 1025 of ROCK I is not sufficient for RhoE interaction (Table 1). Further deletion analysis defined amino acids 1 to 420 on ROCK I (ROCK R1) to be the minimal region required for RhoE binding, as deletion of the first 76 amino acids before the kinase domain in the N terminus (ROCK R2) prevented RhoE from interacting with ROCK I (Fig. 2C). In addition, the region after the kinase domain (amino acids 338 to 420) of ROCK I was important for RhoE binding, as the ROCKΔ5 construct (amino acids 1 to 375) did not interact with RhoE (Fig. 2B). Neither of the domains next to the ROCK I kinase domain (amino acids 1 to 76 or 338 to 420) on its own was able to interact with RhoE (data not shown). The kinase activity of ROCK I was not needed for RhoE binding, as the kinase-dead form of ROCK I (19) was still able to interact with RhoE (data not shown).
FIG. 2.
RhoE and RhoA bind to different domains of ROCK I. (A) Diagrammatic representation of ROCK I mutants used in this study. ROCK I is represented with its structural domains at the top. The region of ROCK I between the kinase domain and the pleckstrin homology (PH) domain is predicted to form a coiled-coil structure. Numbers indicate amino acid residues. CRD, cysteine-rich domain. (B) ROCKΔ3 binds RhoE but not RhoA. COS-7 cells were transiently transfected with expression vectors encoding myc-ROCKΔ1, -Δ3, or -Δ5, and the lysates were incubated with GST, GST-RhoE, or GST-RhoA-V14 on glutathione beads. Bound ROCK I was detected by an immunoblot analysis with anti-myc antibody. (C) The minimal region for RhoE binding is amino acids 1 to 420 in ROCK I. Plasmids encoding myc-tagged ROCK-CC, His-tagged ROCK M2-2, myc-ROCK R1, or myc-ROCK R2 were transfected into COS-7 cells. Cell lysates were incubated with GST, GST-RhoE, or GST-RhoA-V14 on glutathione beads. Bound proteins were detected by immunoblotting.
RhoE and ROCK I colocalize on the Golgi complex.
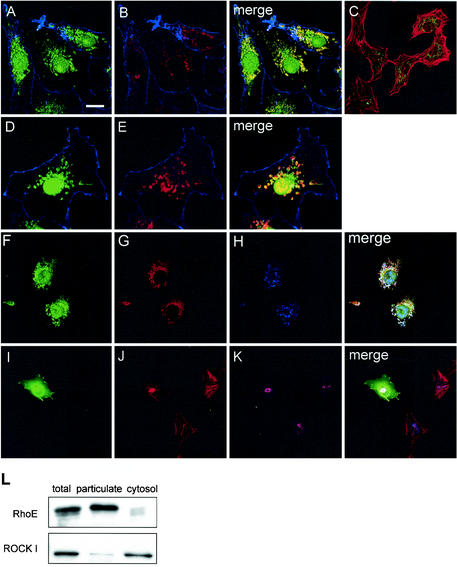
To determine the intracellular localization of RhoE, COS-7 cells were stained with a polyclonal antibody raised against the N terminus of RhoE and with a monoclonal RhoE antibody (see Materials and Methods). RhoE localized primarily in the perinuclear region and on vesicular structures, suggesting localization to the Golgi complex (Fig. 3A and B). In addition, some COS-7 cells showed staining at the plasma membrane. The nuclear staining with the polyclonal, but not the monoclonal, RhoE antibody was due to a cross-reactivity of the antibody with a nuclear protein. A similar perinuclear localization of RhoE was observed in Swiss 3T3 cells (Fig. 3C). However, these cells express lower levels of RhoE than do COS-7 cells (Fig. 4B). RhoE localization to the Golgi complex was verified by double staining of COS-7 cells with an antibody recognizing a marker protein of the trans-Golgi network (TGN38, Fig. 3D and E). Further, dual staining with antibodies to ROCK I and RhoE indicated that there was a significant degree of overlap in the localization of the two proteins in COS-7 cells (Fig. 3F and G), and that considerable amounts of the proteins were localized to the trans-Golgi network (Fig. 3H). RhoE did not significantly colocalize with marker proteins of the early endosomes or lysosomes (data not shown). Overexpression of RhoE in Swiss 3T3 cells disrupts actin stress fibers (15, 30). Costaining of filamentous actin and the trans-Golgi network in RhoE-overexpressing cells revealed that the disrupted actin filaments and the trans-Golgi network localized in the same perinuclear region (Fig. 3I to K).
FIG. 3.
RhoE colocalizes with ROCK I on the Golgi complex. COS-7 cells were stained with polyclonal (A, D, and F) or monoclonal (B) anti-RhoE antibodies, anti-TGN38 antibody (E and H), or polyclonal anti-ROCK I antibody (G). Swiss 3T3 cells were stained with monoclonal RhoE antibody (C), anti-TGN38 antibody (K), or anti-FLAG antibody to stain exogenous FLAG-RhoE (I). Swiss 3T3 cells were microinjected with plasmid encoding FLAG-RhoE (I to K) and fixed after 5 h. Filamentous actin was stained with TRITC-phalloidin in panels A to E and J and is shown in blue in COS-7 cells to indicate the cell shape (A, B, D, and E) and in red in Swiss 3T3 cells (C and J). Bars, 20 μm (A, B, and F to H), 35 μm (C and I to K), and 15 μm (D and E). (L) Intracellular distribution of RhoE and ROCK I by cell fractionation and immunoblotting. RhoE and ROCK I were immunoblotted with polyclonal and monoclonal antibodies, respectively.
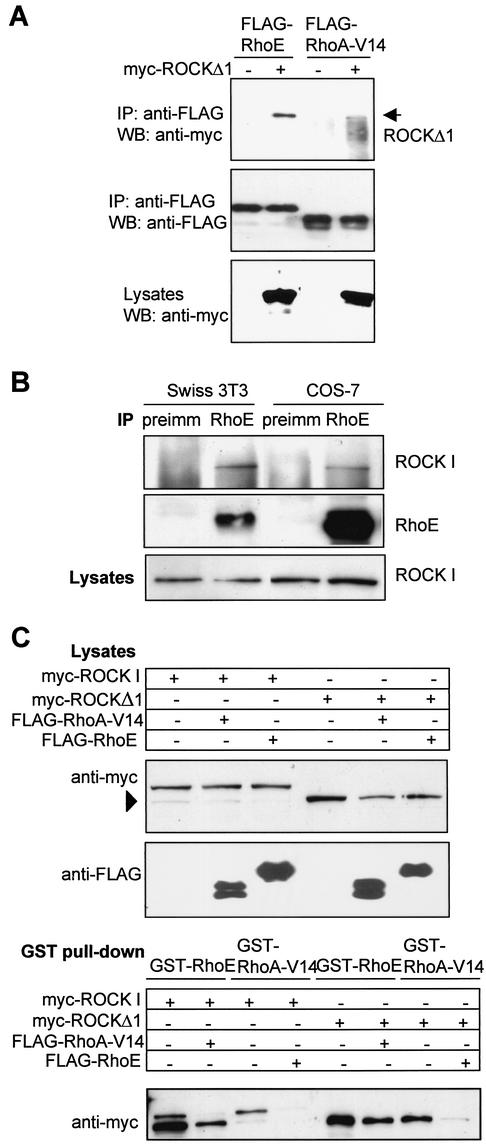
FIG. 4.
RhoE coimmunoprecipitates with ROCK I. (A) Lysates of COS-7 cells expressing myc-ROCKΔ1 and FLAG-RhoE or FLAG-RhoA-V14 were subjected to immunoprecipitation with anti-FLAG antibody. The precipitates were separated by SDS-PAGE and analyzed by immunoblotting. (B) Lysates of Swiss 3T3 cells or COS-7 cells were subjected to immunoprecipitation with polyclonal anti-RhoE antibody or, as a control, with rabbit preimmune serum. Precipitates were immunoblotted with monoclonal RhoE and ROCK I antibodies. (C) COS-7 cells were cotransfected with expression vectors encoding myc-ROCK I wt or myc-ROCKΔ1, together with plasmids encoding FLAG-RhoE or FLAG-RhoA-V14. ROCK proteins were pulled down by GST-RhoA-V14 or GST-RhoE on glutathione beads. After washes the bound proteins were separated by SDS-PAGE and analyzed by immunoblotting. The cleaved form of ROCK I is indicated by an arrowhead on the immunoblot showing the levels of expressed proteins in cell lysates. IP, immunoprecipitation; WB, Western blot.
Fractionation of Swiss 3T3 cells into cytosolic and particulate fractions showed that RhoE was mainly in the particulate fraction, which includes membrane-associated proteins (Fig. 3L). To test if a physical complex of RhoE and ROCK I was possible on the Golgi membranes, the same fractions were subjected to Western blotting for ROCK I. Although a major portion of ROCK I was found to be in the cytosolic fraction, a significant amount of ROCK I was in the particulate fraction. Similar results were obtained with COS-7 cells (data not shown).
RhoE and ROCK I interact in cells.
To study whether RhoE and ROCK I can associate with each other in cells, COS-7 cells were cotransfected with plasmids encoding myc-ROCKΔ1 together with FLAG-RhoE or FLAG-RhoA-V14. myc-ROCKΔ1 coimmunoprecipitated with FLAG-RhoE, and also weakly with FLAG-RhoA-V14 (Fig. 4A), from transfected-cell lysates. In addition, FLAG-RhoE precipitated, although weakly, full-length myc-ROCK I and myc-citron kinase Δ1 from transiently transfected COS-7 cells (data not shown).
To determine if endogenous RhoE and ROCK I can form a complex, Swiss 3T3 and COS-7 cells were subjected to coimmunoprecipitation studies. RhoE specifically precipitated ROCK I from both Swiss 3T3 and COS-7 cell lysates (Fig. 4B).
As RhoE and RhoA bind to different regions of ROCK I, we investigated whether they could bind to ROCK I simultaneously. Expression vectors encoding FLAG-RhoE, myc-RhoA-V14, and myc-ROCK I were cotransfected into COS-7 cells. The complexes of RhoE with ROCK I and of RhoA with ROCK I appeared to be mutually exclusive, as myc-RhoA-V14 did not coimmunoprecipitate with FLAG-RhoE in COS-7 cell lysates (data not shown). To determine if RhoE could inhibit RhoA binding to ROCK I, COS-7 cells coexpressing FLAG-RhoE with myc-ROCK I were subjected to a GST pull-down assay (Fig. 4C). Expression of FLAG-RhoE reduced the binding of myc-ROCK I to GST-RhoA-V14. Similarly, coexpression of FLAG-RhoA-V14 with myc-ROCK I reduced the binding of myc-ROCK I to GST-RhoE (Fig. 4C). The active ROCK I mutant, myc-ROCKΔ1, behaved in the same way as did the wt in this GST pull-down assay (Fig. 4C). Taken together, these results strongly suggest that RhoE and RhoA cannot bind simultaneously to ROCK I, even though they bind to different regions of ROCK I.
RhoE inhibits ROCK I-induced actin stress fiber formation and myosin phosphatase phosphorylation.
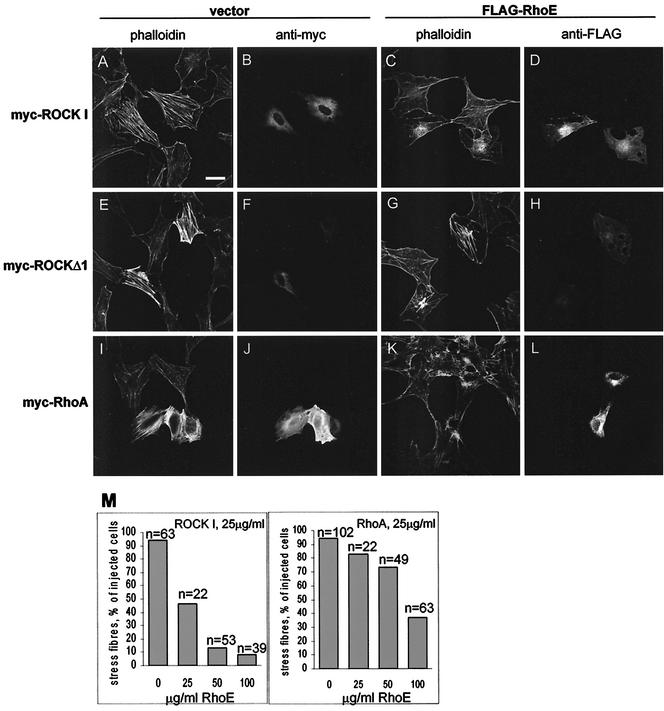
Increased RhoE expression induces rapid loss of stress fibers (15, 30). In contrast, exogenous ROCK I and RhoA can stimulate stress fiber formation (13, 19). To investigate how the interaction of RhoE with ROCK I affected ROCK I-induced responses, we expressed exogenous FLAG-RhoE together with myc-ROCK I or myc-RhoA in Swiss 3T3 cells. In starved Swiss 3T3 cells expression of wt ROCK I or activated ROCKΔ1 induced formation of actin stress fibers without any additional stimulus (Fig. 5A, B, E, and F). Coexpression of RhoE in starved Swiss 3T3 cells inhibited the ability of wt ROCK I to induce stress fibers (Fig. 5C and D). ROCK I protein expression was not detectably altered by FLAG-RhoE coexpression (data not shown). The inhibition of stress fiber formation was concentration dependent, as the number of coinjected cells lacking stress fibers was increased by injecting more of the expression vector encoding RhoE (Fig. 5M). A higher level of RhoE was required to reduce stress fiber formation induced by myc-ROCKΔ1, consistent with the fact that this protein is constitutively active, whereas presumably only a fraction of ROCK I wt is active at any time (Fig. 5G and H). Furthermore, FLAG-RhoE overexpression also inhibited the ability of RhoA to induce stress fibers (Fig. 5I to M), supporting a model where RhoE binding to ROCK I inhibits downstream signaling from ROCK I.
FIG. 5.
RhoE inhibits ROCK I- and RhoA-induced stress fiber assembly. Starved Swiss 3T3 cells were microinjected with plasmids encoding myc-ROCK I (25 μg/ml) (A and B), myc-ROCK I (25 μg/ml) and FLAG-RhoE (50 μg/ml) (C and D), myc-ROCKΔ1 (10 μg/ml) (E and F), myc-ROCKΔ1 (10 μg/ml) and FLAG-RhoE (100 μg/ml) (G and H), myc-RhoA (25 μg/ml) (I and J), or myc-RhoA (25 μg/ml) and FLAG-RhoE (100 μg/ml) (K and L). Cells were stained after 6 h of incubation for filamentous actin with rhodamine-conjugated phalloidin (A, C, E, G, I, and K). For the detection of myc-tagged proteins cells were stained with mouse 9E10 antibody (B, F, and J), and for the detection of FLAG-RhoE they were stained with mouse anti-FLAG antibody (D, H, and L). Bar, 30 μm. The percentages of injected cells that lack stress fibers as determined by using different FLAG-RhoE DNA concentrations are presented in panel M.
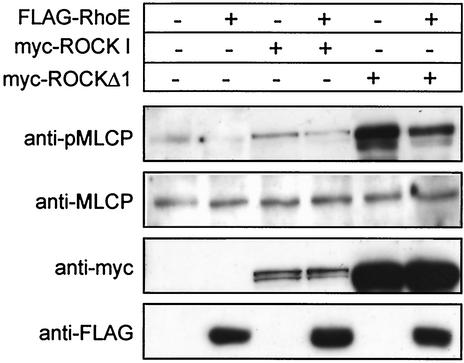
To test whether RhoE affects the ability of ROCK I to phosphorylate downstream targets, we investigated the effect of RhoE expression on MLCP phosphorylation. MLCP is the major known substrate for ROCK I involved in ROCK I-mediated stress fiber formation (13), and as expected COS-7 cells expressing ROCKΔ1 showed a large increase in MLCP phosphorylation compared to that in control cells (Fig. 6). wt ROCK I also induced a small increase in MLCP phosphorylation (Fig. 6). Coexpression of RhoE reduced MLCP phosphorylation induced by both ROCKΔ1 and wt ROCK I, as well as reducing the background level of MLCP phosphorylation in the absence of exogenous ROCK I (Fig. 6). These results indicate that RhoE inhibits stress fiber formation by preventing ROCK I from phosphorylating its substrate, MLCP.
FIG. 6.
RhoE inhibits myosin phosphatase phosphorylation by ROCK I. COS-7 cells were transfected with plasmids encoding FLAG-RhoE, myc-ROCK I, and myc-ROCKΔ1. Equal amounts of proteins of the transfected-cell lysates were separated by SDS-PAGE and analyzed by immunoblotting. The data are representative of three independent experiments.
Increased RhoE expression coincides with PDGF-induced morphological changes in Swiss 3T3 cells.
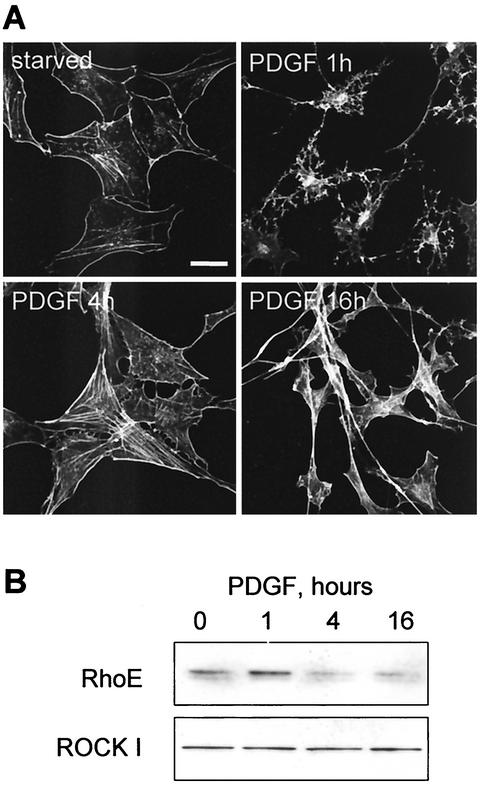
Having shown that increased RhoE expression inhibited ROCK I-induced actin stress fibers, we wanted to determine whether cell morphological changes induced by a physiological stimulus coincided with altered RhoE protein expression levels. PDGF has been shown elsewhere to change Swiss 3T3 cell morphology (32, 34). At low concentrations it primarily induces membrane ruffling and lamellipodium extension, while at higher concentrations it stimulates cell rounding (32). In Swiss 3T3 cells PDGF induced cell rounding and branching by 1 h after addition (Fig. 7A), resembling the morphology of Swiss 3T3 cells microinjected with RhoE (30). This PDGF-induced rounding coincided with upregulated expression of RhoE (Fig. 7B). After 4 h of PDGF the cells started to respread and form actin filaments, and by 16 h the cells had an increased F-actin content and elongated morphology (Fig. 7A). At these time points RhoE protein levels decreased (Fig. 7B). ROCK I levels did not alter during the PDGF treatment (Fig. 7B). Therefore, the RhoE expression and the PDGF-induced cell morphology changes correlated well with the suggested role of RhoE in negatively regulating stress fiber formation.
FIG. 7.
PDGF induces changes in RhoE protein expression level. (A) Starved Swiss 3T3 cells were incubated with 25 ng of PDGF/ml for 1, 4, or 16 h, and the cells were stained for filamentous actin. Bar, 30 μm. (B) Equal amounts of lysates of starved and PDGF-treated Swiss 3T3 cells were separated by SDS-PAGE and immunoblotted with anti-ROCK I antibody or with polyclonal anti-RhoE antibody. The data are representative of three independent experiments.
DISCUSSION
RhoE is known to induce loss of actin stress fibers, but the molecular basis for this effect has not been elucidated. Here, we show that RhoE binds to ROCK I, a kinase required for stress fiber formation and contractility. We found that RhoE binds to the N-terminal region of ROCK I (amino acids 1 to 420) and not to the previously mapped RhoA-binding site (amino acids 934 to 1015, RBD in Fig. 3A) (11). The RhoE-binding region includes the N-terminal amino acids 1 to 76, the kinase domain (amino acids 76 to 338), and amino acids 338 to 420 of ROCK I. This explains why the ROCK I construct used in the yeast two-hybrid analysis (amino acids 349 to 1025), which lacks the N terminus and kinase domain, did not interact with RhoE. Interestingly, deletion of the N-terminal 78 amino acids from ROCK II/Rho-kinase/ROKα has previously been reported to prevent ROCK-induced stress fiber formation (24), suggesting that the N-terminal region upstream of the kinase domain is critical for ROCK function. Even though RhoE does not bind to the C-terminal RhoA-binding site, RhoA and RhoE are unable to bind ROCK I simultaneously and compete for ROCK I binding. This suggests that the binding site for RhoE on ROCK I is masked when RhoA binds (and vice versa), either because of the conformational change induced or because RhoA binds to part of the RhoE-binding site in addition to the C-terminal RBD. Considering that the effector domain sequences of RhoA and RhoE are highly similar, it is perhaps surprising that RhoE does not bind to the RhoA-binding site of ROCK I. However, ROCK I binding to RhoA requires a second region (amino acids 75 to 92) of RhoA in addition to the effector domain (12), and this region is less conserved between RhoE and RhoA.
Both ROCK I and RhoE localize to the trans-Golgi network in COS-7 cells. Recently, ROCK I has been reported to be bound to centrosomes (5), which are localized in the Golgi area. Localization of ROCK II by using different antibodies has yielded contradictory results, but some reports show ROCK II to partially associate with stress fibers (4, 22). COS-7 cells have very few actin stress fibers, and we cannot exclude the possibility that ROCK I would associate with them in other cell types. Unfortunately, immunofluorescence studies with the commercial ROCK I antibodies gave only very weak staining in Swiss 3T3 cells. Nevertheless, other GTPases localize to the Golgi complex, including Cdc42, TC10, and RhoG (3, 8, 20), and Ras proteins have been shown elsewhere to be active on the Golgi complex (6). In addition, it is known that a Golgi complex-associated actin network is important for protein trafficking (10), and myosins can be found on the Golgi complex (40). Golgi complex-to-endoplasmic reticulum trafficking is affected by Cdc42 and TC10 (20, 26). Interestingly, we found that actin filaments were organized around the perinuclear region in the RhoE-overexpressing cells and colocalized with the trans-Golgi network. RhoE may therefore function primarily on the Golgi complex, and one possibility is that it acts to sequester ROCK I, and/or ROCK I may act on Golgi complex-associated myosins.
Of the tested RhoA effectors ROCK I is the only one that binds RhoE. Similarly, Rnd1 did not interact with several Rho/Rac effectors in yeast two-hybrid assays (44). On the other hand, RhoE shows weak interaction with a kinase homologous to ROCK I, namely, citron kinase (27), but this interaction is barely detectable in comparison to RhoA binding. The fact that overexpression of RhoE has a major inhibitory effect on stress fibers is explained by our observation that it binds ROCK I, and prevents ROCK I-induced phosphorylation of MLCP, a major downstream target of ROCKs involved in stress fiber formation (13). In this respect it acts similarly to ROCK I inhibitors, such as Y27632, and our results are therefore in concordance with ROCK playing a central role in stress fiber formation (43). We cannot rule out the possibility that other RhoE-binding proteins also contribute to RhoE-mediated effects on cell morphology, especially as multiple RhoA effectors contribute to stress fiber assembly. Nevertheless, by binding around the kinase domain, RhoE could sterically inhibit ROCK I from interacting with its targets. Recently, Ward et al. (46) reported that overexpressed Gem, another small GTPase, inhibited ROCK-mediated functions, but unlike RhoE, Gem bound adjacent to the RhoA-binding site of ROCK I. Here we have shown that endogenous RhoE interacts with ROCK I, and further studies are required to reveal if other GTPases operate in different areas of the cell to negatively control ROCK I activity.
The kinase domain of inactive ROCK is highly inaccessible, as the protein forms inter- and intramolecular interactions. As the C-terminal region of ROCK can bind to the N-terminal kinase region to form an autoinhibited structure (1, 4), it would be expected that in this inactive conformation the RhoE-binding site around the kinase domain would be masked. In cells transfected with wt myc-ROCK I, we consistently observed a lower-molecular-weight form of ROCK I that interacts much better with RhoE than does full-length ROCK I. C-terminal ROCK I cleavage by caspase 3 has been shown elsewhere to yield a constitutively active ROCK I protein, which is similar in size to the lower-molecular-weight form of ROCK I (7, 39). This implies that ROCK I has to be in an open conformation for RhoE to bind. Although RhoE can bind both full-length ROCK I and a ROCK I mutant that is resistant to caspase cleavage (7), much less full-length ROCK I is pulled down by RhoE than is truncated ROCK I. Possibly the full-length ROCK I pulled down by RhoE represents the small fraction of activated (but not RhoA-bound) ROCK I in cells. Presumably, RhoA-GTP binding to ROCK I induces this open conformation, and RhoE can bind to this once RhoA has dissociated. Our observations that RhoE inhibited stress fiber formation and MLCP phosphorylation induced by either wt or activated ROCK I further support a model where RhoE functions by binding to activated ROCK I.
As RhoE is constitutively in an active, GTP-bound form (9, 15), regulation of RhoE function must be controlled differently from cycling Rho GTPases. Interestingly, we showed that the expression of RhoE is transiently upregulated upon PDGF treatment of Swiss 3T3 cells and that this correlates with the levels of actin stress fibers and cell morphology. In addition, RhoE expression has been shown elsewhere to be upregulated upon Raf activation in MDCK cells (17) and by hepatocyte growth factor, which stimulates cell migration (41). Raf is a Ras effector, and there are several similarities between the phenotypes of Ras-transformed cells and those of cells overexpressing RhoE. RhoE overexpression increases the motility of cells, including hepatocyte growth factor-stimulated MDCK cells, and causes a loss of stress fibers and focal contacts (15). Similarly, Ras-transformed cells are highly motile, and they lack stress fibers and focal contacts. Ras-transformed cells frequently show an upregulation of RhoA activity, which is important for cell proliferation (38, 50). However, to allow the increased motility of Ras-transformed cells, RhoA needs to be uncoupled from inducing actin stress fibers and focal contacts, and this has been attributed to a reduction in Rho to ROCK signaling (38) and extracellular signal-regulated kinase-induced downregulation of ROCK I and/or II expression (31, 38). Reduction in ROCK function by increased RhoE expression could contribute together with ROCK downregulation to the Ras-transformed motile phenotype. Upregulation of RhoE expression may also explain why wt ROCK cannot reverse the phenotype of Ras-transformed cells (31, 38). In the future it will be important to determine whether a sustained increase in RhoE expression alters the level of ROCK I activity.
Our data suggest that the relative expression levels and activities of Rho(A/B/C) and RhoE will determine the final readout of ROCK I. Other studies indicate that similar competition may occur between other Rho family proteins. For example, Xenopus laevis Rnd1 prevents RhoA-induced cell adhesion (48) and RhoH reduces Rho-mediated activation of NF-κB and p38 (25). In addition, RhoD inhibits RhoA-activated stress fiber formation (42) and Rnd1-induced plexin-A1-mediated cytoskeletal collapse (49). The GTPase-deficient Rho family members particularly may employ competition as their mode of action to affect cellular responses. We propose that controlling the expression level of RhoE and thus its binding to ROCK I provides a mechanism to regulate cell behavior in addition to altering the GTP/GDP ratio on conventional Rho GTPases.
Acknowledgments
We are grateful to Shuh Narumiya, Alan Hall, Richard Treisman, Erik Sahai, Mike Olson, and Peter Parker for plasmids; to Xin Lu for assistance with preparing monoclonal antibodies to RhoE; and to Nick Keep for discussions.
This work was supported by a European Commission Marie Curie fellowship (K.R.), a BBSRC research grant (A.J.R.), and the Ludwig Institute for Cancer Research.
REFERENCES
- 1.Amano, M., K. Chihara, N. Nakamura, T. Kaneko, Y. Matsuura, and K. Kaibuchi. 1999. The COOH terminus of Rho-kinase negatively regulates rho-kinase activity. J. Biol. Chem. 274:32418-32424. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, A. L., and A. Hall. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241-255. [PMC free article] [PubMed] [Google Scholar]
- 3.Brunet, N., A. Morin, and B. Olofsson. 2002. RhoGDI-3 regulates RhoG and targets this protein to the Golgi complex through its unique N-terminal domain. Traffic 3:342-358. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X. Q., I. Tan, C. H. Ng, C. Hall, L. Lim, and T. Leung. 2002. Characterization of RhoA-binding kinase ROKα—implication of the pleckstrin homology domain in ROKα function using region-specific antibodies. J. Biol. Chem. 277:12680-12688. [DOI] [PubMed] [Google Scholar]
- 5.Chevrier, V., M. Piel, N. Collomb, Y. Saoudi, R. Frank, M. Paintrand, S. Narumiya, M. Bornens, and D. Job. 2002. The Rho-associated protein kinase p160ROCK is required for centrosome positioning. J. Cell Biol. 157:807-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson II, A. D. Cox, and M. R. Philips. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 7.Coleman, M. L., E. A. Sahai, M. Yeo, M. Bosch, A. Dewar, and M. F. Olson. 2001. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3:339-345. [DOI] [PubMed] [Google Scholar]
- 8.Erickson, J. W., C. Zhang, R. A. Kahn, T. Evans, and R. A. Cerione. 1996. Mammalian Cdc42 is a brefeldin A-sensitive component of the Golgi apparatus. J. Biol. Chem. 271:26850-26854. [DOI] [PubMed] [Google Scholar]
- 9.Foster, R., K. Q. Hu, Y. Lu, K. M. Nolan, J. Thissen, and J. Settleman. 1996. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol. Cell. Biol. 16:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fucini, R. V., J. L. Chen, C. Sharma, M. M. Kessels, and M. Stamnes. 2002. Golgi vesicle proteins are linked to the assembly of an actin complex defined by mAbp1. Mol. Biol. Cell 13:621-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujisawa, K., A. Fujita, T. Ishizaki, Y. Saito, and S. Narumiya. 1996. Identification of the Rho-binding domain of p160ROCK, a Rho-associated coiled-coil containing protein kinase. J. Biol. Chem. 271:23022-23028. [DOI] [PubMed] [Google Scholar]
- 12.Fujisawa, K., P. Madaule, T. Ishizaki, G. Watanabe, H. Bito, Y. Saito, A. Hall, and S. Narumiya. 1998. Different regions of Rho determine Rho-selective binding of different classes of Rho target molecules. J. Biol. Chem. 273:18943-18949. [DOI] [PubMed] [Google Scholar]
- 13.Fukata, Y., M. Amano, and K. Kaibuchi. 2001. Rho-Rho-kinase pathway in smooth muscle contraction and cytoskeletal reorganization of non-muscle cells. Trends Pharmacol. Sci. 22:32-39. [DOI] [PubMed] [Google Scholar]
- 14.Garavini, H., K. Riento, J. P. Phelan, M. S. McAlister, A. J. Ridley, and N. H. Keep. 2002. Crystal structure of the core domain of RhoE/Rnd3: a constitutively activated small G protein. Biochemistry 41:6303-6310. [DOI] [PubMed] [Google Scholar]
- 15.Guasch, R. M., P. Scambler, G. E. Jones, and A. J. Ridley. 1998. RhoE regulates actin cytoskeleton organization and cell migration. Mol. Cell. Biol. 18:4761-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, S. H., M. M. Zegers, M. Woodrow, P. Rodriguez-Viciana, P. Chardin, K. E. Mostov, and M. McMahon. 2000. Induced expression of Rnd3 is associated with transformation of polarized epithelial cells by the Raf-MEK-extracellular signal-regulated kinase pathway. Mol. Cell. Biol. 20:9364-9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihara, K., S. Muraguchi, M. Kato, T. Shimizu, M. Shirakawa, S. Kuroda, K. Kaibuchi, and T. Hakoshima. 1998. Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem. 273:9656-9666. [DOI] [PubMed] [Google Scholar]
- 19.Ishizaki, T., M. Naito, K. Fujisawa, M. Maekawa, N. Watanabe, Y. Saito, and S. Narumiya. 1997. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 404:118-124. [DOI] [PubMed] [Google Scholar]
- 20.Kanzaki, M., R. T. Watson, J. C. Hou, M. Stamnes, A. R. Saltiel, and J. E. Pessin. 2002. Small GTP-binding protein TC10 differentially regulates two distinct populations of filamentous actin in 3T3L1 adipocytes. Mol. Biol. Cell 13:2334-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh, H., A. Harada, K. Mori, and M. Negishi. 2002. Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol. Cell. Biol. 22:2952-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh, K., Y. Kano, M. Amano, H. Onishi, K. Kaibuchi, and K. Fujiwara. 2001. Rho-kinase-mediated contraction of isolated stress fibers. J. Cell Biol. 153:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura, K., M. Ito, M. Amano, K. Chihara, Y. Fukata, M. Nakafuku, B. Yamamori, J. Feng, T. Nakano, K. Okawa, A. Iwamatsu, and K. Kaibuchi. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273:245-248. [DOI] [PubMed] [Google Scholar]
- 24.Leung, T., X. Q. Chen, E. Manser, and L. Lim. 1996. The p160 RhoA-binding kinase ROKα is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol. Cell. Biol. 16:5313-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X., X. Bu, B. Lu, H. Avraham, R. A. Flavell, and B. Lim. 2002. The hematopoiesis-specific GTP-binding protein RhoH is GTPase deficient and modulates activities of other Rho GTPases by an inhibitory function. Mol. Cell. Biol. 22:1158-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luna, A., O. B. Matas, J. A. Martinez-Menarguez, E. Mato, J. M. Duran, J. Ballesta, M. Way, and G. Egea. 2002. Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol. Biol. Cell 13:866-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madaule, P., M. Eda, N. Watanabe, K. Fujisawa, T. Matsuoka, H. Bito, T. Ishizaki, and S. Narumiya. 1998. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature 394:491-494. [DOI] [PubMed] [Google Scholar]
- 28.Maekawa, M., T. Ishizaki, S. Boku, N. Watanabe, A. Fujita, A. Iwamatsu, T. Obinata, K. Ohashi, K. Mizuno, and S. Narumiya. 1999. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285:895-898. [DOI] [PubMed] [Google Scholar]
- 29.Mellor, H., P. Flynn, C. D. Nobes, A. Hall, and P. J. Parker. 1998. PRK1 is targeted to endosomes by the small GTPase, RhoB. J. Biol. Chem. 273:4811-4814. [DOI] [PubMed] [Google Scholar]
- 30.Nobes, C. D., I. Lauritzen, M. G. Mattei, S. Paris, A. Hall, and P. Chardin. 1998. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J. Cell Biol. 141:187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawlak, G., and D. M. Helfman. 2002. Post-transcriptional down-regulation of ROCKI/Rho-kinase through an MEK-dependent pathway leads to cytoskeleton disruption in Ras-transformed fibroblasts. Mol. Biol. Cell 13:336-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rankin, S., and E. Rozengurt. 1994. Platelet-derived growth factor modulation of focal adhesion kinase (p125FAK) and paxillin tyrosine phosphorylation in Swiss 3T3 cells. Bell-shaped dose response and cross-talk with bombesin. J. Biol. Chem. 269:704-710. [PubMed] [Google Scholar]
- 33.Ridley, A. J. 1999. Stress fibres take shape. Nat. Cell Biol. 1:E64-E66. [DOI] [PubMed] [Google Scholar]
- 34.Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389-399. [DOI] [PubMed] [Google Scholar]
- 35.Ridley, A. J., H. F. Paterson, C. L. Johnston, D. Diekmann, and A. Hall. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70:401-410. [DOI] [PubMed] [Google Scholar]
- 36.Riento, K., J. Jantti, S. Jansson, S. Hielm, E. Lehtonen, C. Ehnholm, S. Keranen, and V. M. Olkkonen. 1996. A sec1-related vesicle-transport protein that is expressed predominantly in epithelial cells. Eur. J. Biochem. 239:638-646. [DOI] [PubMed] [Google Scholar]
- 37.Sahai, E., A. S. Alberts, and R. Treisman. 1998. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 17:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahai, E., M. F. Olson, and C. J. Marshall. 2001. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sebbagh, M., C. Renvoize, J. Hamelin, N. Riche, J. Bertoglio, and J. Breard. 2001. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 3:346-352. [DOI] [PubMed] [Google Scholar]
- 40.Stow, J. L., and K. Heimann. 1998. Vesicle budding on Golgi membranes: regulation by G proteins and myosin motors. Biochim. Biophys. Acta 1404:161-171. [DOI] [PubMed] [Google Scholar]
- 41.Tanimura, S., K. Nomura, K. Ozaki, M. Tsujimoto, T. Kondo, and M. Kohno. 2002. Prolonged nuclear retention of activated extracellular signal-regulated kinase 1/2 is required for hepatocyte growth factor-induced cell motility. J. Biol. Chem. 277:28256-28264. [DOI] [PubMed] [Google Scholar]
- 42.Tsubakimoto, K., K. Matsumoto, H. Abe, J. Ishii, M. Amano, K. Kaibuchi, and T. Endo. 1999. Small GTPase RhoD suppresses cell migration and cytokinesis. Oncogene 18:2431-2440. [DOI] [PubMed] [Google Scholar]
- 43.Uehata, M., T. Ishizaki, H. Satoh, T. Ono, T. Kawahara, T. Morishita, H. Tamakawa, K. Yamagami, J. Inui, M. Maekawa, and S. Narumiya. 1997. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389:990-994. [DOI] [PubMed] [Google Scholar]
- 44.Vayssiere, B., G. Zalcman, Y. Mahe, G. Mirey, T. Ligensa, K. M. Weidner, P. Chardin, and J. Camonis. 2000. Interaction of the Grb7 adapter protein with Rnd1, a new member of the Rho family. FEBS Lett. 467:91-96. [DOI] [PubMed] [Google Scholar]
- 45.Vincent, S., and J. Settleman. 1997. The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol. Cell. Biol. 17:2247-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, Y., S. F. Yap, V. Ravichandran, F. Matsumura, M. Ito, B. Spinelli, and K. Kelly. 2002. The GTP binding proteins Gem and Rad are negative regulators of the Rho-Rho kinase pathway. J. Cell Biol. 157:291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe, N., T. Kato, A. Fujita, T. Ishizaki, and S. Narumiya. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136-143. [DOI] [PubMed] [Google Scholar]
- 48.Wunnenberg-Stapleton, K., I. L. Blitz, C. Hashimoto, and K. W. Cho. 1999. Involvement of the small GTPases XRhoA and XRnd1 in cell adhesion and head formation in early Xenopus development. Development 126:5339-5351. [DOI] [PubMed] [Google Scholar]
- 49.Zanata, S. M., I. Hovatta, B. Rohm, and A. W. Puschel. 2002. Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor activity in semaphorin 3A-induced cytoskeletal collapse. J. Neurosci. 22:471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zondag, G. C., E. E. Evers, J. P. ten Klooster, L. Janssen, R. A. van der Kammen, and J. G. Collard. 2000. Oncogenic Ras downregulates Rac activity, which leads to increased Rho activity and epithelial-mesenchymal transition. J. Cell Biol. 149:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]