Abstract
The simian immunodeficiency virus (SIV) transmembrane (TM) protein, gp41, has multiple functions, which include anchoring the glycoprotein complex in the lipid envelope of the virus and mediating fusion of the virus and host cell membranes. Recently, a series of mutants of the SIVmac239 TM protein that have truncations at the carboxyl terminus of the membrane-spanning domain (MSD) have been characterized (J. T. West, P. Johnston, S. R. Dubay, and E. Hunter, J. Virol. 75:9601-9612, 2001). These mutants retained membrane anchorage but demonstrated reduced fusogenicity and infectivity as the MSD length was shortened. We have established a novel three-color fluorescence assay, which allows qualitative confocal and quantitative flow cytometric analyses, to further characterize the nature of the fusion defect in five of the MSD mutants: TM185, TM186, TM187, TM188, and TM189. Our analysis showed that each mutant could mediate complete lipid and aqueous dye transfer at early time points after effector and target cell mixing. No hemifusion with only lipid dye flux was detected. However, another intermediate fusion stage, which appears to involve small-fusion-pore formation that allowed small aqueous dye transfer but prevented the exchange of large cytoplasmic components, was identified infrequently in mutant-Env-expressing cell and target cell mixtures. Quantitative flow cytometric analysis of these mutants demonstrated that the TM187, TM188, and TM189 mutants were significantly more fusogenic than TM185 and TM186 but remained significantly impaired compared to the wild type. Moreover, fusion efficiency showed an increased dependence on the expression level of glycoproteins, suggesting that, for these mutants, formation of an active fusion complex was an increasingly stochastic event.
The envelope glycoprotein of simian immunodeficiency virus (SIV) is synthesized as a glycosylated polypeptide precursor, gp160. During its transport to the plasma membrane, it is cleaved into two subunits, gp130, the surface glycoprotein (SU), and gp41, the transmembrane subunit (TM). Similar to those of human immunodeficiency virus (HIV), the SU proteins of SIV are also involved in the interaction with host cell receptors, CD4, and various chemokine coreceptors (4, 14). The TM protein anchors the glycoprotein complex in the lipid envelope bilayer of the virus and mediates various steps in the fusion of the viral membrane and target cell membrane (8, 19).
The TM protein consists of three domains, an N-terminal ectodomain, a membrane-spanning domain (MSD), and a C-terminal cytoplasmic tail. The N-terminal domain contains the fusion peptide and the heptad repeats which form a coiled-coil structure during membrane fusion (7, 45). As in other lentiviruses, the cytoplasmic tail of the SIV TM protein is long, containing over 150 amino acids. Natural truncation of this C-terminal domain during the passage of SIV in human T-cell lines results in more efficient incorporation of glycoproteins into virions and an expanded host range (18, 20, 22, 47).
The MSDs of type 1 glycoproteins are composed primarily of a stretch of uncharged, hydrophobic amino acids, which is usually more than 20 amino acids in length, and form an α-helix in the lipid bilayer (40). The hydrophobic stretches are often bordered by charged amino acids. The positions of the MSDs for HIV and SIV were initially derived from mutational analyses (reviewed in reference 19). An unusual aspect of these proposed MSDs was the presence of a basic residue within the long stretch of hydrophobic amino acids. However, recent studies on SIV have redefined the topology of the MSD and have suggested that this residue is proximal to the membrane-cytoplasm boundary (46).
In these studies, a series of mutants with C-terminal truncations in the SIV TM were analyzed and virus entry and infectivity were found to be retained in truncation mutants with a minimum of 189 amino acids in the TM (the total length of the TM in the wild type is 354 amino acids). In contrast, infectivity was lost and fusogenicity was progressively reduced as the MSD was shortened in truncation mutants TM188, TM187, TM186, and TM185 (46). For TM185, fusion activity measured in multinuclear activation of galactose indicator (MAGI) cells was reduced by 90%. Nevertheless, all the mutants were stably anchored in the plasma membrane and were competent for incorporation into virus particles. Thus, the MSD plays a key role in mediating membrane fusion that can be dissected from its role as a membrane anchor.
Extensive studies of viral fusion mechanisms and characterization of viral glycoproteins by nuclear magnetic resonance and X-ray crystallography have revealed significant similarities in the structures and functions of molecules as diverse as the influenza virus hemagglutinin (HA), the HIV and SIV TM proteins, the murine leukemia virus TM protein, the Ebola virus TM protein, and paramyxovirus F proteins (13, 44). A common fusion process mediated by these proteins is believed to involve a series of conformational changes in the viral glycoproteins that bring the viral and cellular membranes in close proximity so that lipid merging (perhaps through a hemifusion intermediate) can occur. These initial steps of fusion are believed to be followed by the formation and expansion of fusion pores to yield a fully fused state (5, 6, 42, 43).
Hemifusion has been thought to be a metastable intermediate state in which the two outer leaflets of the membrane bilayer merge and the inner leaflets form a single bilayer diaphragm to separate the two cytoplasmic compartments (27, 33). Hemifusion has been identified in the fusion process mediated by the viral glycoproteins of influenza virus (10, 21, 30, 39), simian virus 5 (SV5) (2, 21), vesicular stomatitis virus (11), and HIV (31). Interestingly, when the two glycine residues in the MSD of vesicular stomatitis virus G protein were replaced by Ala or Leu, the fusion process was arrested at a hemifusion stage (11). The fusion ability of MSD truncation mutants of influenza HA was reduced as the MSD was shortened. In this case, a minimum of 17 amino acids of the 27-amino-acid-long MSD were required for complete membrane fusion while mutant MSDs with lengths of 15 amino acids mediated only hemifusion (1). Similar results were obtained for the SV5 F protein, of which progressive truncations of the cytoplasmic domain and MSD yielded mutants capable primarily of hemifusion (2).
Previous studies on the MSD of SIV employed a fusion assay that required both lipid and content mixing (syncytium assay) (46). We have now characterized further a subset of these SIV MSD truncation mutants to determine the nature of the fusion process that they mediate. In this study, we have established a novel three-color fluorescence assay to study the fusion process mediated by the SIV TM truncation mutants (TM185 to TM189). Combining fluorescence microscopy and flow cytometric analyses, we have determined the nature of the defect in these mutants both phenotypically and quantitatively. We show here that all five mutants can mediate complete membrane fusion and that the efficiency of this process decreases with the length of the MSD. No hemifusion with only lipid dye flux was detected. However, for some of the mutants, the fluorescence assay could detect a rare intermediate fusion stage, apparently involving small-fusion-pore formation that allowed small aqueous dye transfer but prevented the exchange of large cytoplasmic components. Moreover, fusion efficiency showed an increased dependence on the expression level of glycoproteins, suggesting that, for these mutants, formation of an active fusion complex was an increasingly stochastic event.
MATERIALS AND METHODS
Chemicals and cells.
Calcein, acetoxymethyl (AM), and FAST-DiI, a derivative of DiI (1,1-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanide perchlorate) with diunsaturated Δ9,12-C18 alkyl substituents in place of the saturated C18 tail, were from Molecular Probes, Eugene, Oreg. Diluent C and high-purity dimethyl sulfoxide (DMSO) were from Sigma, St. Louis, Mo. Cell dissociation buffer was from Gibco-BRL, Rockville, Md. Glass-bottomed 35-mm-diameter plates were obtained from MatTek Corporation, Ashland, Mass. Fugene6 was from Roche Molecular Biochemicals, Indianapolis, Ind. COS-1 cells were purchased from the American Type Culture Collection. MAGI-CCR5 reagent was obtained from Julie Overbaugh through the National Institutes of Health AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases. JC53-BL cells were kindly provided by Xiaoyun Wu and John Kappes, University of Alabama at Birmingham.
EBFP-SIV Env coexpression vector.
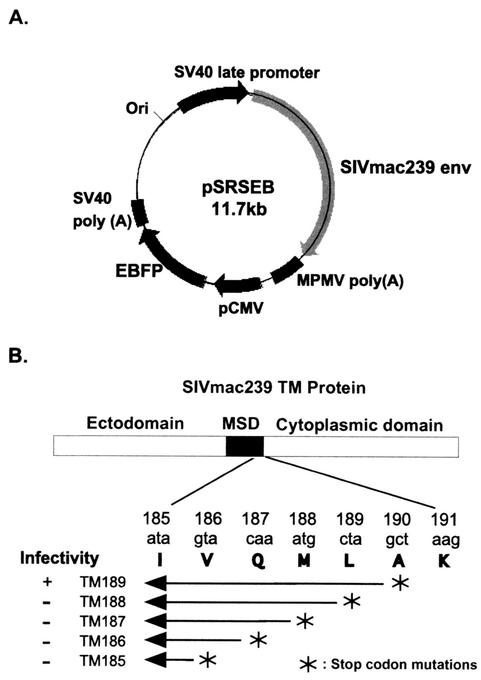
The enhanced blue fluorescent protein (EBFP) expression cassette was excised from pEBFP-N1 (Promega) by double digestion of the plasmid with restriction enzymes AflII and AseI. The purified DNA fragment was inserted into plasmid pSRS at the SacII site by blunt-end ligation. The pSRS vector expresses the wild-type Env protein of SIVmac239 (20). This new pSRSEB construct (Fig. 1A) supports high-level expression of the SIVmac239 wild-type Env proteins from the SV40 late promoter as well as EBFP expression directed by the human cytomegalovirus immediate-early promoter. The pSRS derivative plasmids expressing mutants TM185 to TM189 were engineered in the same way to include the EBFP expression cassette. The pSREB vector is a control plasmid in which the env sequence from XbaI to BspEI has been deleted and thus expresses only EBFP. COS-1 cells were transfected with these vectors (pSREB, pSRSEB, or pTM185EB through pTM189EB) by using the Fugene6 transfection reagent as described by the manufacturer; cells were then used as effector cells in fusion assays.
FIG. 1.
Schematic diagram of SIV TM truncation mutants and map of pSRSEB vector. (A) Map of the expression vector pSRSEB which supports the expression of SIVmac239 env and EBFP genes from two separate promoters. (B) Illustration showing positions of stop codon mutations at the C terminus of the MSD. A short segment of the pertinent amino acid sequence that encompasses the truncations is shown.
Fluorescence labeling of cells and microscopic examination of fusion.
A stock solution of 10 mM calcein-AM (small aqueous dye; excitation, 496 nm; emission, 517 nm) was prepared in high-purity DMSO. This stock solution was then diluted in Dulbecco modified Eagle medium (DMEM) or phosphate-buffered saline (PBS) at a final concentration of 0.2 μM. Transfected COS-1 effector cells or, in some experiments, target cells were incubated with diluted calcein-AM solution for 30 to 45 min at 37°C. Cells were then washed twice with DMEM prior to use.
FAST-DiI (lipid dye; excitation, 550 nm; emission, 565 nm) was prepared as a 10 mM stock solution in DMSO. Immediately prior to use, 1 μl of the stock solution was diluted in 100 μl of Diluent C (Sigma), and after extensive vortexing, 10 μl of the diluted solution was added to 2.5 ml of PBS and vortexed. This solution (final concentration, 0.4 μM) was added to target cells (MAGI-CCR5 or JC53-BL), which were gently rocked continuously at room temperature for 2 min, and then DMEM was added for 5 min at room temperature. Cells were then washed three times with DMEM and two times with PBS. Cells were then dissociated in cell dissociation buffer and resuspended in complete DMEM medium. A total of 2 × 105 labeled target cells were added onto semiconfluent COS-1 effector cells in glass-bottomed plates, and after coincubation at 37°C for the specified periods of time, cells were washed with PBS once and maintained in PBS with Ca2+ and Mg2+ for microscopic examination.
Fluorescent samples were examined with an Olympus IX70 microscope or a Zeiss Axiovert35 microscope. Standard filter sets were used for the different fluorophores: rhodamine filter for FAST-DiI; fluorescein isothiocyanate filter for calcein-AM; and DAPI (4′,6′-diamidino-2-phenylindole) filter for EBFP. Dye concentrations optimized to minimize the leakage of fluorescence signals between filters were used. When necessary, suppression filters were also used to reduce leakage. Images were captured via IPLab Spectrum software and pseudocolored according to their respective emission wavelengths. Phase-contrast images were always taken in parallel to examine cell morphology and cell-cell contacts and to avoid having overlapping cells treated as a positive fusion event.
Flow cytometric analysis of efficiency of membrane lipid transfer during fusion.
COS-1 cells in 60-mm-diameter plates were transfected with the Env-EBFP coexpression plasmids as described above. At 36 to 48 h posttransfection, they were replated into six-well plates at approximately 2 × 105 cells per well. Target cells (JC53-BL or MAGI-CCR5) were labeled with FAST-DiI only or FAST-DiI together with calcein-AM at the same concentration as specified above; the FAST-DiI concentration was increased to 0.8 μM for better separation of cell populations. Approximately 106 fluorescently labeled target cells were added to each well containing transfected COS-1 effector cells for coincubation at 37°C for the specified times. The cocultures were washed with PBS and dissociated with 1 ml of PBS/well with 25 mM EDTA, and the cells were resuspended into fluorescence-activated cell sorter sorting tubes (Falcon). Cells were immediately subjected to flow cytometric analysis. If necessary, cells were fixed with 2% paraformaldehyde. Cell populations were analyzed on a Vantage flow cytometric workstation (Becton Dickson). A UV laser was used for EBFP and a 488-nm-wavelength laser was used for FAST-DiI and calcein-AM. Usually, 10,000 blue cells were gated, and the gated population was analyzed for blue-red or blue-green double-positive cells.
Luciferase assay of fusion activity.
To quantitatively assess cell-cell fusion, an assay based on reporter gene activity was employed: COS-1 cells were transfected with pSRS plasmids expressing wild-type or mutant Env proteins. These plasmids also express the SIV tat gene that is capable of activating the expression of firefly luciferase under the control of a minimal HIV long terminal repeat promoter in JC53-BL cells. Like the parental JC53 cell line (34), the JC53-BL cell line expresses high levels of CD4, CCR5, and CXCR4 on the cell surface (X. Wu, personal communication).
Env-expressing COS-1 cells were mixed with JC53-BL cells at 48 h posttransfection. They were cocultured for 8 to 12 h and then rapidly frozen at −80°C and thawed only immediately before the luciferase assay was conducted. The luciferase assay was performed using a luciferase assay kit (Promega) and the BMG Lumistar program. Triplicate wells were set up for each sample to ensure reliability.
RESULTS
We were interested in further delineating the ability of five C-terminal truncation mutants of the SIV TM protein (46) to mediate membrane fusion. The positions of the truncations relative to the putative MSD are depicted in Fig. 1B. The biological activities of these mutants suggested an involvement of the MSD in the membrane fusion process (46). In summary, all five mutant Env proteins are synthesized, cleaved, and transported normally. They all can be incorporated into virions, but only TM189 retains infectivity. As TM189 was truncated to TM185, a gradual loss of fusion ability was observed in a standard β-galactosidase assay. In particular, the fusion activities of TM185 and TM186 were reduced at least 10-fold from the level of that of wild-type Env (46). In this study, we have further characterized fusion mediated by these truncation mutants by employing the pSRSEB SIV Env-EBFP dual-expression vector system. This vector system allows cells expressing the SIV Env protein after transient transfection to be identified by a high level of blue fluorescence from EBFP. Fusion efficiencies of these mutants were analyzed by identifying the spreading of fluorescence probes between effector cells and target cells in the context of flow cytometry and confocal microscopy.
Flow cytometric analysis of lipid dye transfer to measure fusion efficiency.
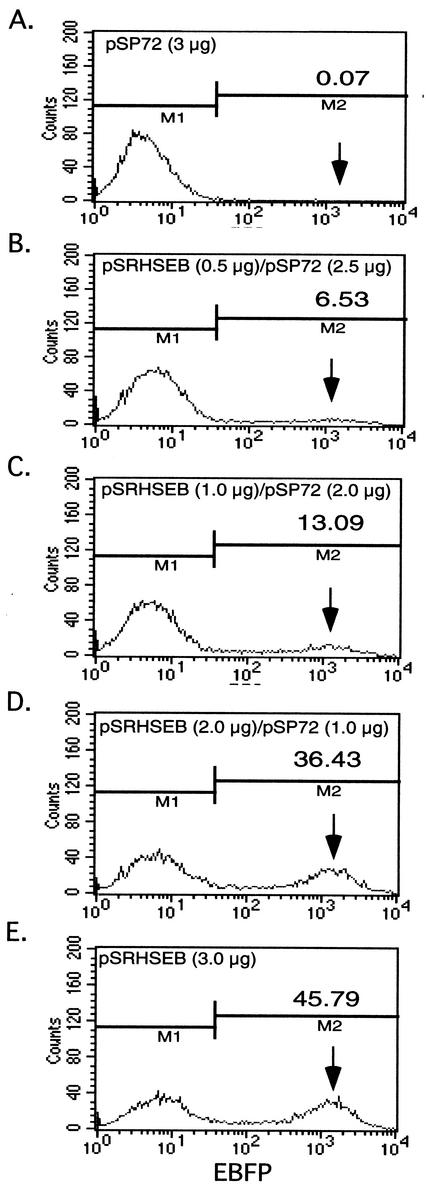
In initial experiments, COS-1 cells were transfected with increasing amounts of the wild-type pSRHSEB plasmid to determine the efficiency of transfection and the effect of increasing DNA on levels of EBFP expression. Figure 2E shows that, following transfection of COS-1 cells with 3 μg of the plasmid, approximately 45% of the cells expressed EBFP and that transfection with increasing amounts of DNA, while increasing the number of expressing cells, did not alter the position of the peak intensity of the EBFP-positive population (Fig. 2B to D). The fact that the EBFP-positive population has a mean fluorescence intensity approximately 30 times higher than the background cutoff enables the quantitative separation of EBFP-positive cells from background cells by flow cytometry.
FIG. 2.
Identification of EBFP-positive cells by flow cytometric analysis. COS-1 cells were transfected with various amounts of the pSRHSEB expression plasmid. The pSP72 plasmid was used to adjust the total amount of DNA to 3 μg in each transfection. Arrows on the right point out the positions of peak populations of EBFP-positive cells. The numbers above the arrows indicate the percentage of EBFP-positive cells obtained for each test transfection. M1 and M2 define the gating parameters for defining negative (M1) and positive (M2) EBFP-expressing cell populations.
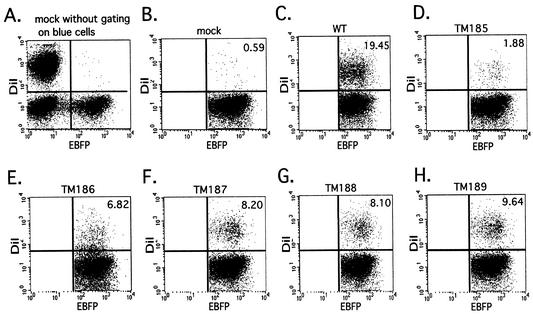
For membrane fusion studies, COS-1 cells were transfected with 3 μg of wild-type pSRSEB, mutants pTM185EB through pTM189EB, or env-negative pSREB. Two days posttransfection, MAGI-CCR5 target cells were labeled with FAST-DiI as described in Materials and Methods. In order to minimize lipid dye internalization, target cells were labeled with FAST-DiI for 2 min immediately prior to coincubation with effector cells. Labeled MAGI-CCR5 cells were mixed with transfected COS-1 cells at a 5:1 ratio for 1.5 h, and then the cells were dissociated with cell dissociation buffer and subjected to flow cytometric analysis. A total of 10,000 blue cells were gated and analyzed for the presence of red dye indicative of lipid transfer or cell fusion. Figure 3A shows that the labeling conditions, in the absence of Env (pSREB), allowed distinct separation of the DiI-labeled target cells and the EBFP-expressing effector cell populations by flow cytometry. Fig. 3B represents the same mock sample after gating of blue cells. Gating of blue cells facilitates the acquisition of quantitative data regarding the percentage of Env-expressing (blue) cells that have undergone lipid dye transfer after fusion with the FAST-DiI-labeled target cells (Fig. 3C to H). This figure demonstrates that, as the length of the TM protein increased, the extent of dye transfer also increased, although TM189 (Fig. 3H) remained significantly less fusogenic than the wild-type Env. Control experiments (data not shown) in which nonfusogenic forms of the HIV or SIV Env protein were expressed from the EBFP coexpression vector yielded background levels of blue-red-positive cells similar to that seen with pSREB. This indicated that Env-induced cell aggregation was not responsible for the detection of double-positive cells.
FIG. 3.
Results of flow cytometric analysis of fusion efficiencies of SIV TM mutants. COS-1 cells were transfected as described in the legend to Fig. 2 to express both EBFP and wild-type (WT) or mutant Env proteins or EBFP alone (mock). Target MAGI-CCR5 cells were labeled with FAST-DiI, coincubated with effector cells for 1.5 h at 37°C, and then subjected to flow cytometric analysis. Flow cytometry panels depict the separation of different populations of cells; the EBFP-DiI-double-positive population comprises cells with fused membranes. The percentages of fused blue cells were calculated to quantitate fusion efficiencies of different mutants and are indicated on panels B to H. In panel A, all cell populations (in the absence of Env expression) are represented without gating of blue cells: nonfluorescent (non-EBFP-expressing) COS-1 are localized to the lower left quadrant, EBFP-positive (blue) COS-1 are in the lower right, and FAST-DiI-labeled MAGI-CCR5 are in the upper left. For simplicity of presentation, panel B represents the population after gating of blue cells. EBFP-FAST-DiI-double-positive cells are located in the upper right quadrant. Panels C through H show results of analyses of COS-1 effector cells and MAGI-CCR5 target cells expressing wild-type and mutant Env.
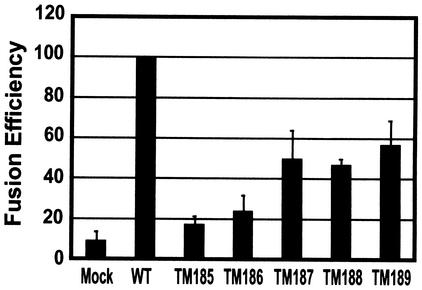
The average fusion efficiencies relative to that of the wild type from three independent flow cytometry experiments were plotted in Fig. 4. Consistent with previous results (46), the percentage of EBFP-expressing cells scoring positive for DiI transfer increased gradually as the length of truncated TM proteins increased. TM189, TM188, and TM187 were the most competent for fusion, with 53% ± 12%, 42% ± 4%, and 45% ± 14% of the dye transfer observed with the wild type, respectively. TM185 and TM186 showed much lower levels of fusion (9% ± 2% and 16% ± 12% that of the wild type, respectively).
FIG. 4.
Relative fusion efficiencies of TM truncation mutants. Flow cytometric analysis of fusion between EBFP-Env-expressing COS-1 cells and FAST-DiI-labeled MAGI-CCR5 target cells was conducted as described in the legend to Fig. 3. The fusion efficiencies for each mutant were normalized to that of the wild type (WT) in three independent experiments, averaged, and plotted.
Confocal microscopy of SIV TM mutants shows complete fusion.
For the Env mutant TM186, the percentage of cells showing lipid dye transfer was consistently higher than the percentage of fused cells previously observed in the standard MAGI fusion assay (46). To investigate this difference, we utilized a three-color confocal microscopy approach to determine whether TM186, or any other of the mutant SIV Env proteins, could induce hemifusion intermediates similar to those observed with influenza HA and SV5 F protein truncation mutants. Hemifusion would result in lipid dye transfer but not complete cytoplasmic mixing as measured by the classic MAGI assay. In the three-color assay, the MAGI-CCR5 target cells were labeled with both FAST-DiI and a cytoplasmic dye, calcein-AM, as described in Materials and Methods. These double-labeled cells were mixed with COS-1 cells expressing wild-type or mutant Env and EBFP as described above and observed by confocal fluorescence microscopy after a 1.5-h coincubation. If hemifusion were to occur, we would expect the lipid label (FAST-DiI) to transfer to the (blue) fusion cell partner while calcein-AM would remain within the target cell.
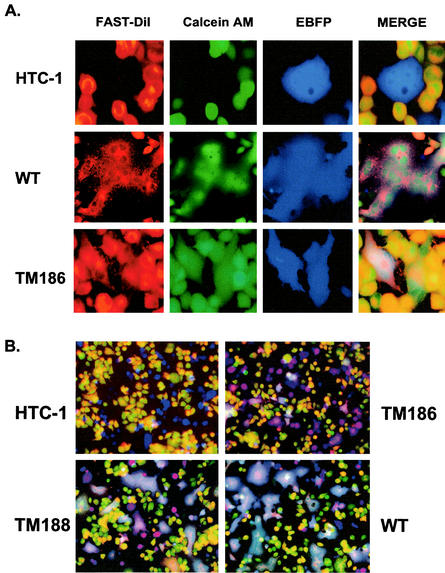
Internalization of DiI from the target cell plasma membrane into intracellular membrane compartments occurred very rapidly, and we thus observed lipid labeling of membranes in intracellular compartments as well as on the plasma membrane (Fig. 5A). Calcein-AM crosses the cell membrane and becomes fluorescent after hydrolysis inside the cell, yielding a diffuse green staining of the cells (Fig. 5A). In the control plate, COS-1 cells were transfected with pHTC-1EB, a dual-expression plasmid that expresses HTC-1, a cleavage-defective, glycosylphosphatidylinositol (GPI)-anchored HIV Env protein (41). This nonfusogenic mutant Env did not show any evidence of mediating fusion even though EBFP-expressing cells were in direct contact with double-labeled target cells (Fig. 5A, HTC-1). In contrast, large syncytia were observed in plates containing COS-1 cells transfected with the pSRS-EB plasmid that expresses wild-type SIV Env. In this case, all three labels were evenly distributed throughout the syncytia (Fig. 5A, WT). Consistent with previous results of the MAGI cell assay (46), we observed that TM185 and TM186 primarily formed fused cell pairs, or small syncytia, with low efficiency (Fig. 5A and B), while TM187, TM188, and TM189 formed more abundant, larger syncytia (Fig. 5B). Wild-type proteins mediated the formation of large syncytia even at 1.5 h following cell mixing.
FIG. 5.
Results of three-color fluorescence assay to monitor fusion process. FAST-DiI (red) and calcein-AM (green) were used to label MAGI-CCR5 target cells, while EBFP (blue) marked Env-expressing cells. (A) Distribution patterns of three different fluorophores were examined individually, and then colocalization was analyzed in the merge field. The HTC-1 mutant provides a negative control for fusion, the TM186 mutant forms smaller and fewer syncytia than the wild type, and the wild-type Env protein (WT) mediates the formation of big syncytia. (B) Low-magnification view of merged fields for HTC-1, TM186, TM188, and wild-type Env.
Under these experimental conditions, all five truncation mutants primarily mediated complete membrane fusion and no evidence was obtained for the presence of unrestricted hemifusion intermediates that would allow the lipid flux without cytoplasmic content mixing.
SIV MSD truncation mutants can induce small-fusion-pore formation—another intermediate fusion event.
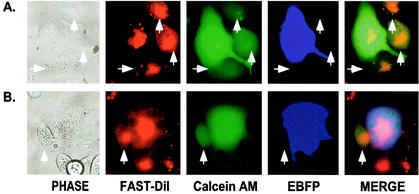
In some experiments, the calcein-AM cytoplasmic marker was loaded into the transfected COS-1 cells so that both calcein-AM and EBFP were in the same cell. This provided an additional approach to monitor the exchange of cytosol components in the fusion process since EBFP (35 kDa) is much larger than calcein-AM (635 Da) and any difference in the redistribution of cytosolic markers would not be the result of intracellular differences in target and effector cells.
Using this labeling approach, we observed essentially the same results as described above in terms of fusion efficiency and size of syncytia for each of the mutants. However, in a few cases, we observed cell pairs that appeared to be at a small-fusion-pore stage of membrane fusion, where the small cytoplasmic marker could transfer to the target cell in the absence of EBFP transfer (Fig. 6A). The observation of these rare events was not consistently associated with a particular mutant; rather, they were observed infrequently with all of the mutants, although they were not observed with cells expressing wild-type Env. Control experiments with pSREB-transfected COS-1 cells, which do not express any Env protein, also did not show this phenotype, ruling out the possibility that it resulted from dye leakage (data not shown).
FIG. 6.
Identification of small-fusion-pore formation. Transfection of COS-1 cells and mixing of effector and target cells were carried out as described in the legend to Fig. 3, with the exception that calcein-AM was loaded into EBFP-expressing COS-1 cells; consequently, the two cytoplasmic markers colocalized to the same cells. Panel A shows small-fusion-pore formation between individual cells, while panel B illustrates small-pore formation between a syncytium and neighboring target cells. Arrowheads point to the cells that allowed the transfer of calcein-AM from an effector COS-1 cell in the absence of EBFP transfer.
Moreover, we also observed small-fusion-pore formation for some syncytia formed by the mutants. Figure 6B shows a typical field that illustrates transfer of calcein-AM to target cells in contact with syncytia while EBFPs are still restricted to the syncytia.
Luciferase-based fusion assays point to inefficient initiation and propagation of fusion.
From previous observations with the MAGI-CCR5 assay (46) and the confocal studies described above, it was clear that the SIV TM truncation mutants exhibited both reduced numbers of syncytia and significantly smaller syncytia. Therefore, in order to quantitatively assess the sum of both fusion initiation and fusion propagation within a target cell population, we have utilized the JC53-BL cell line, which expresses the luciferase enzyme under the control of a minimal HIV long terminal repeat promoter in response to SIV Tat. These cells allow a quantitative assessment of the total number of target cell nuclei recruited into syncytia following cell mixing.
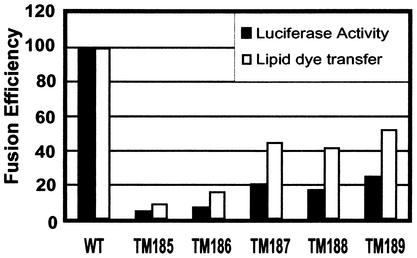
The results of a typical luciferase assay, shown in Fig. 7, indicate that in this assay, while a similar progressive loss of fusion with truncation of Env is observed, all of the truncation mutants are significantly impaired for fusion compared to the wild-type Env protein. TM189, TM188, and TM187, which exhibited fusion efficiencies that were reduced approximately twofold (53, 42, and 45%, respectively) in the flow cytometric analyses, exhibited fusion efficiencies reduced four- to fivefold (25, 18, and 21%, respectively) in this population analysis. TM185 and TM186 showed an even lower ability to mediate cell-cell fusion (5 and 7% of that of the wild type, respectively). Thus, in an assay that measures the sum of the numbers and sizes of syncytia, it is clear that even Env truncation mutants that have the capacity to mediate entry of infectious virus (TM189) are severely impaired for fusion.
FIG. 7.
Results of luciferase assay to determine fusion ability of SIV TM mutants. COS-1 cells were transfected with pSREB (control), pSRSEB (wild type), and pTM185EB to pTM189EB as described in the legend to Fig. 3 except that at 48 h posttransfection, the COS-1 cells were mixed with JC53-BL target cells. After overnight incubation, cells were lysed and measured for luciferase activity. The fusion efficiencies from the luciferase assay and the lipid dye transfer assay were compared for SIV wild-type Env (WT) and TM truncation mutants.
Modulation of fusion efficiency by protein expression levels.
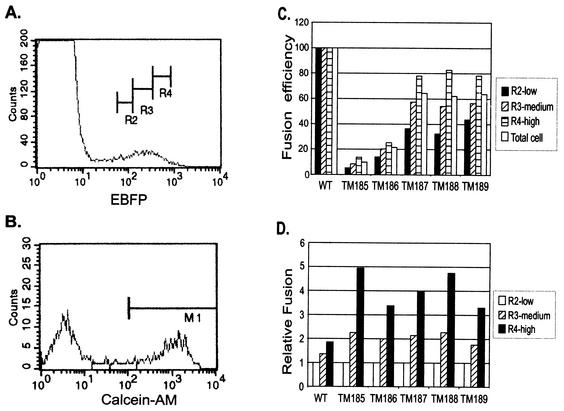
Since in the luciferase assay each of the truncation mutants appeared to be defective in propagating fusion to cells adjacent to an initial fusion event, these assay results raised the possibility that the density of Env on the effector cell surface might additionally modulate the efficiency of fusion—as the size of a syncytium increases, the density of Env on its surface might be expected to decrease. The EBFP-Env dual-expression system allows the classification of cell populations into high, moderate, and low Env expressers based on the intensity of fluorescence of the coexpressed EBFP within the cells (Fig. 8A). Each population can then be quantitated for the percentage of double-labeled fused cells (Fig. 8B). The results of such an experiment in which target cells were labeled with calcein-AM, shown in Fig. 8C, yielded total fusion percentages similar to those from the lipid transfer assays described above. However, when the results were analyzed on the basis of EBFP intensity, it was clear that the expression level modulated the efficiency of fusion (Fig. 8C), with those cells expressing the highest levels yielding the highest percentages of fused cells. In fact, mutants TM187 to TM189 fused almost as efficiently as the wild-type when those cells expressing the highest levels of EBFP were compared—the fusion efficiency was approximately 80% of that of the wild type. In contrast, TM185 and TM186 remained severely impaired for fusion (fusion efficiencies of 12 and 22% of that of the wild type, respectively) at the highest expression levels (Fig. 8C).
FIG. 8.
Modulation of fusion efficiency by expression level. COS-1 cells were transfected as described in the legend to Fig. 3. Target CCR5-MAGI cells were labeled with calcein-AM as described in Materials and Methods. Cells were coincubated for 1.5 h and then subjected to flow cytometric analysis. (A) Definition of low (R2), medium (R3), and high (R4) expression levels for EBFP-positive cells. (B) Separation of fused calcein-AM-labeled cells from nonfused cells for quantitation of fusion efficiency in an R3-gated EBFP-expressing population. M1 defines the gating parameter for quantitating calcein-AM-positive cells. (C) Fusion efficiency of each mutant normalized to wild-type (WT) Env-mediated fusion at each of the three expression levels. (D) Percentages of fusion mediated by cells gated into high and medium expression levels were normalized to that of the low-expression-level-gated cells for the wild type and for each mutant.
When the fusion efficiencies of cells gated on medium and high EBFP expression levels were compared with those of cells expressing low levels of EBFP, it was found that the fusion mediated by each of the truncation mutants was much more dependent on the level of expression of EBFP (and thus Env) than was that mediated by the wild type (Fig. 8D). This was particularly obvious for TM185, with which cells expressing the highest levels of EBFP yielded approximately fivefold higher levels of fusion than those expressing low levels. In contrast, for the wild-type Env, the difference in fusion efficiencies between cells expressing high and low levels of EGFP was less than twofold. This argues strongly that higher expression levels can partially complement the defects in the fusion competency in mutants.
DISCUSSION
Development of a novel three-color fusion assay based on transient expression.
In this study, we have evaluated in more detail the fusion process mediated by five SIV mutant Env proteins with truncations in the MSD or the cytoplasmic tail. To facilitate these studies, we have developed a novel three-color fusion assay that allows the analysis of transiently transfected cells. Previous dye marker-based studies of fusion phenotypes had used stable cell lines expressing the proteins of interest (31). In that approach, effector cell lines were labeled with the dye CMAC (7-amino-4-chloromethylcoumarin), which is quite immobile and serves as a marker for effector cells, and then target cells were loaded with lipid dye and aqueous dyes (31). One advantage of using stable cell lines is that it permits the use of a homogenous cell population with relatively equivalent expression levels of proteins of interest. However, some viral proteins are quite cytotoxic, making it difficult to establish stable cell lines expressing them, and for studies of multiple mutant proteins, the establishment of cell lines for each can be time consuming. In contrast, the three-color fluorescence assay described here, which is based on transient expression of the fusogenic protein, provides an efficient approach to analyze multiple mutants as well as glycoproteins that are cytopathic and are therefore hard to study in stable cell lines. The coexpression of a fluorescence protein from the same plasmid as the glycoprotein provides a powerful tool to identify effector cells and avoids the nonspecific labeling of effector cells with fluorescence dyes.
In addition, previous studies on membrane fusion, either with fluorescence microscopy or electrophysiology, are limited to the analysis of a small number of fusion events. We have successfully applied the dual-expression system for flow cytometric analyses on a population scale in order to obtain quantitative data. Moreover, reporter gene assays can measure enzyme activities only several hours after coincubation of cells, while the flow cytometric approach described here allows the examination of early fusion events mediated on a population scale.
MSD truncation mutants show decreased fusogenicity but are not blocked at a hemifusion intermediate stage.
We have demonstrated that the fusion efficiencies of different SIV TM mutants decrease in parallel with the truncation of the C terminus of the MSD. Since all these mutants can mediate complete cell-cell fusion, albeit with various efficiencies, it appears that none of the C-terminal amino acids removed are critical for this process. The fact that we were unable to detect a mutant that mediated primarily a hemifusion intermediate contrasts with results of studies on the influenza virus HA (1) and the paramyxovirus SV5 F protein (2). In the case of influenza HA MSD truncation mutants, complete fusion was observed until the MSD was shortened to a length of 16 amino acids (Δ11). For this mutant, content mixing was reduced to 20% of that of the wild type even though lipid mixing remained at 100%. Truncation of an additional amino acid (Δ12) reduced complete fusion to 5% of that of the wild type without affecting lipid mixing. In our studies of the SIV truncation mutants, flow cytometric analyses of fusion with target cells labeled with either calcein-AM (contents) or FAST-DiI (lipid) yielded equivalent results (compare Fig. 4 and 8), arguing that for each of the mutants, lipid transfer and content mixing were affected equivalently. This conclusion was supported by microscopic analyses of the triple-labeled cell mixtures, in which we were unable to observe evidence of cells arrested at the hemifusion state.
There are significant differences between the fusion assays described here and those for the influenza virus HA and SV5 F proteins. For SIV Env-mediated fusion, lipid merging occurs between two monolayer cells, while that described for HA and SV5 F involves monolayer cells and labeled erythrocytes. It is conceivable that the structure of the cytoskeleton and the lipid compositions in monolayer cells impose different constraints on the fusion process. It has been demonstrated in several cases that the lipid composition of the membrane can alter fusion capability (9, 16, 17, 27, 36-38), and the involvement of the cytoskeleton in the fusion process has also been demonstrated by Frey et al. (15), who showed that cytochalasins B and D, which disrupt microfilaments, could reduce gp120-CD4-specific fusion. It is interesting that in studies of murine leukemia virus Env truncation mutants, where 293T and NIH3T3 cells were used as effector and target cells, no hemifusion was found for any of the mutants, including one, Env595*, with truncation extending into the C terminus of the MSD (28). HIV-1 and SIV Env proteins also differ from HA when the MSD is replaced by a GPI anchor. GPI-linked HA mediates hemifusion, while GPI-linked Env proteins are unable to mediate either fusion or hemifusion in the assays described here (reference 41 and our unpublished results).
It is also possible that the process of fusion pore formation by the SIV Env might be different from that of viruses found to efficiently mediate hemifusion intermediates. Melikyan et al. (29) found that HIV Env-induced fusion pores did not flicker and rapidly enlarged after formation. The fusion pores formed by murine leukemia virus TM proteins (28) and by HIV-1 TM proteins (29) are 7 and 16 nm in diameter, respectively, and are thus much larger than those of HA (3 nm) but similar in size to those of gp64 of baculovirus (35) and E1/E2 of Semliki Forest virus (25). It is likely then that the SIV TM will also form large fusion pores, and it is possible that with these retroviral fusion proteins the transition from a transient hemifusion intermediate to an open fusion pore would occur more rapidly. Nevertheless, hemifusion intermediates were observed with HIV-1 Env under conditions in which limiting doses of a peptide fusion inhibitor were present (31), and so it is likely that progression to a fusion pore involves such an intermediate.
The most attractive possibility to explain the differences between the results we have obtained here and those obtained with HA is the topological differences between the MSDs of these two proteins. In the case of the hemifusing HA truncation mutants, Δ11 and Δ12, Armstrong et al. postulated that the shortened MSD might be unable to completely span the membrane bilayer and thus might be defective in resolving the hemifusion intermediate into a fusion pore (1). Consistent with this hypothesis, the addition of HA cytoplasmic domain sequences or even of a single arginine residue to the C terminus of the Δ12 mutant resulted in a protein that mediated complete fusion. A model has been proposed for the SIV MSD in which the basic amino acids at residues 164 and 180, which flank a 14-amino-acid hydrophobic region, would be embedded in the bilayer with their charged side chains extending into the polar head groups (46). By this model, truncation of residues 189 to 185 would affect membrane-proximal residues rather than those critical to anchoring the protein in the membrane. Based on the observations with HA, and because each truncation mutant would still span the membrane, we might predict that each would retain the ability to resolve any hemifusion intermediates that formed. Nevertheless, this conclusion is complicated somewhat by the results obtained with SV5 F protein C-terminal truncation mutants (2). In this case, although the exact boundaries of the MSD are not known, the truncations were predicted to merely shorten the cytoplasmic domain of the glycoprotein. The longest (Δ19) terminated immediately before a (predicted) membrane-proximal lysine, but in this case the mutant mediated efficient lipid mixing in the absence of content mixing. Based on this example, we might have predicted the SIV TM mutants with more extensive truncations to mediate hemifusion.
What role then do the residues between 185 and 191 in the SIV TM protein play in the process of membrane fusion? One possibility is that these amino acids facilitate either directly or indirectly the TM-TM interactions that are necessary for the formation of higher-order oligomers assembled to form the fusion pore. For HA, Blumenthal et al. (5) calculated that six trimers must assemble to form an active fusion pore, and Bentz has recently proposed that this number could be eight or higher (3). Similarly, Kuhmann et al. (24) have proposed that for HIV-1 entry, multiple coreceptors are required, which would suggest the participation of multiple Env proteins in the fusion process. The concentration dependence exhibited by each of the truncation mutants is also consistent with inefficient assembly of higher-order multimeric structures, since it is possible that only under conditions of high Env density can sufficient numbers of functionally competent Env trimers assemble into a nascent fusion complex.
Residues proximal to the membrane could also be involved in the expansion of fusion pores. All the mutants described here have reduced levels of fusogenicity in terms of the size and efficiency of syncytia formation. In addition, cases of small-fusion-pore formation were identified for these mutants, especially between syncytia and neighboring target cells, while none were identified for COS-1 cells expressing wild-type Env or EBFP only. This strongly suggests that the expansion of fusion pores was impaired to different degrees for these mutants. Cells expressing HIV TM proteins with mutations in the tryptophan-rich, N-terminal membrane-proximal region were also reported to show formation of small fusion pores in the absence of complete cell fusion (32). Thus it is possible that the membrane-proximal region of the cytoplasmic tail is also involved in the interaction with membranes or in the destabilization of membranes through interaction with cellular factors when the fusion pore enlarges.
This is not the only viral membrane protein system in which mutants of fusion proteins allow fusion pore formation but interfere with the expansion of the fusion pore. Kozerski et al. (23) have shown that chimeric HA proteins with the cytoplasmic tail of the CD4 molecule were strongly impaired in mediating fusion pore expansion but that membrane lipid merging and fusion pore formation were similar to those of wild-type HA. Interestingly, the GPI-anchored HA protein (26) and an SV5 F mutant protein with a 19-amino-acid truncation in the cytoplasmic tail (12) were recently found to mediate fusion pore formation but reduce the expansion of fusion pores. In contrast, earlier observations had suggested that these mutants arrested fusion at the hemifusion stage (2, 21, 30).
The expansion of fusion pores is a critical step in the fusion process that allows the release of nucleocapsids during viral entry and enables the full exchange of contents in cell-cell fusion. The identification of small-fusion-pore formation by our assay system suggests that this fluorescence assay system may provide a powerful tool to further analyze the detailed process of HIV-mediated membrane fusion and the cellular and viral factors involved.
Acknowledgments
We are grateful to Isabel Munoz-Barroso for training in fusion assays and to John West, who constructed and characterized the TM truncation mutants. We also thank Albert Tousson and Tina Rogers for assistance with and teaching of fluorescence microscopy and flow cytometric analysis and Mike Sakalian, Christina Ochsenbauer, and John West for helpful discussions.
This study was supported by grant AI33319 from the National Institutes of Health and by the UAB Center for AIDS Research Flow Cytometry Core through grant P30 AI27767.
REFERENCES
- 1.Armstrong, R. T., A. S. Kushnir, and J. M. White. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagai, S., and R. A. Lamb. 1996. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J. Cell Biol. 135:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentz, J. 2000. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys. J. 78:227-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal, R., D. P. Sarkar, S. Durell, D. E. Howard, and S. J. Morris. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 7.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 9.Chernomordik, L., E. Leikina, M. S. Cho, and J. Zimmerberg. 1995. Control of baculovirus gp64-induced syncytium formation by membrane lipid composition. J. Virol. 69:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chernomordik, L. V., V. A. Frolov, E. Leikina, P. Bronk, and J. Zimmerberg. 1998. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140:1369-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 95:3425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutch, R. E., and R. A. Lamb. 2001. Deletion of the cytoplasmic tail of the fusion protein of the paramyxovirus simian virus 5 affects fusion pore enlargement. J. Virol. 75:5363-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 14.Edinger, A. L., J. E. Clements, and R. W. Doms. 1999. Chemokine and orphan receptors in HIV-2 and SIV tropism and pathogenesis. Virology 260:211-221. [DOI] [PubMed] [Google Scholar]
- 15.Frey, S., M. Marsh, S. Gunther, A. Pelchen-Matthews, P. Stephens, S. Ortlepp, and T. Stegmann. 1995. Temperature dependence of cell-cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type 1. J. Virol. 69:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunther-Ausborn, S., and T. Stegmann. 1997. How lysophosphatidylcholine inhibits cell-cell fusion mediated by the envelope glycoprotein of human immunodeficiency virus. Virology 235:201-208. [DOI] [PubMed] [Google Scholar]
- 17.Hammache, D., G. Pieroni, N. Yahi, O. Delezay, N. Koch, H. Lafont, C. Tamalet, and J. Fantini. 1998. Specific interaction of HIV-1 and HIV-2 surface envelope glycoproteins with monolayers of galactosylceramide and ganglioside GM3. J. Biol. Chem. 273:7967-7971. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., P. Edmondson, M. Murphey-Corb, B. Arbeille, P. R. Johnson, and J. I. Mullins. 1989. SIV adaptation to human cells. Nature 341:573-574. [DOI] [PubMed] [Google Scholar]
- 19.Hunter, E. 1997. gp41, a multifunctional protein involved in HIV entry and pathogenesis, section III, p. 55-73. In B. Korber, B. Hahn, B. Foley, J. W. Mellors, T. Leitner, G. Myers, F. McCutchan, and C. L. Kuiken (ed.), Human retroviruses and AIDS. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 20.Johnston, P. B., J. W. Dubay, and E. Hunter. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J. Virol. 67:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 22.Kodama, T., D. P. Burns, D. P. Silva, F. D. Veronese, and R. C. Desrosiers. 1991. Strain-specific neutralizing determinant in the transmembrane protein of simian immunodeficiency virus. J. Virol. 65:2010-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozerski, C., E. Ponimaskin, B. Schroth-Diez, M. F. Schmidt, and A. Herrmann. 2000. Modification of the cytoplasmic domain of influenza virus hemagglutinin affects enlargement of the fusion pore. J. Virol. 74:7529-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanzrein, M., N. Kasermann, R. Weingart, and C. Kempf. 1993. Early events of Semliki Forest virus-induced cell-cell fusion. Virology 196:541-547. [DOI] [PubMed] [Google Scholar]
- 26.Markosyan, R. M., F. S. Cohen, and G. B. Melikyan. 2000. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol. Biol. Cell 11:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., S. A. Brener, D. C. Ok, and F. S. Cohen. 1997. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melikyan, G. B., R. M. Markosyan, S. A. Brener, Y. Rozenberg, and F. S. Cohen. 2000. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J. Virol. 74:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 131:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nussler, F., M. J. Clague, and A. Herrmann. 1997. Meta-stability of the hemifusion intermediate induced by glycosylphosphatidylinositol-anchored influenza hemagglutinin. Biophys. J. 73:2280-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plonsky, I., and J. Zimmerberg. 1996. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J. Cell Biol. 135:1831-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puri, A., P. Hug, K. Jernigan, J. Barchi, H. Y. Kim, J. Hamilton, J. Wiels, G. J. Murray, R. O. Brady, and R. Blumenthal. 1998. The neutral glycosphingolipid globotriaosylceramide promotes fusion mediated by a CD4-dependent CXCR4-utilizing HIV type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 95:14435-14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puri, A., P. Hug, K. Jernigan, P. Rose, and R. Blumenthal. 1999. Role of glycosphingolipids in HIV-1 entry: requirement of globotriosylceramide (Gb3) in CD4/CXCR4-dependent fusion. Biosci. Rep. 19:317-325. [DOI] [PubMed] [Google Scholar]
- 38.Puri, A., P. Hug, I. Munoz-Barroso, and R. Blumenthal. 1998. Human erythrocyte glycolipids promote HIV-1 envelope glycoprotein-mediated fusion of CD4+ cells. Biochem. Biophys. Res. Commun. 242:219-225. [DOI] [PubMed] [Google Scholar]
- 39.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabatini, B. D., G. Kreibich, T. Morimoto, and M. Adesnik. 1982. Mechanisms for the incorporation of proteins in membranes and organelles. J. Cell Biol. 92:1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzwedel, K., P. B. Johnston, S. J. Roberts, J. W. Dubay, and E. Hunter. 1993. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 67:5279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stegmann, T., J. M. Delfino, F. M. Richards, and A. Helenius. 1991. The HA2 subunit of influenza hemagglutinin inserts into the target membrane prior to fusion. J. Biol. Chem. 266:18404-18410. [PubMed] [Google Scholar]
- 43.Tsurudome, M., R. Gluck, R. Graf, R. Falchetto, U. Schaller, and J. Brunner. 1992. Lipid interactions of the hemagglutinin HA2 NH2-terminal segment during influenza virus-induced membrane fusion. J. Biol. Chem. 267:20225-20232. [PubMed] [Google Scholar]
- 44.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 45.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 46.West, J. T., P. Johnston, S. R. Dubay, and E. Hunter. 2001. Mutations within the putative membrane-spanning domain of the SIV transmembrane glycoprotein define the minimal requirements for fusion, incorporation and infectivity. J. Virol. 75:9601-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zingler, K., and D. R. Littman. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J. Virol. 67:2824-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]