Abstract
p27Kip1 levels increase in many cells as they leave the cell cycle and begin to differentiate. The increase in p27Kip1 levels generally precedes the expression of differentiation-specific genes. Previous studies from our laboratory showed that the overexpression of p27Kip1 enhances myelin basic protein (MBP) promoter activity. This activation is specific to p27Kip1. Additionally, inhibition of cyclin-dependent kinase activity alone is not sufficient to increase MBP expression. In this study, we focused on understanding how p27Kip1 can activate gene transcription by using the MBP gene in oligodendrocytes as a model. We show that the enhancement of MBP promoter activity by p27Kip1 is mediated by a proximal region of the MBP promoter that contains a conserved GC box binding sequence. This sequence binds transcription factors Sp1 and Sp3. Increased expression of p27Kip1 increases the level of Sp1 promoter binding to the GC box but does not change the level of Sp3 binding. The binding of Sp1 to this element activates the MBP promoter. p27Kip1 leads to increased Sp1 binding through a decrease in Sp1 protein turnover. Enhancement of MBP promoter activity by an increase in the level of p27Kip1 involves a novel mechanism that is mediated through the stabilization and binding of transcription factor Sp1.
The mammalian cell cycle is controlled by a family of cyclin-dependent kinases (Cdks) and their partner cyclins. The sequential activation of Cdks triggers cell phase transition. Cdk activity is regulated at many levels, including phosphorylation and dephosphorylation. In addition, Cdk activity can be inhibited by Cdk inhibitor proteins (CKIs). Inhibition of Cdk activity causes cell cycle arrest (for reviews, see references 12 and 26). There are two families of CKIs, one of which is called the INK4 family and includes p15, p16, p18, and p19. These inhibit Cdk4 kinase activity and cause cell cycle arrest in G1 when overexpressed in various cell lines. The other family of CKIs is the Cip/Kip family, which includes p21Cip1, p27Kip1, and p57Kip2 (40). These inhibit the activity of most Cdks by binding with cyclin-Cdk complexes. Among these CKIs, p27Kip1 is essential for cell cycle control and is involved in the response of the cell to environmental cues (for reviews, see references 25 and 33). In addition to its role in cell cycle control, many observations have indicated that p27Kip1 plays a unique role in gene expression and cell differentiation (10, 18, 28).
Oligodendrocytes are the myelinating cells of the central nervous system. Upon differentiation, oligodendrocytes express many myelin-specific proteins, among which is myelin basic protein (MBP). MBP is one of the major protein components of the myelin sheath that surrounds axons to ensure the rapid conduction of nervous impulses.
Oligodendrocytes progress through a series of stages, changing from proliferative, migratory progenitor cells to mature, postmitotic, myelin-membrane-producing cells. The differentiation of oligodendrocytes is accompanied by a striking increase in the level of p27Kip1 protein (37). This accumulation of p27Kip1 causes cell cycle arrest in oligodendrocyte progenitor cells (16), alters the responses of cells to mitogens, and initiates differentiation (11). Miskimins et al. showed previously that p27Kip1 has a role in MBP gene expression in oligodendrocytes, as increased levels of p27Kip1 in CG4 oligodendrocytes dramatically enhance MBP promoter activity (22).
The CG4 oligodendrocyte cell line is derived from primary cultures of rat oligodendrocyte progenitor cells. These cells proliferate in the presence of platelet-derived growth factor (PDGF) and basic fibroblast growth factor (FGF2). Upon withdrawal of PDGF and FGF2, CG4 cells differentiate into mature oligodendrocytes (21). These cells are also capable of myelinating axons after transplantation into myelin-deficient rat brain (13). In this study, CG4 oligodendrocytes were used to investigate the role of p27Kip1 in the activation of the MBP promoter. We show that a conserved GC box located in the proximal region of the MBP promoter is necessary for p27Kip1 to stimulate expression. Increased levels of p27Kip1 during differentiation lead to increased levels of binding of transcription factor Sp1 to this element, contributing to the activation of the MBP promoter. Our data also show that the overexpression of p27Kip1 results in a decrease in Sp1 degradation rates, leading to increased levels of Sp1 promoter binding activity and subsequent activation of the MBP promoter. The results presented here reveal a novel mechanism of p27Kip1-mediated activation of gene transcription.
MATERIALS AND METHODS
Cell culture.
CG4 oligodendrocytes were cultured as previously reported (22). Briefly, cells were grown in growth medium (GM) consisting of Dulbecco's modified Eagle's medium (DMEM) plus 30 nM sodium selenite, 50 ng of insulin/ml, 50 μg of transferrin/ml, 1 mg of bovine serum albumin/ml, 5 ng of PDGF/ml, 10 ng of FGF2/ml, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C in an atmosphere of 5% CO2 and 100% humidity. Fresh PDGF and FGF2 were added to the cultures every other day. To induce differentiation, cells were switched to differentiation medium (DM) consisting of DMEM plus 30 nM sodium selenite, 5 μg of insulin/ml, 50 μg of transferrin/ml, 10 nM biotin, 10 nM hydrocortisone, 30 nM 3,3′,5′-triiodo-l-thyronine, 20 nM progesterone, 2 mM glutamine, 1 mg of bovine serum albumin/ml, 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
Chemicals and plasmids.
Roscovitine was obtained from Calbiochem (San Diego, Calif.). Fascaplysin was purchased from Alexis Biochemicals (San Diego, Calif.). The MBP-luciferase constructs were made by PCR with oligonucleotides that hybridized to the 5′ end of the MBP promoter and ended at the sequences indicated in Fig. 1 and a common 3′ oligonucleotide ending at +30. The PCR fragments were inserted upstream of the luciferase gene in the SmaI site of pGL3Basic (Fig. 1). The p27Kip1 expression plasmid contains the entire mouse p27Kip1 cDNA amplified by PCR and inserted downstream of the cytomegalovirus promoter in pcDNA3.1 (Hygro) (Invitrogen, Carlsbad, Calif.). The Sp1 expression plasmid CMV-Sp1 was a gift from R. Tjian.
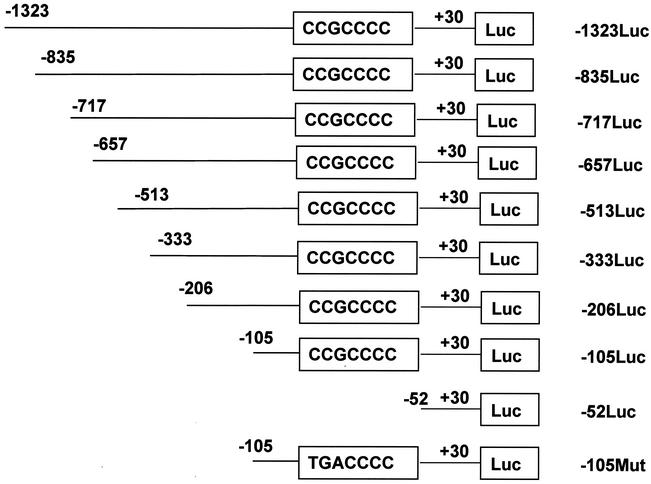
FIG. 1.
Deletion mutant constructs used for transfection assays. The MBP gene from −1323 to +30 was subcloned upstream of the luciferase gene (Luc) in pGL3Basic. The length and deletion point of each construct are shown. The GC box sequence is boxed. The construct −105Mut contains a mutation in the GC box (CCG mutated to TGA).
DNA transfection and luciferase assays.
Cells were plated in 35-mm dishes and cultured as described above. Transfections were done with GM by using GenePorter (Gene Therapy Systems, San Diego, Calif.) according to the manufacturer's protocol. At 4 h after transfection, the medium was changed to GM containing PDGF and FGF2, and the cells were allowed to stand overnight. The medium was changed to DM on the following day. After 48 h in DM, the cells were rinsed once with phosphate-buffered saline (PBS) and lysed by the addition of reporter lysis buffer (Promega, Madison, Wis.). The supernatant was assayed for protein concentration by using Bio-Rad (Hercules, Calif.) protein assay reagent and for luciferase activity by using Steady-Glo luciferase assay reagent (Promega). Luciferase activity was measured by using a Packard TopCount Luminometer and normalized to the protein concentration.
Western blotting.
Cells in 35-mm dishes were rinsed with PBS and lysed by the addition of sodium dodecyl sulfate (SDS) sample buffer (2.5 mM Tris-HCl [pH 6.8], 2.5% SDS, 100 mM dithiothreitol [DTT], 10% glycerol, 0.025% pyronin Y). Equal amounts of protein were separated on SDS-polyacrylamide gels. Proteins were transferred to Immobilon P membranes (Millipore, Bedford, Mass.) by using a Bio-Rad Trans-blot apparatus and transfer buffer (48 mM Tris, 39 mM glycine, 0.0375% SDS, 20% methanol). Membranes were processed as previously described (22). Detection was carried out by using a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) and Super Signal chemiluminescent substrate (Pierce, Rockford, Ill.). The primary antibodies used were anti-p27Kip1 (Transduction Laboratories, Lexington, Ky.; 1:2,500), anti-Sp3 (Santa Cruz Biotechnology; 1:2,500), anti-Sp1 (Santa Cruz Biotechnology;1:1,000), and anti-β-actin (Sigma, St. Louis, Mo.; 1:5,000).
Nuclear extracts.
Nuclear extracts were made as previously described (1) with the following modifications. Cells grown in 100-mm dishes were harvested by scraping, and the pellets were resuspended in 500 μl of buffer A (10 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]). The cells were allowed to swell on ice for 10 min, vortexed (to break the cells), and centrifuged at 14,000 × g for 10 s. The pellets were resuspended in 200 μl of buffer C (20 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 25% glycerol, 0.2 mM EDTA, 420 mM NaCl, 0.2 mM PMSF, 0.5 mM DTT) and incubated on ice for 30 min. Cell debris was removed by centrifugation at 14,000 × g for 15 min at 4°C, and the supernatant fraction was microdialyzed against buffer D2 (20 mM HEPES [pH 7.9], 250 mM sucrose, 50 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 5 mM MgCl2, 0.1 mM ZnCl2, 0.5 mM PMSF). The protein concentration was determined as described above, and samples were stored at −70°C.
Electrophoretic mobility shift assays (EMSAs).
Double-stranded oligonucleotides were end labeled with [γ-32P]ATP (NEN, Boston, Mass.) by using T4 polynucleotide kinase (MBI Fermentis, Hanover, Md.). Approximately 40 ng of 32P-labeled oligonucleotide (106 cpm) was incubated for 15 min on ice with 5 μg of nuclear extract in buffer D2 plus poly(dI-dC) (2 μg) in a total volume of 50 μl. When included as competitors, unlabeled oligonucleotides were added in excess to the nuclear extract together with the labeled probe. For supershift experiments, the labeled probe was incubated with the nuclear extract on ice for 15 min, followed by an additional incubation for 15 min with various amounts of antibodies. Complexes were resolved on 3.5 or 5% nondenaturing polyacrylamide gels in 0.25× Tris-borate-EDTA (0.022 M Tris, 0.022 M boric acid, and 0.5 mM EDTA). Gels were dried and subjected to autoradiography. All oligonucleotide probes were used in double-stranded form. They consisted of mouse MBP promoter sequences between positions −105 and −52 corresponding to the wild-type sequence or containing the mutation detailed in Fig. 1, in which the putative Sp1/Sp3 transcription factor binding site has been mutated. Double-stranded oligonucleotides containing the sequence 5′-CCCGGGTGACGTCACGGGGA-3′ and its complement were used as nonspecific competitor DNAs, while oligonucleotides containing the sequence 5′-GCTATGACCGCCCCATGAT-3′, which harbors the GC box in the MBP promoter, were used as specific competitors for Sp1/Sp3 binding. Antibodies used for supershift assays were obtained from Santa Cruz Biotechnology. The specificities were as follows: Sp1 (PEP 2; sc-59), Sp2 (K-20; sc-643), Sp3 (D-20; sc-644), and Sp4 (V-20; sc-645).
Site-specific mutagenesis.
Site-specific mutagenesis of the MBP promoter was carried out by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's protocol. Primers for mutation of the GC box were the same oligonucleotides as those used for EMSAs. Transformation was done by using One-Shot competent Escherichia coli cells (Invitrogen). The purified plasmid was subjected to sequencing to confirm the mutation.
Pulse-chase labeling.
Cells were grown in 100-mm dishes and transfected with p27Kip1 plasmid or control plasmid as described above. Following transfection, the cells were left in GM or switched to DM. At 2 days after transfection, the cells were washed twice with PBS. The cells were placed in methionine-cysteine-free medium for 1 h, after which 35S-labeled methionine-cysteine was added to 60 μCi/ml (Trans 35S label; ICN). After 1 h, the cells were washed three times with methionine-cysteine-free DMEM and incubated in GM or DM supplemented with 1 mM unlabeled methionine-cysteine. At various times, the cells were washed twice with PBS and lysed for 30 min at 4oC in 1 ml of cell lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 10 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 10 μg of leupeptin/ml, 2 μg of aprotinin/ml, 100 μM PMSF). Lysates were collected by scraping and centrifuged, and the supernatants were precleared by incubation with protein A-conjugated agarose beads (Calbiochem). For immunoprecipitation, protein A-conjugated magnetic beads (Dynal, Lake Success, N.Y.; 30 μl) were incubated with 6 μg of anti-Sp1 antibody for 3 h, followed by two washes with cell lysis buffer. Antibody-conjugated beads were incubated with 1.3 mg of proteins from the precleared extracts overnight. The beads were collected by magnetic separation and washed four times with radioimmunoprecipitation buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.8], 0.5% deoxycholic acid, 0.1% SDS). After the addition of SDS sample buffer to each sample, the samples were heated for 5 min and loaded onto SDS-7% polyacrylamide gels. The gels were dried, and the bands were visualized by autoradiography. Autoradiographs were subjected to densitometry to estimate the level of Sp1 and to determine the half-life of the protein.
RESULTS
Activation of the MBP promoter by p27Kip1 is mediated by a short proximal promoter region.
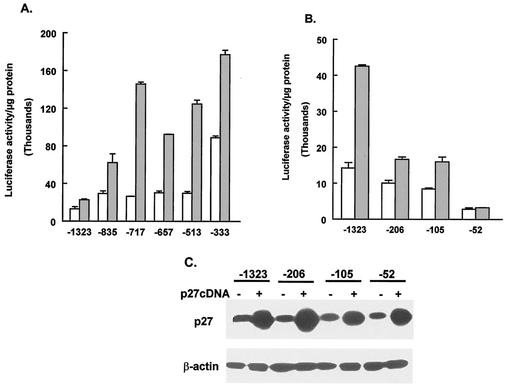
Previous results from our laboratory showed that increased expression of p27Kip1 leads to increased activity of the MBP promoter (22). To explore the mechanism that leads to activation of the MBP promoter by p27Kip1, we sought to determine the sequence elements in the promoter that are involved in this response. For this analysis, a series of plasmid constructs that contained different lengths of the MBP promoter sequence upstream of the luciferase reporter gene were made. These constructs are shown in Fig. 1. CG4 cells were cotransfected with each of the MBP-luciferase constructs and either a control plasmid or a plasmid encoding p27Kip1. Figure 2A and B show that removal of the MBP 5′-flanking sequence between −1323 and −105 does not eliminate the response of the MBP-luciferase constructs to p27Kip1. However, the MBP-luciferase construct containing the 5′-flanking sequence to −52 does not respond to the expression of p27Kip1. The lack of response of this construct is not due to a failure of p27Kip1 to be expressed, as shown by Western blotting of the transfected cells (Fig. 2C). These results suggest that the 5′-flanking sequence between −105 and −52 contains an element that is required for the response of the MBP promoter to increased levels of p27Kip1.
FIG. 2.
Stimulation of MBP promoter activity by p27Kip1 requires sequences between −105 and −52. (A and B) CG4 cells were transfected with the indicated MBP-luciferase constructs together with empty vector (open bars) or a p27Kip1 expression vector (gray bars). Cells were harvested and assayed for luciferase. Results are presented as relative luciferase activity per microgram of protein and are the means and standard errors of the means for at least three replicates. (C) CG4 cells transfected as described for panel B were harvested in SDS sample buffer, and the lysates were subjected to Western blotting to determine p27Kip1 levels. A plus sign indicates the presence of the p27Kip1 expression vector in the transfection. The same blot was probed for β-actin as a loading control.
Expression of p27Kip1 increases binding to the MBP promoter.
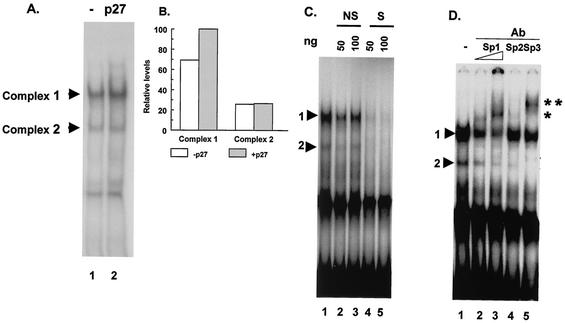
Increased expression of p27Kip1 leads to increased activity of the MBP promoter, and this effect requires sequences between −105 and −52. To determine whether the increased expression of p27Kip1 leads to a change in protein binding to sequences within this region, double-stranded oligonucleotides containing MBP promoter sequences extending from −105 to −52 were used as probes in EMSAs. Nuclear extracts were obtained from CG4 cells transfected with a plasmid encoding p27Kip1 or from control cells transfected with an empty vector. A number of protein-DNA complexes were formed on the probe when extracts from control cells were used (Fig. 3A, lane 1). The amount of one of these complexes (complex 1) was increased when extracts from cells expressing high levels of p27Kip1 were used (Fig. 3A, lane 2, and Fig. 3B).
FIG. 3.
The region of the MBP promoter that is required for a response to p27Kip1 binds the Sp1 family of transcription factors. (A) An EMSA was performed by using an end-labeled probe containing sequences from −105 to −52 of the MBP gene. The probe was incubated with nuclear extract protein from CG4 cells transfected with a control plasmid (lane 1) or a p27Kip1 expression vector (lane 2). The positions of the two specific complexes (complex 1 and complex 2) are indicated. (B) Complexes 1 and 2 from cells transfected with a control plasmid (open bars) or a p27Kip1 expression vector (gray bars) (from panel A) were quantitated by densitometry. (C) An EMSA was performed as described for panel A with extracts from CG4 cells transfected with a control plasmid. The binding reactions were performed in the absence of any competitor DNA (lane 1), in the presence of 50 ng (lane 2) or 100 ng (lane 3) of unlabeled nonspecific oligonucleotide DNA (NS), or in the presence of 50 ng (lane 4) or 100 ng (lane 5) of an oligonucleotide containing the MBP GC box sequence (S). The positions of complex 1 and complex 2 are indicated by arrowheads. (D) An EMSA was performed as described for panel A with extracts from CG4 cells transfected with a control plasmid. Complexes were supershifted by inclusion of the indicated antibodies (Ab) to the binding reactions (lanes 2 to 5). Lane 1 shows binding in the absence of any added antibody. The positions of the two specific complexes are indicated by arrowheads. The positions of the supershifted complexes formed with anti-Sp1 antibody (*) or anti-Sp3 antibody (**) are also indicated.
The Sp1 family of transcription factors binds to the promoter region that responds to p27Kip1.
Analysis of the sequence between −105 and −52 in the MBP promoter reveals the presence of several potential transcription factor binding sites, including an Sp1 family binding site (GC box). To determine whether the GC box is involved in the formation of the observed protein-DNA complexes, competitor oligonucleotides were added to the EMSA reactions. Figure 3C shows that a low concentration of an oligonucleotide containing a consensus GC box strongly competes with the formation of complexes 1 and 2 (Fig. 3C, lane 4). An oligonucleotide containing an unrelated sequence but with the same GC content does not efficiently compete with the formation of either complex 1 or complex 2 (Fig. 3C, lanes 2 and 3). Thus, complexes 1 and 2 most likely represent specific DNA-protein complexes within the GC box of the MBP promoter region.
GC box sequences can be bound by a group of transcription factors belonging to the Sp1 family (8, 35). Indeed, there is evidence that one member of this family, Sp1, can bind to sequences in the MBP promoter (2, 39). In order to determine whether any member(s) of the Sp1 family of transcription factors is involved in forming either complex 1 or complex 2, supershift assays were performed. The results show that complex 1 is most efficiently supershifted by an anti-Sp1 antibody. An anti-Sp3 antibody completely supershifts complex 2 (Fig. 3D). A weak supershifted band is observed when anti-Sp2 antibody is used. A fourth member of the Sp1 transcription factor family, Sp4, does not appear to be involved in the formation of any of the complexes (data not shown). Additionally, the complexes are not supershifted by antibodies to other transcription factors, such as anti-c-Fos or anti-c-Jun (data not shown).
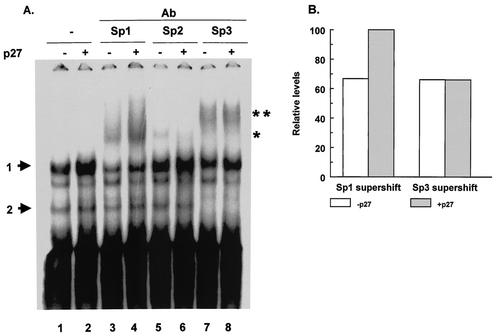
Increased expression of p27Kip1 leads to an increase in binding to the region of the MBP promoter between −105 and −52 (Fig. 3A), and this binding appears to involve Sp1, Sp2, and Sp3. In order to ascertain which of these Sp1 family members is involved in the increased binding to the MBP promoter, supershift assays were performed with extracts from CG4 cells transfected with either an empty vector or a plasmid encoding p27Kip1. With anti-Sp1 antibody, there is a significant increase in the level of the supershifted band in extracts from cells transfected with the p27Kip1 expression construct (Fig. 4). In contrast, there appears to be no major change in the level of the supershifted band with antibody to Sp2 or Sp3 (Fig. 4). These data indicate that the expression of p27Kip1 causes an increase in Sp1 transcription factor binding to the GC box in the MBP promoter.
FIG. 4.
p27Kip1 expression enhances Sp1 binding. (A) An EMSA was performed by using a probe containing sequences from −105 to −52 of the MBP gene and nuclear extracts from CG4 cells transfected with a control plasmid (−) or a p27Kip1 expression vector (+). Lanes 1 and 2 show binding in the absence of added antibody. In lanes 2 to 8, the indicated antibodies (Ab) were added to the binding reactions. The positions of the two specific complexes are indicated by arrows. The supershifted complexes formed with anti-Sp1 antibody (*) or anti-Sp3 antibody (**) are also indicated. The EMSA in this experiment was run with 5% polyacrylamide gels for a longer time than in other experiments in order to better resolve the DNA-protein complexes. All other EMSAs were run with 3.5% polyacrylamide gels. (B) The supershifted complexes from panel A were quantitated by densitometry. The levels of the supershifted complexes from cells transfected with the control vector are indicated by open bars. The levels of the supershifted complexes from cells transfected with the p27Kip1 expression vector are indicated by gray bars
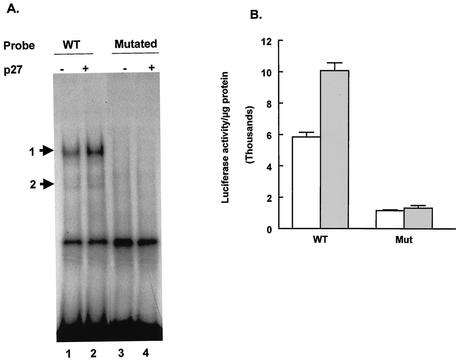
Mutation of the GC box in the MBP promoter eliminates Sp1 binding and disrupts the ability of p27Kip1 to stimulate MBP promoter activity.
The binding of Sp1 to the MBP promoter sequences between −105 and −52 is enhanced by the increased expression of p27Kip1. To determine whether the binding of Sp1 to the GC box correlates with the ability of p27Kip1 to increase MBP promoter activity, the GC box was mutated by site-specific mutagenesis (Fig. 1 shows the sequence). To verify that the mutation eliminated the binding of Sp1 family members to this sequence, a double-stranded oligonucleotide carrying the same mutation was used in EMSAs. Neither complex 1 nor complex 2 was formed when the oligonucleotide containing the mutation was used as a probe (Fig. 5A, lanes 3 and 4). However, with a wild-type probe and the same extracts, both complexes were observed and binding was enhanced by the expression of p27Kip1.
FIG. 5.
Mutation of the GC box in the MBP promoter abolishes the ability of p27Kip1 to activate the promoter. (A) An EMSA was performed by using either a probe containing MBP promoter sequences between −105 and −52 (WT; lanes 1 and 2) or a probe containing a mutation in the GC box (Mutated; lanes 3 and 4) (Fig. 1) and extracts from CG4 cells transfected either with a control plasmid (−) or a p27Kip1 expression plasmid (+). The positions of the two specific complexes are indicated by arrows. (B) CG4 cells were transfected with either a control plasmid (open bars) or a p27Kip1 expression plasmid (gray bars) together with either the −105 MBP-luciferase construct (WT) or the same construct containing a mutation in the GC box (Mut) (Fig. 1). Cells were harvested for luciferase assays after 2 days in DM. Results are presented as luciferase activity per microgram of protein and are the means and standard errors of the means for at least three replicates.
The mutation that disrupted Sp1 binding to the MBP sequence between −105 and −52 was introduced into the MBP-luciferase construct extending to −105 (−105 MBP-luciferase construct). Transfection experiments were then performed with both wild-type and mutant MBP-luciferase constructs. As shown earlier, cotransfection of the wild-type −105 MBP-luciferase construct with a plasmid encoding p27Kip1 leads to increased promoter activity. When the −105 MBP-luciferase construct carrying the mutation in the GC box is transfected into CG4 cells, the luciferase activity in the absence of increased p27Kip1 levels is significantly lower than that in the wild type. Thus, the GC box appears to be required for basal expression from this construct. Additionally, the GC box mutation totally eliminates the ability of the MBP promoter to respond to increased levels of p27Kip1 (Fig. 5B). These results suggest that the Sp1 binding site is required for the MBP promoter to respond to elevated levels of p27Kip1.
The expression of Sp1 has an effect on MBP promoter activity similar to that of p27Kip1.
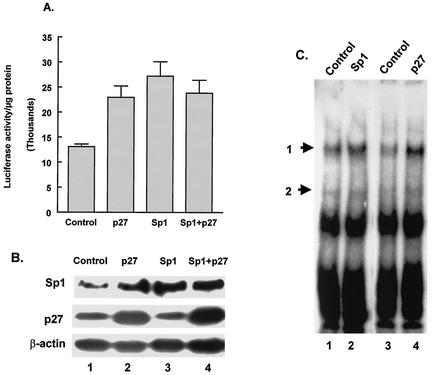
The above data suggest that increased p27Kip1 levels lead to increased binding of Sp1 to the MBP promoter and that this binding results in activation of the promoter. Thus, we would expect that increased expression of Sp1 in CG4 cells would have an effect on the MBP promoter similar to that of increased expression of p27Kip1. To test this idea, CG4 cells were transfected with the MBP-luciferase construct containing the full-length MBP promoter (extending to −1323) together with an Sp1 expression plasmid, a p27Kip1 expression plasmid, or both. When the MBP-luciferase construct is transfected with either the Sp1 expression construct or the p27Kip1 expression construct, there is an increase in luciferase activity over that seen with transfection with the control plasmid (Fig. 6A). The level of the increase is similar when either Sp1 or p27Kip1 is cotransfected. In fact, transfection of both expression constructs with the MBP-luciferase construct did not lead to higher levels of luciferase activity than did transfection of either the Sp1 or the p27Kip1 expression construct alone (Fig. 6A), suggesting that these proteins act through the same pathway. Increased expression of Sp1 and p27Kip1 in transfected cells was confirmed by Western blotting (Fig. 6B). Note that cells transfected with the p27Kip1 expression construct also had higher levels of Sp1 protein, demonstrating that the observed increase in Sp1 binding (Fig. 4) reflects an increase in Sp1 protein levels induced by elevated levels of p27Kip1.
FIG. 6.
Overexpression of Sp1 increases MBP promoter activity. (A) CG4 cells were transfected with −1323Luc plus either an empty vector (Control), a p27Kip1 expression vector (p27), an Sp1 expression vector (Sp1), or both the Sp1 and the p27Kip1 expression vectors (Sp1+p27). Results of luciferase assays are presented as luciferase activity per microgram of protein and are the means and standard errors of the means for at least three replicates. (B) Cells transfected as described for panel A were lysed and subjected to Western blotting for Sp1, p27Kip1, or β-actin. (C) An EMSA was performed by using nuclear extracts from CG4 cells transfected with a control plasmid (Control, lanes 1 and 3), an Sp1 expression vector (Sp1, lane 2), or a p27Kip1 expression vector (p27, lane 4). The positions of the two specific complexes are indicated by arrows.
Corresponding to the increase in MBP promoter activity induced by the expression of Sp1 in transfected cells, nuclear extracts from these cells show an increase in the formation of complex 1 (Fig. 6C). The increased binding seen in the EMSAs is almost identical in extracts from cells expressing Sp1 or p27Kip1 (Fig. 6C, compare lanes 2 and 4). We have also found that the MBP-luciferase construct containing MBP 5′-flanking sequences extending to −52 (without the GC box) does not show increased activity when transfected together with the Sp1 expression vector (data not shown). Additionally, mutation of the GC box disables its ability to respond to increased Sp1 levels (data not shown).
Increased Sp1 levels are a specific response to increased p27Kip1 levels.
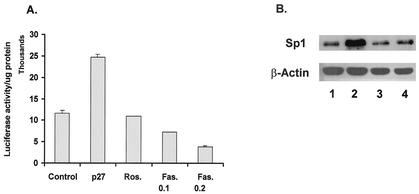
In most cells, including oligodendrocytes, p27Kip1 plays a role in cell cycle regulation through its function as a CKI. Since our data show that the overexpression of p27Kip1 leads to an increase in Sp1 protein levels, it was of interest to determine whether this increase is a response to the cell cycle regulatory function of p27Kip1 or is mediated in a different manner. To address this question, CG4 cells were treated with chemical inhibitors of Cdk activity. For these studies, we used roscovitine, a Cdk2 inhibitor, and fascaplysin, an inhibitor of Cdk4. Using bromodeoxyuridine labeling of S-phase cells, we have demonstrated that the concentrations of roscovitine and fascaplysin used in these studies are sufficient to inhibit the division of CG4 cells (data not shown). Treatment of cells transfected with the −105 MBP-luciferase construct indicates that neither roscovitine nor fascaplysin can enhance MBP promoter activity (Fig. 7A). Additionally, when nuclear extracts made from CG4 cells treated with either roscovitine or fascaplysin are used in EMSAs, the level of binding and the pattern of complexes formed are unaltered compared to those seen with untreated cells (data not shown). Further, there is no increase in endogenous Sp1 levels in cells treated with either roscovitine or fascaplysin (Fig. 7B). Thus, it appears that simply stopping the cell cycle is not sufficient to induce increased Sp1 levels and MBP promoter activity. These results are consistent with previous results showing that cell cycle arrest in the absence of increased p27Kip1 levels does not lead to activation of the MBP promoter (22).
FIG. 7.
The inhibition of Cdk activity is not sufficient to enhance MBP promoter activity or to increase Sp1 binding or Sp1 levels. (A) CG4 cells were transfected with −1323Luc and an empty vector, followed by culturing in DM containing DMSO (Control), 2.5 μM roscovitine (Ros.), or 0.1 or 0.2 μM fascaplysin (Fas.), or were cotransfected with a p27Kip1 expression plasmid (p27). Luciferase activity was determined after 2 days of culturing in DM. Results are presented as luciferase activity per microgram of protein and are the means and standard errors of the means for at least three replicates. (B) Cells transfected with either a control vector (lane 1) or a p27Kip1 expression vector (lane 2) or treated with 2.5 μM roscovitine (lane 3) or 0.1 μM fascaplysin (lane 4) were harvested in SDS sample buffer and subjected to Western blotting for Sp1 or β-actin.
Endogenous Sp1 levels increase with differentiation.
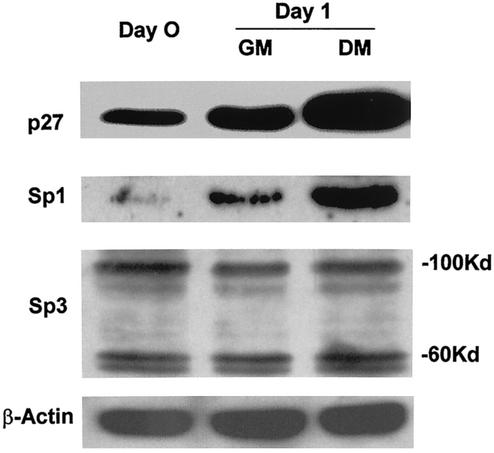
As oligodendrocytes differentiate, the levels of p27Kip1 increase (11, 37). Based on the results presented above, the levels of endogenous Sp1 should also increase when cells are induced to differentiate. Therefore, we measured the levels of Sp1 in primary oligodendrocytes that either had been maintained under growth conditions or had been induced to differentiate. Equal numbers of cells were lysed and loaded onto SDS-polyacrylamide gels, and the levels of Sp1 and p27Kip1 were determined by Western blotting. The results show that Sp1 is present in growing cells (Fig. 8, Day 0). However, as the cells differentiate, the levels of Sp1 are increased compared to those in growing cells maintained in cultures for the same period of time (Fig. 8, compare GM and DM). In differentiating cells, the levels of p27Kip1 are also increased. Thus, the levels of endogenous Sp1 are increased as the cells differentiate, coincident with the increase in the levels of p27Kip1. There are small increases in both Sp1 levels and p27Kip1 levels in growing cells as well. This result is likely due to the nonclonal nature of the primary cells, in which a small percentage of the cells will differentiate even under growth conditions.
FIG. 8.
Sp1 levels increase in primary oligodendrocytes as they differentiate. Equal numbers of primary rat oligodendrocytes were lysed, and the extracts were subjected to Western blotting for p27Kip1, Sp1, Sp3, or β-actin. Day 0 indicates cells that were growing in GM and that were lysed when the cells were transferred to DM. Day 1 indicates either growing cells (GM) or differentiating cells (DM) 1 day after the medium was changed. The molecular masses of the two species of Sp3 detected are indicated.
We also determined the levels of Sp3, the other major GC box binding protein present in the complexes observed with the MBP promoter probe, in differentiating oligodendrocytes. The Sp3 levels remain relatively unchanged as the cells differentiate (Fig. 8). Thus, the increase in the levels of p27Kip1 that occurs when oligodendrocytes differentiate is accompanied by an increase in the levels of Sp1 but not of Sp3.
p27Kip1 increases the levels of Sp1 by decreasing its rate of degradation.
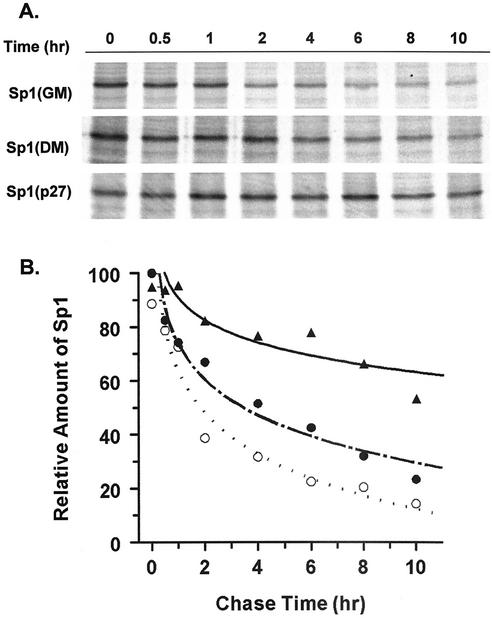
The increase in Sp1 levels induced by the increase in p27Kip1 levels could be caused by an increase in the production of Sp1 or by a decrease in its turnover. To test whether p27Kip1 increased the stability of existing Sp1 protein, a pulse-chase experiment was performed. CG4 cells were transfected with a plasmid encoding p27Kip1 or a control plasmid. At 2 days after transfection, the cells were starved for methionine and cysteine for 1 h, after which 35S-labeled methionine-cysteine was added to the medium. After 1 h, the cells were washed and the medium was replaced. The cells were harvested at different times after the medium was changed and were lysed, and Sp1 was immunoprecipitated. Figure 9A shows that the levels of labeled Sp1 at all time points except for the zero time point are higher in cells transfected with p27Kip1. The autoradiograph was subjected to densitometry, and the results were plotted against time (Fig. 9B). The results show that Sp1 in control transfected cells in DM has a half-life of approximately 4 h, while in p27Kip1-transfected cells, it has a half-life of 10 h or more. Additionally, the transfection results predict that the half-life of Sp1 should be longer in differentiating cells than in growing cells, since the endogenous levels of p27Kip1 are higher in differentiating cells. This prediction appears to be correct, since the half-life of cells maintained in GM is shorter (∼2 h) than that in cells placed in DM (Fig. 9). Thus, it appears that increases in the levels of p27Kip1 can enhance MBP gene expression through the stabilization of transcription factor Sp1.
FIG. 9.
Increased p27Kip1 levels lead to a decrease in the rate of turnover of Sp1. (A) CG4 cells were transfected with either an empty vector (GM or DM) or a p27Kip1 expression vector (p27) and cultured in GM or DM for 2 days. The cells were then pulse-labeled for 1 h with 35S-labeled methionine-cysteine, followed by a chase for the times indicated above the lanes. Sp1 was immunoprecipitated and separated by SDS-polyacrylamide gel electrophoresis, and the resulting gels were dried and autoradiographed. (B) The results from panel A for cells transfected with the empty vector and maintained in GM (○) or DM (•) or transfected with the p27Kip1 expression vector (▴) were quantitated by densitometry and plotted against the chase time. The zero-hour time point for cells in DM was set to 100%. All values shown are relative to this value.
DISCUSSION
The most widely recognized function of p27Kip1 in mammalian cells is to regulate the cell cycle by inhibiting the activity of Cdk2 (29, 31). p27Kip1 is also a key element in cell cycle progression in many cell types, including oligodendrocytes (36, 38). This role is consistent with the major role played by the Cdk2-cyclin E complex in oligodendrocyte progenitor cell cycle progression (15). In addition to its role in cell cycle arrest, evidence is now accumulating that p27Kip1 plays other roles in different cell types. These include a role in neuronal differentiation (32), triggering of apoptosis in breast cancer cells (20), and lengthening of the G1 phase in central nervous system cells (23).
Previous results indicated that the MBP promoter could be activated in response to an increase in p27Kip1 protein levels (22). The response of the MBP promoter to increased p27Kip1 levels could not be mimicked by inhibiting the activities of Cdk2 and Cdk4, as indicated by the results presented here, in which the inhibition of Cdk2 or Cdk4 with chemical inhibitors did not lead to an increase in MBP promoter activity. Additionally, it was shown previously that MBP promoter activity is not increased when other CKIs, such as p21, p57, and p16, are coexpressed with −1323Luc (22). Other studies also indicated that p27Kip1 is a multifunctional protein. In addition to its function as a cell cycle repressor, p27Kip1 plays a role in transcription in other systems. In some cells, p27Kip1 can repress transcription. In anergic T cells, p27Kip1 associates with the c-Jun coactivator JAB1, resulting in defective transactivation of AP-1 and interleukin 2 transcription (6). In mouse fibroblasts, p27Kip1 downregulates the P4 promoter of parvovirus through a cyclic AMP response element that interacts with complexes containing transcription factor E2F (9). In intestinal cells, the overexpression of p27Kip1 enhances sucrase-isomaltase gene expression by activating its promoter, while the overexpression of other CKIs, including p21 and p57, has no significant effect (10). Thus, p27Kip1 has specific effects on gene expression through mechanisms which at present are poorly understood.
By exploring the elements in the MBP promoter that are involved in the response to the increased expression of p27Kip1, we identified a GC box in the proximal region of the MBP promoter that is essential for the response to p27Kip1. Transcription factor Sp1 also was identified as interacting with the GC box. Other regions of the MBP promoter show differential responses to increased p27Kip1 levels. Computer analysis of these regions shows several additional potential Sp1 binding sites upstream of the one explored here. Thus, it is possible that these sites are responsible for the responses of other regions of the MBP promoter to increased p27 levels. Indeed, the increased expression of Sp1 mimics the effect of p27Kip1 on the MBP promoter and, when coexpressed, they activate the MBP promoter to the same level as either does alone. This finding suggests that they are a part of the same pathway of activation. The binding of Sp1 to the GC box is required for the effect of p27Kip1, as the disruption of binding through site-specific mutation of the GC box abolishes the ability of p27Kip1 to activate the MBP promoter. Disruption of the Sp1 binding site leads to a decrease in the basal promoter activity of MBP-luciferase constructs, indicating the importance of this site for promoter activity. However, we believe that the above results indicate a role for the binding of Sp1 to this site in mediating the effect of p27Kip1 on the MBP promoter. This effect of p27Kip1—increasing binding of Sp1 and binding to the GC box—is not simply the result of inhibiting Cdk2 activity, as treatment with chemical inhibitors of Cdk2 (or Cdk4) (Fig. 7) or the expression of p21, p57, or p16 (22) does not lead to increased Sp1 binding.
Both Sp1 and Sp3 binding activities appear to be present in CG4 cells, and both factors are capable of binding the GC box in the MBP promoter. The activation of promoters containing Sp1 binding sites can be regulated by the ratio of Sp1 to Sp3 in a cell at any given time. For example, in primary keratinocytes, Sp3 levels are higher than Sp1 levels. When these cells differentiate, the ratio becomes inverted, suggesting that the transcription of several genes required for cell type specification in keratinocytes can be regulated by a change in the ratio of Sp1 to Sp3 (3). A similar mechanism may occur in oligodendrocyte differentiation. As the cells enter the differentiation pathway, p27Kip1 levels increase and, subsequently, genes involved in the production of myelin are expressed. Both Sp3 and Sp1 are capable of binding the GC box in the MBP promoter. With an increase in p27Kip1 levels, Sp1 levels appear to increase, while there is no apparent change in Sp3 binding to the promoter. Thus, it is possible that the activation of the MBP promoter is achieved by an alteration in the ratio of Sp3 to Sp1.
Increased expression of p27Kip1 leads to increased Sp1 expression and binding to the MBP promoter. Prior to the increase in endogenous p27Kip1 levels, MBP expression is low, suggesting that regulation of the abundance of cellular Sp1 is one mechanism through which MBP transcription may be regulated. Changes in the abundance of Sp1 may occur through the regulation of Sp1 gene expression, mRNA stability, or posttranslational events that result in increased affinity for its binding site or decreased protein turnover. Interestingly, we found that the overexpression of p27Kip1 increases Sp1 levels by decreasing its turnover. The turnover of Sp1 in other systems has been investigated. For example, in the green monkey kidney cell line CV-1, Sp1 is susceptible to a specific protease, SPase, purified from cell nuclear extracts (27). Human Sug1 (also known as p45 or thyroid hormone receptor-interacting protein), an ATPase subunit of the 26S proteasome and a putative transcriptional modulator, is also able to stimulate the proteasome-dependent degradation of Sp1 (34). The role of these or other proteins in p27Kip1-mediated Sp1 stabilization in oligodendrocytes remains to be determined.
Additionally, Sp1 is phosphorylated by a number of protein kinases, including the cyclin A-Cdk2 complex (14), DNA-dependent protein kinase (19), casein kinase II (4), protein kinase A (30), and an unidentified cell cycle-regulated kinase (5). While the phosphorylation of Sp1 is mostly involved in the regulation of its transactivation activity, the phosphorylation of some specific sites also has an effect on its degradation. In a rat pituitary cell line, GH4, sustained stimulation by epidermal growth factor initiates a cascade of phosphorylation events that promotes Sp1 proteolysis and decreases the levels of Sp1 in the nucleus (24). In human tumor cells, blocking epidermal growth factor receptor tyrosine kinase activity upregulates the expression of p27Kip1 (7). These findings raise the possibility that the expression of p27Kip1 may inhibit the phosphorylation of Sp1 at a specific site or sites, leading to its stabilization. The effect of increased p27Kip1 expression on the phosphorylation status of Sp1 in oligodendrocytes remains to be determined.
In addition to phosphorylation, Sp1 can be modified through covalent linkage of the monosaccharide N-acetylglucosamine to serine and threonine residues (O-linked glycosylation). Reduced O-linked glycosylation of Sp1 is associated with increased proteasome susceptibility (17). Through an as-yet-undetermined mechanism, p27Kip1 may function to decrease the level of phosphorylation of Sp1 or increase its level of glycosylation, thus increasing Sp1 stability.
We have shown here a novel mechanism by which p27Kip1 can activate the MBP promoter. Elevated p27Kip1 expression causes an increase in the level of Sp1 protein by decreasing the rate of Sp1 degradation. Sp1 acts as a transcriptional activator of the MBP promoter through binding to the conserved GC box in the proximal region of the MBP promoter. In this way, p27Kip1 enhances MBP promoter activity. The mechanism by which p27Kip1 leads to a decrease in Sp1 degradation remains to be determined.
Acknowledgments
This work was supported by grants from the National Multiple Sclerosis Society (3293-A-5) and the NIH (CA84325).
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoyama, A., T. A. Tamura, and K. Mikoshiba. 1990. Regulation of brain-specific transcription of the mouse myelin basic protein gene: function of the NFI-binding site in the distal promoter. Biochem. Biophys. Res. Commun. 167:648-653. [DOI] [PubMed] [Google Scholar]
- 3.Apt, D., R. M. Watts, G. Suske, and H. U. Bernard. 1996. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology 224:281-291. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. [DOI] [PubMed] [Google Scholar]
- 5.Black, A. R., D. Jensen, S. Y. Lin, and J. C. Azizkhan. 1999. Growth/cell cycle regulation of Sp1 phosphorylation. J. Biol. Chem. 274:1207-1215. [DOI] [PubMed] [Google Scholar]
- 6.Boussiotis, V. A., G. J. Freeman, P. A. Taylor, A. Berezovskaya, I. Grass, B. R. Blazar, and L. M. Nadler. 2000. p27kip1 functions as an anergy factor inhibiting interleukin 2 transcription and clonal expansion of alloreactive human and mouse helper T lymphocytes. Nat. Med. 6:290-297. [DOI] [PubMed] [Google Scholar]
- 7.Busse, D., R. S. Doughty, T. T. Ramsey, W. E. Russell, J. O. Price, W. M. Flanagan, L. K. Shawver, and C. L. Arteaga. 2000. Reversible G(1) arrest induced by inhibition of the epidermal growth factor receptor tyrosine kinase requires up-regulation of p27(KIP1) independent of MAPK activity. J. Biol. Chem. 275:6987-6995. [DOI] [PubMed] [Google Scholar]
- 8.Cook, T., B. Gebelein, and R. Urrutia. 1999. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci. 880:94-102. [DOI] [PubMed] [Google Scholar]
- 9.Deleu, L., F. Fuks, D. Spitkovsky, R. Horlein, S. Faisst, and J. Rommelaere. 1998. Opposite transcriptional effects of cyclic AMP-responsive elements in confluent or p27KIP-overexpressing cells versus serum-starved or growing cells. Mol. Cell. Biol. 18:409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deschenes, C., A. Vezina, J. F. Beaulieu, and N. Rivard. 2001. Role of p27(Kip1) in human intestinal cell differentiation. Gastroenterology 120:423-438. [DOI] [PubMed] [Google Scholar]
- 11.Durand, B., Gao, F. B., and Raff, M. C. 1997. Accumulation of the cyclin-dependent kinase inhibitor p27/kip1 and the timing of oligodendrocyte differentiation. EMBO J. 16:306-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekholm, S. V. a. R., S.I. 2000. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12:676-684. [DOI] [PubMed] [Google Scholar]
- 13.. Espinosa de los Monteros, A., P. Zhao, C. Huang, T. Pan, R. Chang, R. Nazarian, D. Espejo, and J. de Vellis. 1997. Transplantation of CG4. oligodendrocyte progenitor cells in the myelin-deficient rat brain results in myelination of axons and enhanced oligodendroglial markers. J. Neurosci. Res. 50:872-887. [DOI] [PubMed] [Google Scholar]
- 14.Fojas de Borja, P., N. K. Collins, P. Du, J. Azizkhan-Clifford, and M. Mudryj. 2001. Cyclin A-CDK phosphorylates Sp1 and enhances Sp1-mediated transcription. EMBO J. 20:5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghiani, C., and V. Gallo. 2001. Inhibition of cyclin E-cyclin-dependent kinase 2 complex formation and activity is associated with cell cycle arrest and withdrawal in oligodendrocyte progenitor cells. J. Neurosci. 21:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghiani, C. A., A. M. Eisen, X. Yuan, R. A. DePinho, C. J. McBain, and V. Gallo. 1999. Neurotransmitter receptor activation triggers p27(Kip1)and p21(CIP1) accumulation and G1 cell cycle arrest in oligodendrocyte progenitors. Development 126:1077-1090. [DOI] [PubMed] [Google Scholar]
- 17.Han, I., and J. E. Kudlow. 1997. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 17:2550-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauser, P. J., D. Agrawal, M. Flanagan, and W. J. Pledger. 1997. The role of p27kip1 in the in vitro differentiation of murine keratinocytes. Cell Growth Differ. 8:203-211. [PubMed] [Google Scholar]
- 19.Jackson, S. P., J. J. MacDonald, S. Lees-Miller, and R. Tjian. 1990. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell 63:155-165. [DOI] [PubMed] [Google Scholar]
- 20.Katayose, Y., M. Kim, A. N. Rakkar, Z. Li, K. H. Cowan, and P. Seth. 1997. Promoting apoptosis: a novel activity associated with the cyclin-dependent kinase inhibitor p27. Cancer Res. 57:5441-5445. [PubMed] [Google Scholar]
- 21.Louis, J. C., E. Magal, D. Muir, M. Manthorpe, and S. Varon. 1992. CG-4, a new bipotential glial cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J. Neurosci. Res. 31:193-204. [DOI] [PubMed] [Google Scholar]
- 22.Miskimins, R., R. Srinivasan, M. Marin-Husstege, W. K. Miskimins, and P. Casaccia-Bonnefil. 2002. p27Kip1 enhances myelin basic protein gene promoter activity. J. Neurosci. Res. 67:100-105. [DOI] [PubMed] [Google Scholar]
- 23.Mitsuhashi, T., Y. Aoki, Y. Z. Eksioglu, T. Takahashi, P. G. Bhide, S. A. Reeves, and V. S. Caviness, Jr. 2001. Overexpression of p27Kip1 lengthens the G1 phase in a mouse model that targets inducible gene expression to central nervous system progenitor cells. Proc. Natl. Acad. Sci. USA 98:6435-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortensen, E. R., P. A. Marks, A. Shiotani, and J. L. Merchant. 1997. Epidermal growth factor and okadaic acid stimulate Sp1 proteolysis. J. Biol. Chem. 272:16540-16547. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, K. 1998. Cip/Kip cyclin-dependent kinase inhibitors: brakes of the cell cycle engine during development. Bioessays 20:1020-1029. [DOI] [PubMed] [Google Scholar]
- 26.Nigg, E. A. 1995. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays 17:471-480. [DOI] [PubMed] [Google Scholar]
- 27.Nishinaka, T., Y. H. Fu, L. I. Chen, K. Yokoyama, and R. Chiu. 1997. A unique cathepsin-like protease isolated from CV-1 cells is involved in rapid degradation of retinoblastoma susceptibility gene product, RB, and transcription factor SP1. Biochim. Biophys. Acta 1351:274-286. [DOI] [PubMed] [Google Scholar]
- 28.Quaroni, A., J. Q. Tian, P. Seth, and C. Ap Rhys. 2000. p27(Kip1) is an inducer of intestinal epithelial cell differentiation. Am. J. Physiol. Cell Physiol. 279:C1045-C1057. [DOI] [PubMed] [Google Scholar]
- 29.Rank, K. B., D. B. Evans, and S. K. Sharma. 2000. The N-terminal domains of cyclin-dependent kinase inhibitory proteins block the phosphorylation of cdk2/Cyclin E by the CDK-activating kinase. Biochem. Biophys. Res. Commun. 271:469-473. [DOI] [PubMed] [Google Scholar]
- 30.Rohlff, C., S. Ahmad, F. Borellini, J. Lei, and R. I. Glazer. 1997. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J. Biol. Chem. 272:21137-21141. [DOI] [PubMed] [Google Scholar]
- 31.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki, K., S. Tamura, H. Tachibana, M. Sugita, Y. Gao, J. Furuyama, E. Kakishita, T. Sakai, T. Tamaoki, and T. Hashimoto-Tamaoki. 2000. Expression and role of p27(kip1) in neuronal differentiation of embryonal carcinoma cells. Brain Res. Mol. Brain Res. 77:209-221. [DOI] [PubMed] [Google Scholar]
- 33.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 34.Su, K., X. Yang, M. D. Roos, A. J. Paterson, and J. E. Kudlow. 2000. Human Sug1/p45 is involved in the proteasome-dependent degradation of Sp1. Biochem. J. 348:281-289. [PMC free article] [PubMed] [Google Scholar]
- 35.Suske, G. 1999. The Sp-family of transcription factors. Gene 238:291-300. [DOI] [PubMed] [Google Scholar]
- 36.Tang, X. M., J. S. Beesley, J. B. Grinspan, P. Seth, J. Kamholz, and F. Cambi. 1999. Cell cycle arrest induced by ectopic expression of p27 is not sufficient to promote oligodendrocyte differentiation. J. Cell. Biochem. 76:270-279. [DOI] [PubMed] [Google Scholar]
- 37.Tikoo, R., P. Casaccia-Bonnefil, M. V. Chao, and A. Koff. 1997. Changes in cyclin-dependent kinase 2 and p27kip1 accompany glial cell differentiation of central glia-4 cells. J. Biol. Chem. 272:442-447. [DOI] [PubMed] [Google Scholar]
- 38.Tikoo, R., D. J. Osterhout, P. Casaccia-Bonnefil, P. Seth, A. Koff, and M. V. Chao. 1998. Ectopic expression of p27Kip1 in oligodendrocyte progenitor cells results in cell-cycle growth arrest. J. Neurobiol. 36:431-440. [PubMed] [Google Scholar]
- 39.Tretiakova, A., A. Steplewski, E. M. Johnson, K. Khalili, and S. Amini. 1999. Regulation of myelin basic protein gene transcription by Sp1 and Puralpha: evidence for association of Sp1 and Puralpha in brain. J. Cell. Physiol. 181:160-168. [DOI] [PubMed] [Google Scholar]
- 40.Vidal, A., and A. Koff. 2000. Cell-cycle inhibitors: three families united by a common cause. Gene 247:1-15. [DOI] [PubMed] [Google Scholar]