Abstract
Two members of the signal transducer and activator of transcription family, STAT1 and STAT2, form, together with interferon regulatory factor 9 (IRF-9), the ISGF3 complex that activates the expression of the interferon-stimulated genes (ISG). The ISGF3 complex also participates in the virus-induced alpha/beta interferon (IFN-α/β) gene amplification cascade by up-regulating IRF-7 gene expression. Here, we show that treatment of cells with trichostatin A (TSA), a deacetylase inhibitor, inhibits the virus-induced activation of IFN-α/β promoters and dramatically reduces the ability of different ISG promoters to respond to IFN stimulation. Impairment of IFN-α/β and ISG expression by TSA in infected cells is due to the blockage of interferon-stimulated ISGF3 complex formation, which leads to the abolition of IRF-7 gene expression. We also show that the TSA-dependent inhibition of ISGF3 is related to impaired nuclear accumulation of STAT2. Our data suggest that an acetylation/deacetylation mechanism participates in the regulation of cellular distribution and function of STAT2 in IFN-α/β signaling.
Infection by viruses is in challenge with the antiviral defense mechanisms triggered by different types of interferon (mainly alpha interferon [IFN-α], IFN-β, and IFN-γ) and various proteins encoded by IFN-stimulated genes (ISGs) (16, 28). During viral infection, different members of the interferon regulatory factor (IRF) and signal transducer and activator of transcription (STAT) families participate in the regulation of IFN-α/β genes (encoding different IFN-α subtypes and IFN-β) and ISGs (38, 39). In mice, IFN-β and IFN-α4 are the only members of the IFN-α/β gene family to be transactivated by IRF-3 during the early stage of infection (21, 29). According to a multistage induction model, secreted IFN-β and IFN-α4 proteins bind to their common cellular receptor and activate a specific Jak-STAT pathway. Following receptor activation, the transcription factors STAT1 and STAT2 are phosphorylated by Janus protein tyrosine kinases Jak1 and Tyk2 and released from their docking sites on the receptor (14). They associate with IRF-9 and form the ISGF3 complex, which stimulates IFN-α/β-dependent gene transcription by binding to the IFN-stimulated response element (ISRE) sequences located in the promoter of target genes (34). ISRE sequences are also found in the promoter of IRF-7, and ISGF3 has been shown to activate the IRF-7 gene (18). In the late stage of infection, once translated following stimulation by IFN-α/β and activated by virus-induced phosphorylation, IRF-7 participates in the amplification of IFN-α4 and IFN-β expression and in the transcription of the multigenic IFN-α family members (30). This biphasic regulation, which defines the specific IFN and ISG expression patterns in virus-infected cells and in the neighboring uninfected cells, determines also the extent of cell growth inhibition, impairment of protein synthesis, and initiation of innate and adaptive responses of antiviral immunity in host cells.
Treatment of murine cells by trichostatin A (TSA), an inhibitor of deacetylase activity (20, 23), was suggested to mimic the effect of virus infection for the activation of IFN-β promoter transcription (26, 36). Induction of IFN-β gene transcription by TSA or virus correlates with an increase in the acetylation levels of histone H4 bound to the IFN-β promoter. The effect of TSA was suggested to be essentially mediated by the negative regulatory domain (NRD II) located between −220 and −110 from the start site of transcription and not by factors interacting with the virus-responsive element of the IFN-β promoter. The effect of TSA on the expression of IFN-α4 or other delayed IFN-α genes induced by virus or on ISG transcription is unknown. In the present study, we show that TSA treatment negatively affects the induction of IFN-α genes and ISG expression in virus-induced cells by blocking formation of ISGF3 complex.
TSA affects IFN-α4 and ISG-15 transcription induced by virus.
To test the effect of TSA treatment on the induction of IFN-α and ISG gene expression in virus-induced cells, we performed transient transfections with plasmids carrying the mouse IFN-α4 and IFN-β gene promoters, pIF4T-CAT and pIFNB-CAT, described previously (6), or with a construction containing the ISRE from the ISG-15 gene obtained by insertion of this fragment at the HindIII and BamHI sites upstream of the herpes simplex virus thymidine kinase promoter fused to the CAT gene in pBLCAT2 (19) (Fig. 1A). The first two constructs were shown to respond to IRF-3, alone or together with IRF-7, during virus induction (17, 24). The reporter plasmid containing the ISRE, also responsive to IRF-3, is essentially induced by IFN stimulation via ISGF3 in the late phase of virus induction. L929 cells transfected by DEAE-Dextran, (200 μg/ml per 2 × 105 cells) were treated only with TSA for 40 h; infected at a multiplicity of infection of 5:1 by Newcastle disease virus (NDV) (La Jolla strain) for 24 h; or treated with TSA for 16 h and further infected by NDV for 24 h in the presence of TSA (Fig. 1A, left panel, lanes 1 to 4). We also tested the effect of TSA on virus-stimulated expression of IFN-α4 and ISG15 gene promoters by simultaneous addition of the deacetylase inhibitor during viral infection (lane 5). Infection of transfected cells by NDV led to a more than 100-fold induction of IFN-α4 and IFN-β promoters and to a 10-fold induction of pISRE-tkCAT (Fig. 1A, right panel, lane 3). The 10-fold inducibility is due to high constitutive levels of this construct due to the presence of the tk promoter. TSA treatment reduced considerably the virus inducibility of IFN-α4 and ISG-15 promoters and decreased by twofold the virus-induced transcription of the IFN-β promoter (lane 4). Inhibition of IFN-α/β- and ISRE-dependent transcription is not due to a long-term treatment with TSA, since a similar decrease was observed when cells were simultaneously infected by virus and treated with TSA (lane 5). The severe impairment of IFN-α4 gene promoter activity and ISRE-dependent transcription by TSA during virus infection suggested a defect in the pathways mediated by different IRFs, especially IRF-3 and IRF-7, or the ISGF3 complex, which is specifically involved in the IFNα/β-stimulated transcription. The modest twofold effect of TSA on virus-induced transcription of the IFN-β gene promoter may be explained by the fact that the transcription of this promoter is also dependent on NF-κB- or ATF2/c-Jun-mediated pathways (1), which might be less affected by TSA in the presence of virus.
FIG. 1.
TSA down-regulates virus-induced and IFN-stimulated gene expression. (A) Mouse L929 cells were transfected with 1 μg of pIF4T-CAT, pIFNB-CAT, or pISRE-CAT plasmid carrying the −470 to +19 fragment of the mouse IFN-α4, the −110 to +20 fragment of the mouse IFN-β gene promoter, or the −115 to −89 promoter region of the ISG-15 gene in the presence of 0.25 μg of pRSV-LacZ. Transfected cells were left untreated (NT), treated only with TSA (50 ng/ml) for 40 h (TSA), infected by NDV for 24 h (NDV), or treated with TSA for 16 h and further infected by NDV for 24 h in the presence of TSA (TSA+NDV), as schematized on the left side of the figure. (B) L929 cells were transfected with 1 μg of pISG15-luc, pISG54-luc, or p6-16-luc plasmid carrying the human ISG15, ISG54, or IFI6-16 gene promoters, respectively, in the presence of 0.25 μg of pRSV-LacZ. Transfected cells were left untreated or treated with TSA (100 ng/ml) for 8 h or were stimulated by 1,000 IU of recombinant mouse IFN-α11/ml for 8 h in the absence or presence of TSA. CAT or luciferase values obtained in at least three independent transfection experiments were normalized according to the β-galactosidase levels. Inducibility (Ind. or IFN inducibility) corresponds to the ratio between normalized CAT or luciferase values obtained under virus-induced, IFN-stimulated, or TSA-treated conditions in comparison to mock-induced values.
TSA down-regulates the IFN-stimulated transcription of different ISG promoters.
We tested the effects of TSA treatment on the IFN-α-stimulated transcription of three typical IFN-α/β-inducible genes, ISG15, ISG54, and IFI6-16 (7, 25) by transfection experiments with mouse L929 cells. The pISG54-luc plasmid contains the −423 to +27 fragment of the human ISG54 gene. Construct p6-16-luc (gift from S. Pellegrini) contains the IFI6-16 promoter (1.8 kb) cloned upstream of the luciferase reporter gene. TSA treatment of L929 cells dramatically reduced the capacity of all these promoters to respond to IFN stimulation without affecting their constitutive transcription levels (Fig. 1B). The TSA-dependent decrease observed with ISG15 is underestimated because of the low inducibility (3.3-fold) due to the constitutive levels of the tk promoter. TSA-mediated inhibition of the promoter activation of the IFI6-16 gene, shown to be unresponsive to expression of a constitutively activated IRF-3 in microarray analyses (10), argues in favor of a defect in the ISGF-3 activation pathway rather than a default in IRF-3-dependent transcription. Similar experiments were performed with 2fTGH cells lacking the IRF-7 protein expression due to hypermethylation of the IRF-7 gene promoter (18). The TSA-mediated inhibition of IFN-α2-dependent transcription observed in this case (data not shown) confirmed that the effect of TSA altered the ISGF3 pathway, rather than signaling by IRF-7, and further indicated that this inhibition was not restricted to mouse cell lines.
Impairment of IFN-induced IRF-7 gene transcription by TSA during viral infection.
We then tested if the decrease of virus-induced IFN-α/β and ISG expression by TSA was due to a deficiency in ISGF-3-dependent transactivation stimulated by IFN-α/β during viral infection. Since IRF-7 gene expression was shown to be mediated by ISGF3 (2, 18), we therefore compared endogenous IRF-7 gene expression in virus-infected cells in the presence or absence of TSA. We have also determined the effect of TSA on IRF-3 gene expression, which is shown to be independent of ISGF3 in cells infected by virus or treated with IFN (21, 31). To determine the mRNA expression levels of murine IRF-3 and IRF-7 genes, a 282-bp HindIII-EcoRI antisense fragment and a 522-bp HindIII-BamHI antisense fragment were obtained by PCR amplification of murine IRF-3 and IRF-7 cDNA-containing plasmids, respectively (a kind gift from I. Marié) and cloned into plasmid pcDNA3. [α-32P]UTP-labeled antisense RNA probes were generated, and hybridization and RNase treatments were performed with 5 μg of total RNA, using the RiboQuant RPA kit (Pharmingen). Protected fragments were quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), mouse GAPDH expression levels being used as an internal control. RNase protection experiments were performed with L929 cells that were left untreated, treated with TSA for 24 h, infected with NDV for 8 h, or treated with TSA for 16 h and further infected with NDV for 8 h in the presence of freshly added TSA.
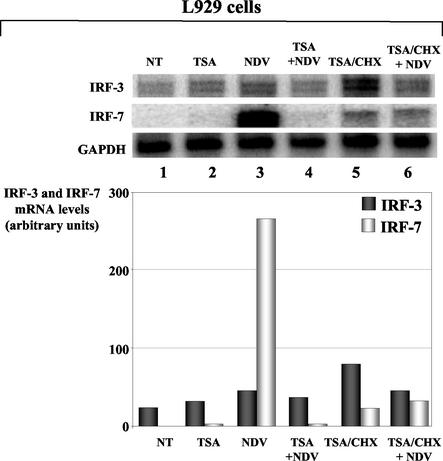
With a probe specific to mouse IRF-3 mRNA, we detected two IRF-3 transcripts in untreated cells, with the upper band corresponding to the full-length mRNA of the IRF-3 gene according to its size (Fig. 2, lane 1). The transcript of smaller size could correspond to the mouse homolog of the human IRF-3a isoform (13), although such a spliced form has not yet been described for mice. Quantification of the IRF-3 mRNA performed by taking into account the full-length form or both forms of transcripts and normalized relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA did not show any variation of IRF-3 gene expression by TSA treatment in mock- or virus-infected cells (lanes 2 and 4). Nor have we observed significant variations of IRF-3 gene expression when cells were cotreated with TSA and cycloheximide (CHX) in the presence or absence of viral infection (lanes 5 and 6).
FIG. 2.
Constitutive and virus-inducible expression of mouse IRF-3 and IRF-7 genes in TSA-treated L929 cells. L929 cells pretreated (conditions 2 and 4) or not (conditions 1 and 3) with TSA were left untreated (conditions 1 and 2) or infected by NDV for 1 h (conditions 3 and 4) and cultured in 5% serum for 7 h. For experiments performed in the absence of de novo protein synthesis, CHX was added during TSA pretreatment (conditions 5 and 6). Transcripts from IRF-3, IRF-7, and GAPDH genes, detected by the RNase protection assay (upper panels) and quantified by a PhosphorImager, are presented in arbitrary units relative to GAPDH mRNA levels (graph).
Following virus infection of L929 cells, a single band corresponding to the full-length IRF-7 mRNA is detected with a probe specific for mouse IRF-7. Quantification of the detected signal revealed a more than 200-fold increase of mouse IRF-7 gene expression levels (Fig. 2, lanes 1 and 3), comparable to those detected in HeLa cells or mouse embryonic fibroblasts by other groups (3, 21). Strikingly, treatment of L929 cells with TSA completely abolished the IFN-induced expression of the IRF-7 gene during viral infection (compare lanes 3 and 4). TSA impairment of NDV-induced IRF-7 gene expression was independent of de novo protein synthesis, since CHX treatment of cells in the presence of TSA did not restore the IRF-7 gene induction following virus infection (lane 6). The residual expression observed in this case was similar to the low levels of IRF-7 transcripts detected in the control cells treated with TSA in the presence of CHX (lane 5). Similar results obtained with murine NIH 3T3 cells indicated that the TSA-mediated inhibition of virus-induced IRF-7 gene expression was not restricted to L929 cells and that this inhibition was maintained from 1 to 16 h of infection (data not shown).
ISGF3 complex formation is abolished by TSA.
The ISGF3 complex that is formed and activated following IFN-α/β stimulation binds on and activates the interferon-stimulated responsive elements identified in the human or murine IRF-7 gene promoter (18, 31). Since TSA inhibited IFN-α/β-induced IRF-7 gene expression in virus-infected cells, we examined whether formation of the ISGF3 complex was impaired by TSA. Electrophoretic mobility shift assays (EMSA) were performed as previously described (9), using the high-affinity 5′-(GAAACC)4-3′ sequence for ISGF3 as the DNA probe. Nuclear and cytoplasmic extracts, prepared according to the method of Dignam et al. (8), were obtained from L929 cells infected by NDV for 8 h in the absence and in the presence of TSA. The extracts were preincubated for 1 h on ice with anti-STAT2 (sc-950X; Santa Cruz Biotechnology, Santa Cruz, Calif.) or with polyclonal antisera at a 1:10 dilution for the human STAT1 or IRF-9/ISGF3γ proteins (32, 40) before the addition of the labeled probe.
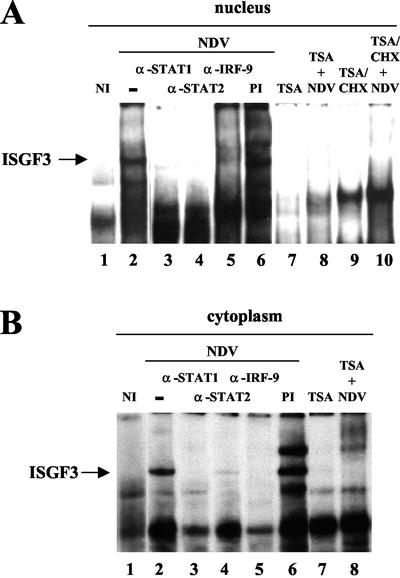
As expected, NDV induction of L929 cells led to the formation and binding of the ISGF3 complex (Fig. 3A, lanes 1 and 2) that reacted with STAT1, STAT2, and IRF-9 antibodies but not with preimmune serum (lanes 3 to 6). Pretreatment with TSA did not significantly affect the EMSA pattern of uninfected cells (lane 7), but it completely inhibited ISGF3 binding activity induced by IFN in virus-infected cells (lane 8). Addition of CHX during the TSA pretreatment did not prevent inhibition of IFN-induced ISGF3 complex formation, indicating that this inhibition does not require de novo protein synthesis (lane 10). Similar EMSA taken out with cytoplasmic extracts revealed that the ISGF3 complex was barely detected in the cytoplasm of virus-infected cells and was not detected when cells were pretreated with TSA (Fig. 3B, lanes 1 to 6 and 8). This result indicated that TSA also prevented ISGF3 complex assembly in the cytoplasm. By using these different extracts, we have also shown that the virus-inducible binding of IRF-3 or the formation of GAF (IFN-γ-activated factor) consisting of activated STAT1 homodimers was not affected by TSA treatment of virus-infected L929 cells (data not shown). These results suggested that TSA specifically inhibited ISGF3 complex formation in IRF-mediated signaling.
FIG. 3.
Effect of TSA on ISGF3 complex formation. EMSA was performed with nuclear (A) or cytoplasmic (B) extracts from L929 cells left untreated (lane 1), infected by NDV (lanes 2 to 6), or treated with TSA (lane 7) or in cells pretreated with TSA and infected by NDV (lane 8). For de novo protein synthesis inhibition experiments, cycloheximide was added on TSA-treated cells (lane 9) or on TSA-treated and virus-infected cells (lane 10). The ISGF3 complex, indicated by an arrow, reacted with antibodies directed against STAT1, STAT2, and IRF-9/p48 (lanes 3 to 5), whereas preimmune antiserum did not affect its formation (lane 6).
TSA treatment inhibits nuclear accumulation of STAT2.
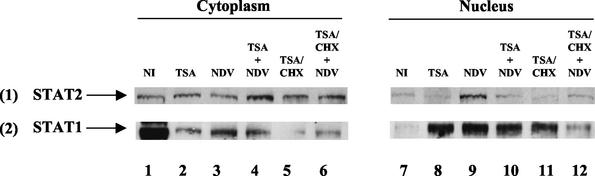
The kinetics of nuclear import of STAT1 and STAT2 has been correlated with the stimulated DNA-binding activity of ISGF3, and STAT1-STAT2 heterodimer formation has been shown to be determinant for ISGF3 complex formation (22). We therefore examined the protein expression patterns of STAT1 and STAT2 in both nuclear and cytoplasmic extracts from NDV-infected cells pretreated or not with TSA in order to identify the ISGF3 component affected by TSA treatment. Western blot analysis performed using standard procedures (11), with 40 μg of nuclear or cytoplasmic extracts and anti-STAT2 or anti-STAT1 antibodies (sc-950X and sc-417X, respectively; Santa Cruz Biotechnology) at a 1:5,000 dilution, allowed detection of STAT2 protein in the cytoplasm of uninfected, TSA-treated and NDV-infected cells (Fig. 4, row 1, columns 1 to 3). Consistent with nuclear accumulation following cell stimulation, a nuclear pool of STAT2 was clearly detected only after virus infection (row 1, columns 7 to 9). Interestingly, STAT2 was barely detected in the nuclear extracts of infected cells pretreated with TSA (column 10), and the STAT2 signal obtained in this case was similar to that obtained under uninduced conditions (column 7). These observations indicated that TSA severely impaired or completely blocked the IFN-induced nuclear accumulation of STAT2 in virus-infected L929 cells. Accordingly, the amount of the cytoplasmic pool of STAT2 was higher in virus-infected cells treated with TSA than in NDV-infected cells, in agreement with an accumulation of the STAT2 cytoplasmic pool (columns 3 and 4). Consistent with the absence of ISGF3 formation when infected cells were treated with TSA and CHX, inhibition of de novo protein synthesis did not affect the TSA-mediated inhibition of STAT2 nuclear accumulation in infected cells (Fig. 4, row 1, compare columns 6 and 12 with 5 and 11, respectively).
FIG. 4.
Effect of TSA on the nuclear-cytoplasmic localization of STAT1 and STAT2. STAT1 (row 1) and STAT2 (row 2) proteins, components of the ISGF3 complex, were visualized by Western blotting in cytoplasmic (columns 1 to 6) and nuclear (columns 7 to 12) extracts obtained from L929 cells left untreated (columns 1 and 7), treated with TSA (columns 2 and 8), or infected by NDV in the absence (columns 3 and 9) or in the presence (columns 4 and 10) of TSA. For de novo protein synthesis inhibition experiments, cycloheximide was added on TSA-treated cells (columns 5 and 11) or on TSA-treated and virus-infected cells (columns 6 and 12).
STAT1 protein that was detected in the cytoplasm but not in the nucleus of uninfected cells (Fig. 4, row 2, columns 1 and 7) translocated to the nucleus following virus infection (row 2, columns 3 and 9). In contrast to STAT2 blockage, TSA pretreatment of cells did not affect IFN-induced STAT1 nuclear accumulation in infected cells (compare columns 4 and 10 to 3 and 9, respectively). Interestingly, TSA treatment alone promoted STAT1 nuclear accumulation in uninfected cells (columns 7 and 8) in contrast to STAT2 (row 1, columns 7 and 8), suggesting that TSA differentially affected STAT1 and STAT2 subcellular localization. The mechanism of the TSA effect on constitutive STAT1 protein remains unclear. However, we concluded that the TSA-mediated inhibition of ISGF3 formation in virus-infected cells, responsible for the impairment of NDV-induced IRF-7 gene transcription, was due to the blockage of virus-induced STAT2 nuclear accumulation.
TSA-dependent impairment of STAT2 nuclear accumulation may be related to STAT2 acetylation.
Impairment of IFN-α/β gene expression in virus-induced cells treated by TSA and down-regulation of different ISG promoters in mouse and human cells stimulated with exogenous IFN-α in the presence of TSA are due to a defect in ISGF3 complex formation, especially caused by a blockage of STAT2 nuclear accumulation. Our finding is based on the following: (i) ISGF3 is not detected in the nucleus and cytoplasm of infected cells pretreated with TSA; (ii) STAT2 protein is very poorly detected in the nuclear extracts of virus-infected cells pretreated with TSA, in comparison to large amounts of nuclear pool of STAT2 observed following virus infection; (iii) higher amounts of STAT2 are present when cytoplasmic extracts from virus-infected L929 cells are pretreated with TSA; (iv) in contrast, TSA treatment of infected cells does not affect STAT1 nuclear transport, indicating that inhibition of ISGF3 complex formation by TSA relies rather on the inability of STAT2 to accumulate in the nucleus.
ISGF3 transcriptional activity is essentially regulated by the active shuttling of ISGF3 components between cellular compartments and its ability to communicate with the RNA polymerase II enzyme complexes and coactivators of transcription. Yet little is known about how STAT1 and STAT2 proteins enter the nucleus and return to the cytoplasm once the transient transcriptional response to IFN has subsided (15). Several hypotheses have been proposed concerning ISGF3 complex formation. Thus, STAT2 has been proposed to bring along IRF-9 to the IFNAR1 chain of IFN-α/β receptor for oligomerization with STAT1 (15). Alternatively, formation of phosphorylated STAT1-STAT2 heterodimers at the receptor intracellular domain has been proposed to constitute the first step in ISGF3 formation (14). Upon release from the receptor, STAT1 and STAT2 heterodimers become competent for nuclear import and join IRF-9 at the ISRE-containing promoters. However, according to these sequential recruitment models of ISGF3 components, ISGF3 is predominantly located in the nucleus. Our results suggest that STAT2-IRF-9 or STAT1-STAT2 complexes, proposed as the ready protein pools for rapid ISGF3 complex assembly, might lose their ability to recruit the third association partner in the presence of TSA. Interestingly, EMSA presented in this study show that TSA inhibits the formation of nuclear ISGF3 but also the small amounts of ISGF3 detectable in the cytoplasm.
The common feature of the different models concerning the ISGF3 activation pathway resides in the phosphorylation of STAT1 and STAT2 on tyrosines 701 and 690, respectively, upon receptor activation by IFN-α/β. Within minutes after ligand-dependent tyrosine phosphorylation and dimerization, STAT1 and STAT2 migrate to the nucleus, accumulate during 1 to 2 h, and are exported to the cytoplasm within 3 h (22, 33). In the IFN-γ signaling pathway, STAT1 homodimers are recycled within hours of their inactivation by a yet-unknown nuclear phosphatase (4, 12). Although this model has not yet been extended to IFN-α/β signaling, STAT2 protein is suggested to exhibit a similar activation-inactivation cycle following IFN-α treatment, suggesting that the recycling of STAT2 to the cytoplasm is an essential component of IFN-α/β signaling (15). TSA-mediated inhibition of STAT2 nuclear accumulation presented in this study suggests the existence of a posttranslational mechanism that regulates its subcellular localization and the transcriptional activity of ISGF3. Actually, we show that CHX neither prevents the inhibition of ISGF3 formation by TSA nor restores virus-mediated nuclear accumulation of STAT2 in the presence of TSA. This indicates that TSA-dependent inhibition of STAT2 nuclear accumulation involves posttranslational modifications rather than de novo protein synthesis. TSA reversibly inhibits histone deacetylases, altering the acetylation/deacetylation equilibrium, leading to hyperacetylation of the chromatin (41). In addition, TSA inhibits the deacetylase activities targeting various transcription factors by altering their transcriptional activity (37). Since TSA affects STAT2 nuclear-cytoplasmic distribution, it is tempting to speculate that STAT2 might be directly targeted by acetylation/deacetylation activities. The fact that STAT1 and STAT2 recruit the histone acetyltransferase CBP, p300, or GCN5-TAFII130 complex via their carboxyl-terminal transactivation domains suggested that STAT1 and STAT2 may be substrates for nuclear acetylation (5, 27, 42). This modification may regulate the duration of ISGF3-mediated transcription. In the case of STAT6 protein, acetylation by CBP/p300 is required for its transcriptional activity induced by interleukin 4 (35). Our results suggest that acetylation of STAT2 may lead to impaired nuclear accumulation of STAT2 and down-regulation of ISGF3 complex formation.
Acknowledgments
We thank Isabelle Marié for plasmids containing murine cDNA for IRF-3 and IRF-7. We are also indebted to Peter Howley and Tom Maniatis for providing us with monoclonal anti-IRF3 SL12 antibodies.
This work was supported by the Centre National de la Recherche Scientifique, the Université Paris V, and grants from the Association pour la Recherche sur le Cancer (Contrat 5828) and from the Ligue Régionale contre le Cancer (Contrat 75/01-RS/44).
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Au, W.-C., P. A. Moore, W. Lowther, Y.-T. Juang, and P. M. Pitha. 1995. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc. Natl. Acad. Sci. USA 92:11657-11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au, W. C., P. A. Moore, D. W. LaFleur, B. Tombal, and P. M. Pitha. 1998. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J. Biol. Chem. 273:29210-29217. [DOI] [PubMed] [Google Scholar]
- 4.Begitt, A., T. Meyer, M. van Rossum, and U. Vinkemeier. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 97:10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP by interferon-α. Nature 383:344-347. [DOI] [PubMed] [Google Scholar]
- 6.Bragança, J., P. Génin, M.-T. Bandu, N. Darracq, M. Vignal, C. Cassé, J. Doly, and A. Civas. 1997. Synergism between multiple virus-induced-factor-binding elements involved in the differential expression of IFN-A genes. J. Biol. Chem. 272:22154-22162. [DOI] [PubMed] [Google Scholar]
- 7.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genin, P., J. Braganca, N. Darracq, J. Doly, and A. Civas. 1995. A novel PRD I and TG binding activity involved in virus-induced transcription of IFN-A genes. Nucleic Acids Res. 23:5055-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 12.Haspel, R. L., and J. E. Darnell, Jr. 1999. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. USA 96:10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpova, A. Y., L. V. Ronco, and P. M. Howley. 2001. Functional characterization of interferon regulatory factor 3a (IRF-3a), an alternative splice isoform of IRF-3. Mol. Cell. Biol. 21:4169-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kisseleva, T., S. Bhattacharya, J. Braunstein, and C. W. Schindler. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1-24. [DOI] [PubMed] [Google Scholar]
- 15.Lau, J. F., and C. M. Horvath. 2002. Mechanisms of type I interferon cell signaling and STAT-mediated transcriptional responses. Mt. Sinai J. Med. 69:156-168. [PubMed] [Google Scholar]
- 16.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 17.Lin, R., P. Genin, Y. Mamane, and J. Hiscott. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 20:6342-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, R., W. C. Au, W. S. Yeow, N. Hageman, and P. M. Pitha. 2000. Regulation of the promoter activity of interferon regulatory factor-7 gene. Activation by interferon and silencing by hypermethylation. J. Biol. Chem. 275:31805-31812. [DOI] [PubMed] [Google Scholar]
- 19.Luckow, B., and G. Schutz. 1987. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acid Res. 15:5490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 21.Marié, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melen, K., L. Kinnunen, and I. Julkunen. 2001. Arginine/lysine-rich structural element is involved in interferon-induced nuclear import of STATs. J. Biol. Chem. 276:16447-16455. [DOI] [PubMed] [Google Scholar]
- 23.Minucci, S., V. Horn, N. Bhattacharyya, V. Russanova, V. V. Ogryzko, L. Gabriele, B. H. Howard, and K. Ozato. 1997. A histone deacetylase inhibitor potentiates retinoid receptor action in embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 94:11295-11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin, P., J. Bragança, M.-T. Bandu, R. Lin, J. Hiscott, J. Doly, and A. Civas. 2002. Preferential binding sites for interferon regulatory factors 3 and 7 involved in interferon-A gene transcription. J. Mol. Biol. 316:1009-1022. [DOI] [PubMed] [Google Scholar]
- 25.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 26.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 27.Paulson, M., C. Press, E. Smith, N. Tanese, and D. E. Levy. 2002. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140-147. [DOI] [PubMed] [Google Scholar]
- 28.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 30.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 31.Sato, M., N. Tanaka, N. Hata, E. Oda, and T. Taniguchi. 1998. Involvement of IRF family transcription factor IRF-3 in virus-induced activation of IFN-β gene. FEBS Lett. 425:112-116. [DOI] [PubMed] [Google Scholar]
- 32.Schindler, C., X. Y. Fu, T. Improta, R. Aebersold, and J. E. Darnell, Jr. 1992. Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. USA 89:7836-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindler, C., K. Shuai, V. R. Prezioso, and J. E. Darnell. 1992. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257:809-813. [DOI] [PubMed] [Google Scholar]
- 34.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 35.Shankaranarayanan, P., P. Chaitidis, H. Kuhn, and S. Nigam. 2001. Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. J. Biol. Chem. 276:42753-42760. [DOI] [PubMed] [Google Scholar]
- 36.Shestakova, E., M. T. Bandu, J. Doly, and E. Bonnefoy. 2001. Inhibition of histone deacetylation induces constitutive derepression of the beta interferon promoter and confers antiviral activity. J. Virol. 75:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda, K., and S. Akira. 2000. STAT family of transcription factors in cytokine-mediated biological responses. Cytokine Growth Factor Rev. 11:199-207. [DOI] [PubMed] [Google Scholar]
- 39.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 40.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Sato, K. Ozato, and T. Fujita. 1996. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3). J. Biochem. 120:160-169. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 42.Zhang, J. J., U. Vinkemeier, W. Gu, D. Chakravarti, C. M. Horvath, and J. E. Darnell. 1996. Two contact regions between STAT1 and CBP/p300 in interferon gamma signalling. Proc. Natl. Acad. Sci. USA 93:15092-15096. [DOI] [PMC free article] [PubMed] [Google Scholar]