Abstract
In this study, we investigate the role of the apex nucleotides of the two turns found in the intramolecular “paperclip” type triplex DNA formed by 5′-TCTCTCCTCTCTAGAGAG-3′. Our previously published structure calculations show that residues C7-A18 form a hairpin turn via Watson-Crick basepairing and residues T1-C6 bind into the major groove of the hairpin via Hoogsteen basepairing resulting in a broad turn of the T1-T12 5′-pyrimidine section of the DNA. We find that only the C6C7/G18 apex triad (and not the T12A13/T1 apex triad) is required for intramolecular triplex formation, is base independent, and occurs whether the purine section is located at the 5′ or 3′ end of the sequence. NMR spectroscopy and molecular dynamics simulations are used to investigate a bimolecular complex (which retains only the C6C7/G18 apex) in which a pyrimidine strand 5′- TCTCTCCTCTCT-3′ makes a broad fold stabilized by the purine strand 5′-AGAGAG-3′ via Watson Crick pairing to the T8-T12 and Hoogsteen basepairing to T1-T5 of the pyrimidine strand. Interestingly, this investigation shows that this 5′-AGAGAG-3′ oligo acts as a new kind of triplex forming oligonucleotide, and adds to the growing number of triplex forming oligonucleotides that may prove useful as therapeutic agents.
INTRODUCTION
DNA has the ability to adopt a wide variety of single-stranded and multi-stranded conformations including hairpin, triplex, and quadraplex configurations (1). The appearance of triplex structures was first reported in RNA by Felsenfeld et al. in 1957 (2). Triplex DNA is formed through the binding of a third DNA strand into the major groove of a double helix via Hoogsteen hydrogen bonding to the purine residues (3–11). The ability of triplex structure formation in cell nuclei has been demonstrated (12). In addition, a number of studies have demonstrated the ability of triplex forming oligonucleotides (TFOs) to inhibit transcription or translation in vitro as well as elicit a variety of responses in vivo, including mutagenesis and recombination, as well as inhibition of transcription of specifically targeted genes (13–20). The sequence specificity of TFO binding makes triplex formation an attractive tool for both gene therapy and molecular biology (21–23).
The thorough understanding of the structural features of triplex DNA is a critical piece in our overall understanding of the biological functions and the potential therapeutic uses of triplex-forming DNA. A number of triplex structures have been investigated by NMR spectroscopy (5,24–36). More recently, studies revealed that the sequence 5′-TCTCTCCTCTCTAGAGAG-3′ (ccag) was able to form an intramolecular triple helix in natural or low pH solutions by folding into a “paperclip” structure with no loop residues (Fig. 1 A) (24,25). This paperclip formation can be thought of as being comprised of two turns: 1), residues C7-A18 form a hairpin turn via Watson-Crick basepairing (with T12 and A13 at the apex of the turn) and, 2), residues T1-C6 fold back and bind into the major groove of the hairpin via Hoogsteen basepairing resulting in a broad turn of the 5′ pyrimidine section (with C6 and C7 at the apex of the turn). In these studies, nuclear Overhauser effect (NOE)-based structure calculations of ccag resulted in a structure that suggested the G18 at the 3′ end of the oligomer forms hydrogen bonding with the C6 and C7 apex residues, resulting in a distorted triad (CC/G) at the apex of the pyrimidine turn (pypy turn) (24). This NOE-based structure calculation, however, was not able to clearly identify if the T1 at the 5′ end of the oligomer forms hydrogen bonding with the T12 and A13 residues at the apex of the hairpin turn, leading us to question the role of the first pyrimidine residue in the triad formation with T12 and A13 nucleotides.
FIGURE 1.
(A) Paperclip configuration of 5′-TCTCTCCTCTCTAGAGAG-3′ (ccag), (B) 5′-CTCTCTTCTCTCGAGAGA-3′ (ttga), (C) 5′-GAGAGATCTCTCCTCTCT-3′ (gacc), (D) 5′-AGAGAGCTCTCTTCTCTC-3′ (agtt), and (E) bimolecular complex of 5′-TCTCTCCTCTCT-3′ (cc) and 5′-AGAGAG-3′ (ag). (|) Indicates Watson Crick basepairing and (*) indicates Hoogsteen basepairing.
In this study, we investigate the effect of modifications of the oligomer ccag on triplex formation to establish the role of the pypy/pu and pypu/py triads at the apexes of the paperclip triplex. These studies included the removal of either the first pyrimidine residue T, or the last purine residue G from ccag, and rearrangement of the position of the pyrimidine rich or purine rich section in the sequence. In addition, this study investigates the triplex formation of a pyrimidine 12mer 5′-TCTCTCCTCTCT-3′ (cc) with a separate purine 6mer 5′-AGAGAG-3′ (ag), a triplex arrangement that retains only the pypy/pu apex triad (Fig. 1 E).
From the result of this study, we observed a similar triplex formation of the oligomer sequence without the first thymine residue as for ccag, under similar experimental conditions. But when the last guanine residue was removed from the ccag, no triplex formation is detected. Triplex formation was also observed for the mixture of cc and ag under similar condition as ccag (Fig. 2 E). The result of NOE-based structure calculations combined with molecular dynamics simulations showed similar triad formation of CC/G at the apex of the pyrimidine turn. These observations indicate that the pypy/pu triad at the apex of the pyrimidine turn is essential for triplex formation, and a pypu/py triad at the apex of the hairpin turn is not required for triplex formation.
FIGURE 2.
(A) CD spectra of ccag (solid), ccag-t (dashed), and ccag-g (dotted) at pH 4.5, 25°C. (B) CD spectra of ttga (solid), ttga-c (dashed) and ttga-a (dotted) at pH 4.5, 25°C. (C) CD spectra of gacc (solid), gacc-g (dashed) and gacc-t (dotted) at pH 4.5, 25°C. (D) CD spectra of agtt (solid), agtt-a (dashed) and agtt-c (dotted) at pH 4.5, 25°C. (E) CD spectra of ccag (solid), 6ag +12cc (dashed), and 6ag+12tt (dotted), at pH 4.5, 5°C.
Interestingly, ag constitutes a new kind of TFO, a poly-purine sequence (TFOpur) that essentially bends single stranded pyrimidine rich sequences and stabilizes the bend by forming Watson-Crick basepairing to one arm and Hoogsteen basepairing to the other arm of the pyrimidine-rich sequence. This configuration looks remarkably like naturally occurring DNA structures such as H-DNA, which demonstrate similar formations (37). Thus, our increasing knowledge of the turn regions of triplex DNA provides insight into naturally occurring DNA structures and adds to the growing number of TFOs that may prove useful as therapeutic agents targeting specific pyrimidine sequences.
MATERIALS AND METHODS
Synthesis of oligodeoxyribonucleotides
All oligodeoxyribonucleotides were purchased commercially (TriLink Technologies, San Diego, CA) with double reverse phase high-performance liquid chromatography purification. All purchased oligodeoxyribonucleotides showed a single peak in an analytical Hypersil BDS-C18 column (Agilent Technologies, Santa Clara, CA) high-performance liquid chromatography. Thus, these oligodeoxyribonucleotides, listed in Table 1, were used directly without further purification.
TABLE 1.
List of the oligodeoxyribonucleotides, their symbols used in the article, and extinction coefficients
| Symbol | Oligodeoxyribonucleotides (18 mers) | Extinction coefficient |
|---|---|---|
| ccag | 5′-TCTCTCCTCTCTAGAGAG | 165,680 |
| gacc | 5′-GAGAGATCTCTCCTCTCT | 165,220 |
| ttga | 5′-CTCTCTTCTCTCGAGAGA | 165,420 |
| agtt | 5′-AGAGAGCTCTCTTCTCTC | 164,760 |
| (AG)9 | 5′-AGAGAGAGAGAGAGAGAG | 216,940 |
| Oligodeoxynucleotides (17 mers) | ||
| ccag-t | 5′-CTCTCCTCTCTAGAGAG | 156,980 |
| ccag-g | 5′-TCTCTCCTCTCTAGAGA | 155,440 |
| gacc-g | 5′-AGAGATCTCTCCTCTCT | 154,720 |
| gacc-t | 5′-GAGAGATCTCTCCTCTC | 157,500 |
| ttga-c | 5′-TCTCTTCTCTCGAGAGA | 158,800 |
| ttga-a | 5′-CTCTCTTCTCTCGAGAG | 151,740 |
| agtt-a | 5′-GAGAGCTCTCTTCTCTC | 151,340 |
| agtt-c | 5′-AGAGAGCTCTCTTCTCT | 157,160 |
| Oligodeoxynucleotides (others) | ||
| 12cc | 5′-TCTCTCCTCTCT | 92,740 |
| 12tt | 5′-CTCTCTTCTCTC | 92,720 |
| 6ag | 5′-AGAGAG-3′ | 73,420 |
| (AG)18 | 5′-AGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAG-3′ | 432,220 |
Experimental conditions
The concentrations of all oligodeoxyribonucleotides were determined by the ultraviolet (UV) molar extinction coefficients at 260 nm based on the Fasman method (Table 1) (38). Buffer solutions contain 100 mM NaCl with 20 mM acetate (pH 4.5-5) or 20 mM phosphate (pH 6.5–8) except when otherwise stated. The concentration of single-stranded oligodeoxyribonucleotides used for spectroscopic methods are in 4 μM and 2 μM of each component used in the 1:1 mixtures. However, the concentrations of 6ag and 12cc for NMR were 3 mM. The concentrations of oligodeoxyribonucleotides for gel were 2 μM.
CD spectroscopy
Circular dichroism (CD) spectra were obtained on a Jasco-720 CD spectropolarimeter (Jasco, Tokyo, Japan). The temperature was controlled by a water-circulated jacketed cell. Dry nitrogen gas was blown through the cell compartment to expel oxygen and to prevent moisture condensation. Cells with 1 cm path lengths were used.
UV thermal melting experiment
The UV absorbance versus temperature profiles were measured by a Jasco V-560 UV/VIS and Varian (Palo Alto, CA) Cary 3E spectrophotometer at 260 nm. The temperature was controlled by a Peltier-type thermostatic cell holder. The UV melting curves were recorded with heating and cooling rates of 0.5 C/min. Cells with 1 and 0.1 cm path lengths were used.
Native gel electrophoresis
The gel concentration is 20% polyacrylamide and run in solution contains 100 mM NaCl, 5 mM MgCl2 with 20 mM acetic buffer, pH 5, at 25°C. The voltage is 200 V for 3 h.
NMR spectroscopy
A 1:1 mixture of cc and ag was made using 0.5 ml of 90% H2O/10% D2O or 100% D2O containing 100 mM NaCl, 50 μM EDTA, and 20 mM phosphate buffer, pH 6.0 or pH 4.5.
NMR data were collected on Bruker Avance-500, DRX-600, or Varian Unity+ 600 spectrometers. All data were recorded in phase-sensitive mode. The discriminated time domain free induction decays (fids) of two-dimensional experiments performed on Bruker spectrometers were recorded in TPPI (39) format and further processed to the readable frequency domain spectra by XWIN-NMR (version 2.6, Bruker BioSpin, Karlsruhe, Germany). The fids performed on Varian spectrometer were recorded in hypercomplex (40) format and were processed by FELIX (version 2000, Accelrys, San Diego, CA).
One-dimensional jump-return spectra, for suppressing the intense H2O signal of samples in 90% H2O, were acquired at 10°C, 20°C, 30°C, 40°C, and 50°C on a Bruker Avance-500 spectrometer. The spectral width was set to 13 kHz. The carrier frequency was positioned on the H2O resonance. The τ-delay was set to be 55 μs, thus optimizing maximum excitation in the Hoogsteen hydrogen-bond imino proton region around 14.5 ppm. The fid was apodized with Gaussian window function (−0.3 Hz line broadening and 0.03 Gaussian broadening) before Fourier transformation. The residual solvent signal of all jump-return related experiments was suppressed by applying a low pass digital filter (41).
All phase sensitive DQF-COSY (42) experiments were performed on a Bruker Avance-500 spectrometer at 35°C with a 4006.4 Hz spectral width. T1-increments of 512 were collected and 64 scans were accumulated for each fid. Fids in both dimensions were apodized with 45°-shifted sine square window function and the processed matrix size was 2048 × 2048 real points.
Phase-sensitive D2O-NOESY experiments and Clean TOCSY (43) were performed on Bruker Avance-500 and DRX-600 spectrometers with 4006.4 and 4595.6 Hz spectral width, respectively. For assignment purposes, a D2O-NOESY spectrum with 240 ms mixing time was performed on a 600 MHz spectrometer using the same acquisition parameters as those performed on a 500 MHz spectrometer. The fids were apodized with a 45°-shifted sine square window function and the processed matrix size was 2048 × 2048 real points. A series of D2O-NOESY spectra with various mixing times (60, 120, 240, and 360 ms) were collected at 500 MHz with 512 t1-increments at 35°C and 64 scans. The fids were apodized with 90°-shifted sine square window function and the processed matrix sizes were 2048 × 2048 real points.
The H2O signal in NOESY spectra of samples in H2O was suppressed by jump-return and spectra were acquired on a 500 MHz spectrometer with spectral width 11574.1 Hz. The mixing time was set to be 100 ms and 60 ms at 10°C, 20°C, respectively. Spectra were acquired with 128 scans, 700 t1-increments, and 1024 TPPI complex data points. For assignment purpose, the fids were apodized with a 45°-shifted sine square window function. For NOE intensity measurement, the fids were apodized with 90°-shifted sine square window function. Both processed matrix sizes were 2048 × 2048 in real points.
Resonance assignments were accomplished by the NOE-observable 1H-1H distances between neighboring nucleotides in the sequence (44-47). Nonexchangeable proton assignment was carried out on D2O-NOESY spectra. Before starting sequential assignment, the correlations of cytosine-H5 to cytosine-H6 and thymine-CH3 to thymine-H6 were first characterized by a DQF-COSY spectrum. Assignment of all H5, H6, and CH3 chemical shifts of the pyrimidine strand can then be traced by the NOE connectivity between T(n)CH3↔T(n)H6, T(n)H6↔C(n+1)H5, C(n+1)H5↔C(n+1)H6, C(n+1)H6↔T(n+2)CH3, T(n+2)CH3↔T(n+2)H6. Then the analysis of the entire H1′-H6/H8 region of the NOESY spectrum was accomplished. The other chemical shifts of H2′, H2″, H3′, H4′, H5′, H5″ are obtained by their correlation to the H6/H8 resonances. Chemical shift assignment of exchangeable protons was carried out by H2O-NOESY spectrum. Resonance of imino protons can be sequentially assigned by the imino-imino NOE connectivity. In this complex, imino-imino connectivity can be traced between purine-imino and pyrimidine-imino of the Watson-Crick pairings, and sequentially between the pyrimidine-imino protons of the Hoogsteen pairings (27). The imino protons can also be identified by their correlations with intra- or interresidual amino protons. For cytosine, amino protons can be also assigned by the NOE correlations with intrabase H5 and H6 protons (46).
Constraint determination
For distance constraints of nonexchangeable proton pairs, the NOE intensities were compared to NOE of cytosine H5↔H6 and divided into three levels, strong (2.0–4.0 Å), medium (3.0–5.0 Å), and weak (4.0–6.0 Å). For distance constraints of the imino protons, the NOE intensities were compared to the NOE of the intrabase NOE of cytosine imino to amino proton (set to be strong) and loosely divided into strong (2.0–5.0 Å) and weak (3.0–6.0 Å). Thymine and guanine imino proton-related NOE intensities were classified by the same method, except the standard NOE intensity used was the interbase NOE of imino to the hydrogen bonded amino proton of the paired base.
Molecular dynamics simulation
The structure was determined by using Distance-Geometry (DGII, Accelrys) calculations followed by restrained molecular dynamics (MD) simulation (Discover, Accelrys). A template was established that contains two independent B-form DNA single strands. The N3 proton of cytosines participating in Hoogsteen basepairing was added and all carbon and nitrogen atoms of these cytosines were set to sp2 hybridized orbitals. Besides the NOE constraints, some distance constraints between proton-donor and proton acceptor of Watson-Crick and Hoogsteen basepairs were also added to the triads of the stem part. Thus a total of 22 distance constraints, 251 NOE-distance constraints, 88 dihedral angle constraints, and 55 chiral constraints were applied during the distance-geometry calculation and restrained MD simulation with force constants of 32 kcal for distance constraints and 100 kcal for dihedral constraints. Before the DGII calculation, the template molecules were energy minimized for 200 steps with steepest descents. DGII calculations resulted in 20 initial structures for MD simulation. Molecules were heated to 27°C and dynamics resumed for 10 ps. Finally, molecules were energy minimized again for 2000 steps by using conjugate gradient method and 200 steps of steepest descent. All calculations were performed on a Silicon Graphics O2 workstation with DGII and Discover programs utilizing the AMBER force field. Helical parameters of cc+ag and ccag were analyzed using the program Curves5.3 (48). Only minor variations in helical parameter values were found between the two structures. Values of basepair parameters reflect that the orientation of the bases of the pyrimidine turn is similar in both structures.
RESULTS
A complete pypy/pu apex triad is required for a paper-clip type triplex
CD and UV melting data were compared for three oligomers: 5′-TCTCTCCTCTCTAGAGAG-3′ (ccag), 5′-CTCTCCTCTCTAGAGAG-3′ (ccag-t), and 5′-TCTCTCCTCTCTAGAGA-3′ (ccag-g). Fig. 2 A shows the CD spectra of these three oligomers in acidic condition (pH 4.5). The CD spectra of ccag and ccag-t have troughs at 210 nm, which are characteristic of triplex formation. The fact that ccag-t (missing the T1 at the 5′ end of the oligomer) is still able to form a triplex indicates that the 5′-T is not required for triplex formation and, therefore, a complete TA/T (a pypu/py apex turn) is not required for triplex formation. This further supports the original model of ccag, which indicated that the T1 at the 5′ end of the oligomer may not be involved in triad formation (24). In the CD spectrum of ccag-t at pH 8 (data not shown), the trough at 210 nm disappears and a trough at 240nm (typical of B-DNA) was observed. On the other hand, the CD spectrum of ccag-g at pH 4.5 (as well as at pH 7) has a trough at 240 nm only, typical of B-DNA, not a triplex as the other two oligomers. This suggests that the G18 at the 3′ end of the oligomer is involved in the triplex formation and a complete CC/G is required to form the apex bending site.
The stability of ccag, ccag-t, and ccag-g has been studied by UV thermal melting temperature (Tm) experiments (Table 2). The similar high Tms of ccag (54.0°C) and ccag-t (59.0°C) support the conclusion that a triplex forms with or without the T1 at the 5′ end of the oligomer. The lower Tm of ccag-g (41.9°C) reflects duplex formation, consistent with the trough at 240 nm in its CD spectrum. Tm measurements were also taken of ccag-t and ccag-g at 10-fold excess and found to be 59°C and 49°C, respectively. This concentration-dependent change in Tm of 7.1°C for ccag-g indicates that ccag-g is a bimolecular complex consistent with B-DNA, whereas the lack of any such change for ccag-t indicates a unimolecular complex consistent with the paperclip type triplex model.
TABLE 2.
Melting temperatures of all DNAs (°C)
| Oligo | pH = 4.5 | pH = 7 | |
|---|---|---|---|
| ccag | 54.0 | 22.0 | 56.3 |
| ccag-t | 59.0 | 26.7 | 58.8 |
| ccag-g | 41.9 | 44.2 | |
| ttga | 61.5 | 24.2 | 56.0 |
| ttga-c | 57.6 | 28.7 | 53.5 |
| ttga-a | 39.8 | 48.4 | |
| gacc | 53.0 | 11.7 | 48.7 |
| gacc-t | 53.1 | 13.8 | 46.1 |
| gacc-g | 36.0 | 44.0 | |
| agtt | 54.2 | 18.4 | 52.3 |
| agtt-c | 45.1 | 20.9 | 50.4 |
| agtt-a | 42.5 | 46.5 | |
Requirement for a complete pypy/pu is independent of base identity
The three oligomers 5′-CTCTCTTCTCTCGAGAGA-3′ (ttga) (Fig. 1 B), 5′-TCTCTTCTCTCGAGAGA-3′ (ttga-c), and 5′-CTCTCTTCTCTCGAGAG-3′ (ttga-a) were analyzed by CD and UV melting curves. These DNA oligomers are similar to the ccag, ccag-t, and ccag-g set of DNAs, except that the pyrimidine bases and the purine bases are switched. The pypy/pu turn is now TT/A and the pypu/py turn is now CG/C. CD spectra of ttga at pH 4.5 show the characteristic triplex trough at 210nm (Fig. 2 B). The removal of a single base at either end of the ttga (ttga-c and ttga-a) resulted in the same profile as for the removal of either end basepair of ccag. The DNA ttga-c was able to form triplex, indicating that the pypu/py complete apex triad is not required for triplex formation. However, ttga-a was not able to form a triplex, indicating that the complete pypy/pu apex triad is required for triplex formation. The melting temperatures of ttga, ttga-c, and ttga-a (Table 2) follow the same trend as the melting temperatures of ccag, ccag-t, and ccag-g. These data indicate that both TT/A or CC/G may act as the pypy/pu apex nucleotides. In all cases under this investigation, the complete pypy/pu triad is necessary for triplex formation, and the complete pypu/py triad is not. In addition, it is interesting to point out that the melting temperature of ttga is higher than that of ccag. This is consistent with that the stability of CGC+ is higher than that of TAT (9).
Triplex formation is independent of the location of the CC/G pypy/pu turn
The DNA oligomers 5′-GAGAGATCTCTCCTCTCT-3′ (gacc) (Fig. 1 C), 5′-GAGAGATCTCTCCTCTC-3′ (gacc-t), and 5′-AGAGATCTCTCCTCTCT-3′ (gacc-g) were analyzed by CD and UV melting curves. These DNA oligomers are the same as the ccag, ccag-t, and ccag-g set of DNAs except that the sense of the strand is reversed thus moving the purine section to the 5′ end of the DNA. This places the CC turn toward the 3′ end of the DNA. CD spectra of gacc at pH 4.5 show the characteristic triplex trough at 210nm (Fig. 2 C). In addition, the removal of either end of gacc (gacc-t and gacc-g) resulted in the same profile as for the removal of either end basepair of ccag. The DNA gacc-t was able to form triplex, indicating that the complete pypu/py apex triad is not required for triplex formation. However, gacc-g was not able to form a triplex, indicating that the complete pypy/pu apex triad is required for triplex formation. The melting temperatures of gacc, gacc-t, and gacc-g (Table 2) follow the same trend as the melting temperatures of ccag, ccag-t, and ccag-g. In addition, this experiment was repeated with the DNA oligomers 5′-AGAGAGCTCTCTTCTCTC-3′ (agtt) (Fig. 1 D), 5′-AGAGAGCTCTCTTCTCT-3′ (agtt-c), and 5′-GAGAGCTCTCTTCTCTC-3′ (agtt-a) (Fig. 2 D and Table 2) with similar results indicating that base identity is not important. These data indicate that the pypy/pu turn can be located at the 5′ or 3′ end of the DNA, that either base can be present, but that the complete pypy/pu is necessary for triplex formation.
Triplexes are more stable than duplexes
The average Tm of all triplexes and duplexes at pH 4.5 was compared. The average Tm of the triplexes (ccag, gacc, ttga, agtt, ccag-t, gacc-t, ttga-c, and agtt-c) is 53.1°C and of the duplexes (ccag-g, gacc-g, ttga-a, and agtt-a) is 42.6°C. The difference in average Tm of triplexes to duplexes is 10.5°C. This supports the previously suggested conclusion that the triplex is not only a unique molecule but also more stable at slight acidic conditions (49).
Formation of a bimolecular triplex cc+ag
To evaluate the degree to which the pypu turn of the pypu/py apex residues is necessary for triplex formation, two DNA oligomers were designed that mimic ccag but are discontinuous between T12 and A13. This resulted in the DNA oligomers 5′-TCTCTCCTCTCT-3′ (cc) and 5′-AGAGAG-3′ (ag). The CD spectrum of a 1:1 mixture of cc and ag (cc+ag) is very similar to the CD profile of ccag under similar conditions and thus indicates the presence of triplex formation (Fig. 2 E). Similar results are seen when the bimolecular complex is formed by a 1:1 mixture of 5′-CTCTCTTCTCTC-3′ (tt) and 5′-AGAGAG-3′ (ag) (tt+ag) at pH 4.5 (Fig. 2 E). The CD spectrum of cc+ag shows B-DNA features as the pH is increased to 7 reflecting the transition from triplex to duplex (data not shown).
The Tms of cc+ag at pH 4.5 were 12°C and 50°C. The lower Tm of 12°C is due to a weak association of (12cc)2 via C+C basepairing (50). The Tm of 50°C is typical of triplex formation.
Gel retardation studies
Native gel electrophoresis was performed to verify that the triplex formation seen in the CD data of cc+ag and tt+ag was due to 1:1 complexes of the DNA oligomers instead of association of three linear strands. The native gel electrophoresis of ag (I), cc (II), cc+ag (III), tt+ag (IV), (AG)9 (V), and (AG)18 (VI) are shown in Fig. 3. The single strands ag (I), cc (II), (AG)9 (V), and (AG)18(VI) were used as markers of 6, 12, 18, and 36 nucleotidyl-unit oligomers, respectively. Both cc+ag and tt+ag have similar mobility to the (AG)9, indicating that they are a 1:1 complex resulting in an 18 nucleotide complex. No higher order species were detected.
FIGURE 3.
Native gel electrophoresis of ag (column I from left), cc (II), cc+ag (III), tt+ag (IV), (AG)9 (V), and (AG)18 (VI). The gel concentration was 20% polyacrylamide, in 100 mM NaCl, 5 mM MgCl2 with 20 mM acetic buffer, pH 5, at 25°C. The voltage was 200 V for 3 h.
NMR assignment and conformation of cc+ag
The one-dimensional 1H NMR spectrum of cc+ag in H2O at pH 6.0 was performed on a 500 MHz spectrometer. Fig. 4 shows the resonances of the hydrogen-bonded imino proton region from 10°C to 40°C. Triplex structure formation is evident in all spectra. Besides the resonance of all hydrogen-bonded imino protons of thymines and guanines (12.5 ppm–14.5 ppm), there are three additional signals between 14.5 ppm and 16 ppm. These three signals disappeared when the pH was increased to 8, indicating that they are the imino protons of the protonated cytosines, C3H+, that participate in Hoogsteen hydrogen-bonding under acid buffer conditions. Such one-dimensional-1H NMR resonances are indicative of triplex formation (34–36).
FIGURE 4.
Downfield region of the 1H NMR spectra of cc+ag at 500 MHz.
Expanded regions of the NOESY spectrum of cc+ag in D2O at pH 6.0 acquired at 600 MHz with a 240 ms mixing time are shown in Fig. 5. Assignment was accomplished with the sequential assignment method as described in the Materials and Methods section. We can identify the sequential connectivities T(n)CH3↔T(n)H6↔C(n+1)H5↔C(n+1)H6↔T(n+2)CH3 of the pyrimidine strand, connectivities of pyrimidine-H1′/H3′↔ H6 and purine-H1′/H3′↔H8. This then allows for assignment of the resonance of the nonexchangeable protons. Full assignment is available in Table 3.
FIGURE 5.
Expanded regions of the NOESY spectrum of cc+ag in D2O at pH 6.0, 35°C acquired at 600 MHz with a 240 ms mixing time. (a) A3H8↔T3H1′, (b) G4H8↔C4H1′, (c) A5H8↔T5H1′, (d) G2H8↔T3H1′, (e) A3H8↔C4H1′, (f) G4H8↔T5H1′, and (g) A5H8↔C6H1′.
TABLE 3.
Proton chemical shifts (ppm) of (AG)3/(TC)3(CT)3 at 293 K
| H1′ | H2′ | H2′′ | H3′ | H4′ | H5′/H5′′ | H6/H8 | H2/H3/CH3 | NH2 | NH | |
|---|---|---|---|---|---|---|---|---|---|---|
| 5′-T1 | 6.16 | 2.56 | 2.38 | 4.79 | 4.19 | 3.83/3.79 | 7.63 | 1.75 | 13.62* | |
| C2 | 6.12 | 2.71 | 2.24 | 4.77 | 4.31 | 4.23/4.15 | 8.05 | 5.96 | 9.06/9.85 | 15.66 |
| T3 | 6.27 | 2.63 | 2.41 | 4.90 | 4.32 | 4.21/4.16 | 7.71 | 1.72 | 13.61 | |
| C4 | 5.87 | 2.63 | 2.19 | 4.51 | 4.36 | 4.39/4.19 | 7.67 | 5.67 | 9.14/9.86 | 14.89 |
| T5 | 6.33 | 2.64 | 2.32 | 4.91 | 4.37 | 4.12/4.18 | 7.74 | 1.67 | 13.37 | |
| C6 | 5.87 | 2.53 | 2.08 | 4.75 | 4.40 | 4.31/4.19 | 7.57 | 5.78 | 8.16/9.79 | 14.66 |
| C7 | 6.14 | 2.53 | 2.14 | 4.88 | 4.20 | 4.13/4.02 | 7.65 | 5.53 | 7.42/8.24 | |
| T8 | 5.93 | 2.57 | 2.27 | 4.87 | 4.15 | 4.21/4.12 | 7.45 | 1.66 | 14.07 | |
| C9 | 5.97 | 2.54 | 2.07 | 4.66 | 4.15 | (4.14)/4.22 | 7.51 | 5.48 | 7.14/8.19 | |
| T10 | 5.97 | 2.55 | 2.51 | 4.86 | 4.15 | 4.08/(4.15) | 7.57 | 1.71 | 14.20 | |
| C11 | 6.07 | 2.52 | 2.11 | 4.69 | 4.16 | 4.17/4.10 | 7.55 | 5.57 | 7.07/8.25 | |
| T12-3′ | 6.16 | 2.21 | 2.21 | 4.50 | 3.96 | 4.14/4.02 | 7.41 | 1.59 | 14.15* | |
| 5′-A1 | 5.96 | 2.69 | 2.56 | 4.91 | 4.20 | 3.66/3.63 | 7.65 | 8.01 | ||
| G2 | 6.03 | 2.88 | 2.53 | 5.02 | 4.29 | 4.12/4.05 | 7.52 | 12.73 | ||
| A3 | 5.83 | 2.76 | 1.94 | 4.75 | 4.56 | 3.99/4.36 | 7.08 | 7.47 | 7.32/7.75 | |
| G4 | 5.91 | 2.84 | 2.36 | 4.87 | 4.29 | 4.19/4.05 | 7.33 | 12.69 | ||
| A5 | 5.71 | 2.66 | 1.87 | 4.65 | 4.50 | 4.32/3.91 | 7.00 | 7.38 | 7.72/7.35 | |
| G6-3′ | 5.85 | 2.47 | 2.33 | 4.49 | 4.09 | 4.15/4.02 | 7.54 | 12.85 |
Measured at 283 K.
Several NOE correlations typical of triple-strand DNA structures (27,51–54) are present, including interstrand NOE correlations G2H8↔T3H1′, A3H8↔C4H1′, G4H8↔T5H1′, A5H8↔C6H1′, A3H8↔T3H1′, G4H8↔C4H1′, and A5H8↔T5H1′ (crosspeaks d, e, f, g, a, b, and c on Fig.5). In addition, that the CH3 of thymines and H5 of cytosines were found to have NOE correlations with H6 of their 5′ neighboring base indicates that the triplex adopts a right-handed helical structure.
The expanded imino-imino proton region of the NOESY spectrum of cc+ag in H2O at pH 6.0 acquired at 600 MHz at 20°C with a 120 ms mixing time is shown in Fig. 6. The cross-strand NOE correlation of imino protons between the purine-rich strand and the 3′ end of the pyrimidine strand can be traced, typical of B-form DNA. This indicates that the two antiparallel strands adopt Watson-Crick basepairing. The sequential connectivity from T1 to 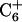 indicates that the 5′ region of the pyrimidine strand is paired to the purine strand by Hoogsteen hydrogen bonds. All chemical shifts of imino protons but T12 have been assigned and are listed in Table 3.
indicates that the 5′ region of the pyrimidine strand is paired to the purine strand by Hoogsteen hydrogen bonds. All chemical shifts of imino protons but T12 have been assigned and are listed in Table 3.
FIGURE 6.
Imino-imino proton region of the NOESY spectrum of cc+ag in H2O at pH 6.0 acquired at 600 MHz at 20°C with a 120 ms mixing time. The detail descriptions are in the text.
Fig. 7 shows the expanded regions of the exchangeable protons of the NOESY spectrum of cc+ag in H2O at pH 6.0 acquired at 500 MHz at 20°C with 60 ms mixing time. Fig. 7 shows the imino protons of thymines have strong NOE correlations with the amino protons of the adenines within the paired triad, e.g., T8/5H3↔A5NH2 (peaks h1, h2, l2, and l3) and T10/3H3↔A3NH2 (peaks f1, f2, j2, and j3). The imino protons on the 3′ end of the pyrimidine strand also have NOEs with adenine H2, e.g., T10H3↔A3H2 and T8H3↔A5H2 (peaks f3 and h3) and the imino protons on the 5′ end of the pyrimidine strand also have NOEs with adenine H8, e.g. T3H3↔A3H8, T5H3↔A5H8 (peaks j1 and l1). The imino protons of guanines have strong NOE correlations with the amino protons of cytosines on the 3′ end of cc, e.g., G2H1↔C11NH2, G4H1↔C9NH2 and G6H1↔C7NH2 (peaks p1, p2, r2, r3, m2, and m3) and weak NOE intensities with protonated cytosine on the 5′-end of cc, e.g., G4H1↔C4NH2 and G6H1↔C6NH2 (peaks o and u). The imino proton of protonated cytosines on the 5′ end of cc have NOE with guanine H8, C4H3↔G4H8, and C6H3↔G6H8 (peaks c and d). Such NOE patterns reveal that the pyrimidines of the 5′ end of cc adopt Hoogsteen basepairing with the purines of ag, and that the pyrimidines on the 3′ end of cc adopt Watson-Crick basepairing with the purines of ag.
FIGURE 7.
Exchangeable proton region of NOESY spectrum of cc+ag in H2O at pH 6.0 acquired at 500 MHz at 20°C with 60ms mixing time. (a) C4H3↔A3H8, (b1 and b2) C4H3↔A5NH2, (c) C4H3↔G4H8, (d) C6H3↔G6H8, (el) T10H3↔C11H5, (e2 and e3) T10H3↔C11NH2, (f1 and f2) T10H3↔A3NH2, (f3) T10H3↔A3H2, (gl) T8H3↔C9H5, (g2 and g3) T8H3↔C9NH2, (h1 and h2) T8H3↔A5NH2, (h3) T8H3↔A5H2, (il) T3H3↔C4H5, (i2 and i3) T3H3↔C4NH2, (j1) T3H3↔A3H8, (j2 and j3) T3H3↔A3NH2, (kl) T5H3↔C6H5, (k2 and k3) T5H3↔C6NH2, (l1) T5H3↔A5H8, (l2 and l3) T5H3↔A5NH2, (m1) G6H1↔C7H5, (m2 and m3) G6H1↔C7NH2, (n) G6H1↔C6H5, (o) G4H1↔C4NH2, (p1 and p2) G2H1↔C11NH2, (q) G4H1↔A5H2, (r1) G4H1↔C9H5, (r2 and r3) G4H1↔C9NH2, (s) G4H1↔C4H5, (t) G2H1↔A3H2, and (u) G6H1↔C6NH2.
Intrastrand weak intensity NOE correlations of each thymine imino protons to the H5 protons of its 3′ neighboring cytosine, e.g., T10H3↔C11H5, T8H3↔C9H5, T5H3↔C6H5, T3H3↔C4H5 (peaks el, gl, kl, and il) and the intrastrand weak intensity NOE correlations of each imino proton on the 5′ end of cc (but not C6) to H2′ and H2″ of it's 5′ neighboring base are further evidence of a right-handed triple helical structure.
Additional interstrand and interbase medium intensity NOE correlations, C4H3↔A5NH2, G2H1↔A3H2, G4H1↔A5H2 (peaks b1, b2, t, and q) and T10H3↔C11NH2, T8H3↔C9NH2, T3H3↔C4NH2, T5H3↔C6NH2 (peaks e2, e3, g2, g3, i2, i3, k2, and k3) provide more information about the triplex structure.
In addition to the gel retardation studies, a number of unusual chemical shifts and NOE correlations indicate that triplex formation of cc+ag is composed of a single bent pyrimidine strand and a single purine strand instead of three linear strands. First of all, the appearance of several NOEs, T5CH3↔C7NH2, T5CH3↔C9NH2, 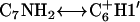 , and
, and 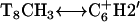 , are not expected in linear DNA. Second, the chemical shift of the nonhydrogen bonded amino proton of
, are not expected in linear DNA. Second, the chemical shift of the nonhydrogen bonded amino proton of 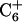 shifts up field by ∼1 ppm relative to those of
shifts up field by ∼1 ppm relative to those of 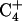 and
and 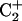 . Third, the chemical shift difference of the two amino protons on C7 is ∼0.9 ppm, whereas those of
. Third, the chemical shift difference of the two amino protons on C7 is ∼0.9 ppm, whereas those of  and
and 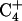 are ∼1.2 ppm (typical of linear DNA). Fourth, there is no NOE between
are ∼1.2 ppm (typical of linear DNA). Fourth, there is no NOE between 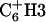 and the amino proton of its 3′ neighboring paired base, an NOE that is typically found in linear DNA. Fifth, the intensity of intrabase NOE correlation,
and the amino proton of its 3′ neighboring paired base, an NOE that is typically found in linear DNA. Fifth, the intensity of intrabase NOE correlation, 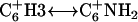 is weaker than
is weaker than  and may be caused by increased chemical exchange between the exposed exchangeable protons of
and may be caused by increased chemical exchange between the exposed exchangeable protons of 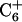 and solvent.
and solvent.
Structure analysis
The NOE distance constraint data were mainly extracted from D2O-NOESY spectra with various mixing times. Due to high chemical exchange rate of the exchangeable protons with H2O at 35°C, NOE distance constraints were extracted from H2O-NOESY performed at 20°C and bounded loosely at 2–6 Å. Triplex structure is apparent from 10°C to 40°C (Fig. 4).
The total relevant constraints used in structural refinement are summarized in Table 4. DGII embedding calculation gave 30 initial structures for molecular dynamics simulation. The initial structure determined by NMR data revealed the Watson-Crick and Hoogsteen basepairing of the stem region. Additional calculations included Watson-Crick and Hoogsteen hydrogen bonding constraints for the triads on the stem region. Fig. 8 A shows the superposition of the 10 lowest energy structures with an average heavy atom root mean-square deviation of 0.58 (±0.05) Å.
TABLE 4.
Input and results of structure calculations
| Calculation included: | |
|---|---|
| Total NOE constraints | 298 |
| Intranucleotide* | 75 |
| Sequential internucleotide | 126 |
| Nonsequential internucleotide | 11 |
| Interstrand | 64 |
| Hydrogen bonds | 22 |
| Violations of experimentally determined distance constraints (>0.6Å) | 1 |
| All atom root mean-square deviation of the 10 lowest energy structures | 0.58 (±0.05) Å |
Trivial intranucleotide sugar NOE distances were not included.
FIGURE 8.
(A) Stereo view of the superimposition of the 10 low energy structures of cc+ag. (B) Stereo view of cc+ag.
In the final model (Fig. 8 B), the 3′ end of the pyrimidine strand and the purine strand are antiparallel to each other and form Watson-Crick basepairing. Simultaneously, the 5′ end of the pyrimidine strand is positioned in the major groove formed by the 3′ end of the pyrimidine strand with the purine strand and parallel to the purine strand. All bases on the 5′ end of the pyrimidine strand pair to the purine bases with Hoogsteen hydrogen bonds except the C6. Residue C6 shows hydrogen bonding potential between C6H3 and 3′-G6O6, and C6NH2 and 3′-G6N7. The C7 base of the pyrimidine strand and 3′-G6 of the purine strand incline toward each other, resulting in potential hydrogen bonding between C7NH2 and 3′-G6O6, C7N3 and 3′-G6H1, and C7O2 and 3′-G6NH2, typical of Watson-Crick hydrogen bonding. No hydrogen bond constraints between C6 and 3′-G6 or C7 and 3′-G6 were applied during the molecular modeling calculations.
Comparison of the final structure of cc+ag and the previously determined structure of ccag (24) shows remarkable similarity in the pypy/pu region. NOE data from both ccag and cc+ag resulted in pypy/pu turn structures in which the C6, C7 and T8 bases stack onto one another and similar potential hydrogen bonding patterns exist between C6, C7 and the 3′-G6 (3′-G18 in ccag) residues.
DISCUSSION
Recent studies revealed that the DNA sequence 5′-TCTCTCCTCTCTAGAGAG-3′ (ccag) forms an intramolecular triple helix at natural or low pH by folding over into a paperclip structure (Fig. 1 A) (24,25). This paperclip formation can be thought of as being comprised of two turns: 1), residues C7-A18 form a hairpin (pypu turn) turn via Watson-Crick basepairing, and 2), residues T1-C6 fold back and bind into the major groove of the hairpin via Hoogsteen basepairing, resulting in a broad turn (pypy turn) of the T1-T12 pyrimidine section.
Previously, NOE-based structure calculations of ccag suggested that the G18 at the 3′ end of the oligomer forms hydrogen bonding with the C6 and C7 residues of the pypy turn, resulting in a distorted triad involving C6, C7, and G18 (CC/G) at the apex of the pyrimidine turn (24). The UV and Tm results indicate that removal of the G18 nucleotide at the 3′ end of the oligomer results in loss of triplex formation, indicating that the G18 at the 3′ end of the oligomer is critical for triplex formation (24). It remained unclear, however, the degree to which the T1 at the 5′ end of the oligomer forms hydrogen bonding with the T12 and A13 residues at the apex of the hairpin turn. When a DNA oligomer was designed that eliminated the T1 at the 5′ end of the oligomer (ccag-t), triplex formation occurred similar to ccag, indicating that the T1 at the 5′ end of the oligomer was not required for triplex formation; therefore, a complete pypu/py triad at the apex of the hairpin turn was not required.
In this study, we evaluate what role the triads at the apex of these two turns plays in the formation of this intramolecular paperclip DNA. A number of DNAs were designed to test which apex nucleotides were necessary for triplex formation, if it mattered what pyrimidines or purines were present at the pypy/pu or pypu/py turns, or if it mattered where in the sequences these turns appeared. We find that the presence of a complete pypy/pu triad (the distorted triad at the apex of the pyrimidine fold) being necessary for intramolecular triplex formation and a complete pypu/py (the triad at the apex of the hairpin turn) not being necessary, is a general profile. Experiments in which the pyrimidines were switched such that the apex triad of the pypy/pu turn was now TT/A and the purines were switched such that the apex of the pypu/py turn was CG/C (ttga) displayed the above profile; when the purine which would complete the pypy/pu turn was removed (ttga-a) triplex formation was not achieved, but when the pyrimidine that would complete the pypu/py turn was removed (ttga-c) triplex formation occurred. To find out if it mattered where in the sequence the pypy/pu triad appears, we designed another DNA by reversing the sense of the sequence, thus placing the purine section at the 5′ end of the DNA (gacc and agtt). These DNA do indeed form pH dependent paperclip type triplexes and show the same pattern of behavior when the 5′ or 3′ nucleotide is not present; when the purine that would complete the pypy/pu turn was removed (gacc-g and agtt-a) triplex formation was not achieved, but when the pyrimidine that would complete the pypu/py turn was removed (gacc-t and agtt-c), triplex formation occurred.
In addition, comparison of the Tm values obtained for all DNA oligomers at pH 4.5 revealed that triplex formation is more stable than duplex formation in these DNA oligomers. The average Tm for triplex forming DNA oligomers was found to be 53.1°C, whereas the average Tm of duplex forming DNA oligomers in this study was found to be 42.6°C. This agrees with previous findings that the triplex is not only a unique molecule but also, at slightly acidic conditions, more stable than duplex structures (49).
As discussed above, we find that a complete pypu/py (triad at the apex of the hairpin turn) is not necessary for triplex formation. Removal of the 5′ nucleotide of ccag required to complete the pypu/py triad fails to inhibit triplex formation. Indeed it is possible to completely disrupt the pypu/py by breaking the DNA between T12 and A13 and still achieve triplex formation. This creates a bimolecular complex (verified by gel electrophoresis to be a 1:1 complex) of 5′-TCTCTCCTCTCT-3′ (cc) and 5′-AGAGAG-3′ (ag). Similar results were seen when the bimolecular complex is formed by a 1:1 mixture of 5′-CTCTCTTCTCTC-3′ (tt) and 5′-AGAGAG-3′ (ag) (Fig. 3). Thus, in general, it appears as if the requirement for the paperclip′ triplex formation is only that a complete pypy/pu triad is formed.
Previous examples of bimolecular triplex formation have been reported; however, these complexes have been of polypyrimidine/polypurine hairpin DNA with an additional pyrimidine strand binding into the major groove of the hairpin via Hoogsteen hydrogen bonding to the poly-purine residues (55). In the case of cc+ag and tt+ag, the palindromic pyrimidine strand is not capable of hairpin formation on its own. Thus triplex formation would require both arms of the pyrimidine strand to form hydrogen bonding with the purine strand, essentially bending the pyrimidine strand. The three-dimensional structure for the bimolecular triplex cc+ag determined herein supports this model. The structure of cc+ag was determined based on data obtained from two-dimensional nuclear magnetic resonance studies and molecular dynamics simulations. We found that ag binds parallel to the 5′-end of cc with Hoogsteen hydrogen bonding and antiparallel to the 3′-end of cc with Watson-Crick hydrogen bonding. The backbone at the triplex stem is similar to that of B-form DNA. The backbone and base stacking at the pypy bend of the cc+ag structure allows C6, C7 and T8 to stack and C6 and C7 to form hydrogen bonding to 3′-G6 of the purine strand in a similar fashion as seen in ccag (24) (Fig. 8 B). Inspection of the overall structure shows that the presence of the 3′-G6 of the pypy/pu triad at the apex of the pyrimidine turn results in an extensive hydrogen bonding network with C6 and C7, thus suggesting that stability of the bases associated with the apex of the pyrimidine turn is critical for triplex formation.
Interestingly, this cc+ag formation adds to a growing number of TFOs. In general, TFOs have been defined as being poly-pyrimidine oligonucleotides that bind in the major groove of duplex DNA by making Hoogsteen basepairing to the purine strand (Fig. 9 A). TFOs have sparked considerable interest recently due to their potential use in sequence-specific targeting of DNA for both basic research and antigene therapy applications (21-23). Indeed, a number of studies have demonstrated the ability of TFOs to inhibit transcription or translation (13–20). Since a TFO binds to a poly-purine strand, these studies have used TFOs that target double-stranded poly-purine/poly-pyrimidine. In a desire to widen the range of potential DNA targets, considerable interest has developed toward designing TFOs that will bind poly-pyrimidine sequences (19,56–58). The 6mer ag of the cc+ag bimolecular complex defined in this study constitutes a poly-pyrimidine binding TFO; ag is a TFOpur, capable of binding a single stranded pyrimidine rich stretch of DNA, bending it and forming both the Watson-Crick and Hoogsteen basepairing necessary to stabilize triplex formation (Fig. 9 B).
FIGURE 9.
(A) Traditional TFO binds in the major groove of duplex DNA forming Hoogsteen basepairing, (B) TFOpur bends pyrimidine rich regions forming W-C pairing to one arm of pyrimidine section and Hoogsteen pairing to the other arm, and (C) H-DNA.
In addition, this cc+ag structure bears great similarity to naturally occurring intramolecular triplex structures first found in plasmids and called H-DNA (in which a pyrimidine section of double-stranded DNA folds back and binds into the major groove of adjacent DNA via Hoogsteen basepairing to purine residues) (Fig. 9 C) (37).
Thus, additional insights developed through the further understanding of these paperclip and bimolecular triplex structures add to our increasing knowledge of naturally occurring DNA structures and poly-pyrimidine targeting TFOs that will, no doubt, prove useful for gene therapy and molecular biology applications.
Acknowledgments
Thanks are due to the High-Field Biomacromolecular NMR Core Facility, which is supported by the National Research Program for Genomic Medicine and Academia Sinica, Taiwan, ROC, for assistance of the NMR observations. This work is partially supported by the Academia Sinica and National Science Council, The Executive Yuan of Taiwan (92-2113-M-001-030). The Scripps Research Institute Manuscript number is 17812-CH.
This article is dedicated to deceased colleague Yu-Yu Tseng.
References
- 1.Gilbert, D. E., and J. Feigon. 1999. Multistranded DNA structures. Curr. Opin. Struct. Biol. 9:305–314. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld, G., D. R. Davies, and A. Rich. 1957. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 79:2023–2024. [Google Scholar]
- 3.Frank-Kamenetskii, M. D., and S. M. Mirkin. 1995. Triplex DNA structures. Annu. Rev. Biochem. 64:65–95. [DOI] [PubMed] [Google Scholar]
- 4.Callahan, D. E., T. L. Trapane, P. S. Miller, P. O. P. Ts'o, and L.-S. Kan. 1991. Comparative circular dichroism and fluorescence studies of oligodeoxyribonucleotide and oligodeoxyribonucleoside methylphosphonate pyrimidine strands in duplex and triplex formation. Biochemistry. 30:1650–1655. [DOI] [PubMed] [Google Scholar]
- 5.Kan, L.-S., D. E. Callahan, T. L. Trapane, P. S. Miller, P. O. P. Ts'o, and D. H. Huang. 1991. Proton NMR and optical spectroscopy studies on the DNA triplex formed by d-A(G-A)7-A and d-C-(T-C)7-T. J. Biomol. Struct. Dyn. 8:911–933. [DOI] [PubMed] [Google Scholar]
- 6.Ono, A., C.-N. Chen, and L.-S. Kan. 1991. DNA triplex formation of oligonucleotide analogues consisting of linker groups and octamer segments that have opposite sugar-phosphate backbone polarities. Biochemistry. 30:9914–9921. [DOI] [PubMed] [Google Scholar]
- 7.Lin, S.-B., C.-F. Kao, S.-C. Lee, and L.-S. Kan. 1994. DNA triplex formed by d-A-(G-A)7-G and d-mC-(T-mC)7-T in aqueous solution at neutral pH. Anticancer Drug Des. 9:1–8. [PubMed] [Google Scholar]
- 8.Ono, A., and L.-S. Kan. 1994. Triplex formation of oligonucleotides containing 2′-O-methyluridine, 5-mromo-2′-O-methyluridine and 2′-O-methylcytidine. J. Chin. Chem. Soc. 41:857–864. [Google Scholar]
- 9.Kan, L.-S., and A. Ono. 1994. Triplex formation as functions of variation of sequence and chain length of deoxyoligonucleotides at varied concentrations of NaCl and MgCl2. J. Chin. Chem. Soc. 41:865–869. [Google Scholar]
- 10.Trapane, T. L., R. I. Hogrefe, M. A. Reynold, L.-S. Kan, and P. O. P. Ts'o. 1996. Interstrand complex formation of purine oligonucleotides and their nonionic analogs: the model system of d(AG)8 and its complement, d(CT)8. Biochemistry. 35:5495–5508. [DOI] [PubMed] [Google Scholar]
- 11.Wei, M.-T., A. Walter, A. Gabrielian, H. Schütz, E. Birch-Hirschfeld, S.-B. Lin, W.-C. Lin, H. Fritzsche, and L.-S. Kan. 2005. Studies on the extension of sequence-independence and the enhancement of DNA triplex formation. J. Chin. Chem. Soc. 52:375–381. [Google Scholar]
- 12.Besch, R., C. Giovannangeli, T. Schuh, C. Kammerbauer, and K. Degitz. 2004. Characterization and quantification of triple helix formation in chromosomal DNA. J. Mol. Biol. 341:979–989. [DOI] [PubMed] [Google Scholar]
- 13.Barre, F.-X., S. Ait-Si-Ali, C. Giovannangeli, R. Luis, P. Robin, L. I. Pritchard, C. Helene, and A. Harel-Bellan. 2000. Unambiguous demonstration of triple-helix-directed gene modification. Proc. Natl. Acad. Sci. USA. 97:3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone, G. M., S. Napoli, A. Valentini, F. Cavalli, D. K. Watson, and C. V. Catapano. 2004. Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Res. 32:4358–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catapano, C. V., E. M. McGuffie, D. Pacheco, and G. M. R. Carbone. 2000. Inhibition of gene expression and cell proliferation by triplex helix-forming oligonucleotides directed to the c-myc gene. Biochemistry. 39:5126–5138. [DOI] [PubMed] [Google Scholar]
- 16.Faria, M., C. D. Wood, L. Perrouault, J. S. Nelson, A. Winter, M. R. H. White, C. Helene, and C. Giovannangeli. 2000. Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc. Natl. Acad. Sci. USA. 97:3862–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovannangeli, C., S. Diviacco, V. Labrousse, S. Gryaznov, P. Charneau, and C. Helene. 1997. Accessibility of nuclear DNA to triplex-forming oligonucleotides: the integrated HIV-1 provirus as a target. Proc. Natl. Acad. Sci. USA. 94:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, T., M. A. Macris, F. Faruqi, and P. M. Glazer. 2000. High-frequency intrachromosomal gene conversion induced by triplex-forming oligonucleotides microinjected into mouse cells. Proc. Natl. Acad. Sci. USA. 97:9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds, M. A., L. J. Arnold Jr., M. T. Almazan, T. A. Beck, R. I. Hogrefe, M. D. Metzler, S. R. Stoughton, B. Y. Tseng, T. L. Trapane, P. O. P. T-so, and T. M. Woolf. 1994. Triple-strand-forming methylphosphonate oligodeoxynucleotides targeted to mRNA efficiently block protein synthesis. Proc. Natl. Acad. Sci. USA. 91:12433–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song, J., Z. Intody, M. Li, and J. H. Wilson. 2004. Activation of gene expression by triplex-directed psoralen crosslinks. Gene. 324:183–190. [DOI] [PubMed] [Google Scholar]
- 21.Casey, B. P., and P. M. Glazer. 2001. Gene targeting via triple-helix formation. Prog. Nucleic Acid Res. Mol. Biol. 67:163–192. [DOI] [PubMed] [Google Scholar]
- 22.Knauert, M. P., and P. M. Glazer. 2001. Triplex forming oligonucleotides: sequence-specific tools for gene targeting. Hum. Mol. Genet. 10:2243–2251. [DOI] [PubMed] [Google Scholar]
- 23.Seidman, M. M., and P. M. Glazer. 2003. The potential for gene repair via triple helix formation. J. Clin. Invest. 112:487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasternack, L. B., S.-B. Lin, T.-M. Chin, W.-C. Lin, D.-H. Huang, and L.-S. Kan. 2002. Proton NMR studies of 5′-d-(TC)3(CT)3(AG)3-3′-A paperclip triplex: the structural relevance of turns. Biophys. J. 82:3170–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin, T.-M., S.-B. Lin, S.-Y. Lee, M.-L. Chang, A. Y.-Y. Cheng, F.-C. Chang, L. Pasternack, D.-H. Huang, and L.-S. Kan. 2000. “Paper-Clip” type triple helix formation by 5′-d-(TC)3Ta(CT)3Cb(AG)3 (a and b = 0–4) as a function of loop size with and without the pseudoisocytosine base in the Hoogsteen strand. Biochemistry. 39:12457–12464. [DOI] [PubMed] [Google Scholar]
- 26.Rajagopal, P., and J. Feigon. 1989. NMR studies of triple-strand formation from the homopurine-homopyrimidine deoxyribonucleotides d(GA)4 and d(TC)4. Biochemistry. 28:7859–7870. [DOI] [PubMed] [Google Scholar]
- 27.Macaya, R., E. Wang, P. Schultze, V. Sklenar, and J. Feigon. 1992. Proton nuclear magnetic resonance assignments and structural characterization of an intramolecular DNA triplex. J. Mol. Biol. 225:755–773. [DOI] [PubMed] [Google Scholar]
- 28.de los Santos, C., M. Rosen, and D. Patel. 1989. NMR studies of DNA (R+)n-(Y-)n-(Y+)n triple helices in solution; imino and amino proton markers of T-A-T and C-G-C+ base-triple formation. Biochemistry. 28:7282–7289. [DOI] [PubMed] [Google Scholar]
- 29.Warmlander, S., K. Sandstrom, M. Leijon, and A. Graslund. 2003. Base-pair dynamics in an antiparallel DNA Triplex measured by catalyzed imino proton exchange monitored via 1H NMR spectroscopy. Biochemistry. 42:12589–12595. [DOI] [PubMed] [Google Scholar]
- 30.Koshlap, K. M., P. Schultze, P. B. Brunar, P. B. Dervan, and J. Feigon. 1997. Solution structure of an intramolecular DNA triplex containing an N7-glycosylated guanine which mimics a protonated cytosine. Biochemistry. 36:2659–2668. [DOI] [PubMed] [Google Scholar]
- 31.Macaya, R., E. Wang, P. Schultze, V. Sklenar, and J. Feigon. 1992. Proton nuclear magnetic resonance assignments and structural characterization of an intramolecular DNA triplex. J. Mol. Biol. 225:755–773. [DOI] [PubMed] [Google Scholar]
- 32.Radhakrishnan, I., C. de los Santon, and D. J. Patel. 1991. Nuclear magnetic resonance structural studies of intramolecular purine-purine-pyrimidine DNA triplexes in solution. J. Mol. Biol. 221:1403–1418. [PubMed] [Google Scholar]
- 33.Radhakrishnan, I., and D. J. Patel. 1994. a. Solution structure and hydration patterns of pyrimidine·purine·pyrimidine DNA triplex containing a novel T·CG base-triple. J. Mol. Biol. 241:600–619. [DOI] [PubMed] [Google Scholar]
- 34.Radhakrishnan, I., and D. J. Patel. 1994. b. Solution structure of pyrimidine·purine·pyrimidine DNA triplex containing T*AT, C+*GC and G*TA triples. Structure. 2:17–32. [DOI] [PubMed] [Google Scholar]
- 35.Tarkoy, M., A. K. Phipps, P. Schultz, and J. Feigon. 1998. Solution structure of an intramolecular DNA triplex lined by hexakis(ethyleneglycol) units: d(AGAGAGAA-(EG)6-TTCTCTCT-(EG)6-TCTCTCTT). Biochemistry. 37:5810–5819. [DOI] [PubMed] [Google Scholar]
- 36.Wang, E., K. M. Koshlap, P. Gillespie, P. B. Dervan, and J. Feigon. 1996. Solution structure of a pyrimidine-purine-pyrimidine triplex containing the sequence-specific intercalating non-natural base D3. J. Mol. Biol. 257:1052–1069. [DOI] [PubMed] [Google Scholar]
- 37.Mirkin, S. M., and M. D. Frank-Kamenetskii. 1994. H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct. 23:541–576. [DOI] [PubMed] [Google Scholar]
- 38.Fasman, G. D., Ed. 1975. CRC Handbook of Biochemistry and Molecular Biology, 3rd ed., Vol. 1. CRC Press, Cleveland, OH.
- 39.Marion, D., and K. Wüthrich. 1983. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113:967–974. [DOI] [PubMed] [Google Scholar]
- 40.States, D. J., R. A. Haberkorn, and D. J. Ruben. 1982. A two dimensional nuclear Overhauser experiments. J. Magn. Reson. 48:286–292. [Google Scholar]
- 41.Mario, D., M. Ikura, and A. Bax. 1989. Improved solvent suppression in one and two-dimensional NMR spectra by convolution of time-domain data. J. Magn. Reson. 84:425–430. [Google Scholar]
- 42.Rance, M., O. W. Sørensen, G. Bodenhausen, G. Wagner, R. R. Ernst, and K. Wüthrich. 1983. Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117:479–485. [DOI] [PubMed] [Google Scholar]
- 43.Griesinger, C., G. Otting, K. Wüthrich, and R. R. Ernst. 1988. Clean TOCSY for 1H spin identification in macromolecules. J. Am. Chem. Soc. 110:7870–7872. [Google Scholar]
- 44.Wijmenga, S. S., M. M. W. Mooren, and C. W. Hilbers. 1993. NMR of Macromolecules, A Practical Approach. G. C. K. Roberts, editor. Oxford University Press, New York. 217–288.
- 45.Wuthrich, K. 1986. NMR of Proteins and Nucleic Acids. John Wiley & Sons, New York.
- 46.Tseng, Y.-Y., and S.-H. Chou. 1999. Systematic NMR assignment of DNA exchangeable protons. J. Chin. Chem. Soc. 46:699–706. [Google Scholar]
- 47.Clore, G. M., H. Oschkinat, L. W. McLaughlin, F. Benseler, C. S. Happ, E. Happ, and A. M. Gronenborn. 1988. Refinement of the solution structure of the DNA dodecamer 5′-d(CGCGPATTCGCG)2 containing a stable purine-thymine base pair: combined use of nuclear magnetic resonance and restrained molecular dynamics. Biochemistry. 27:4185–4197. [DOI] [PubMed] [Google Scholar]
- 48.Lavery, R., and H. Sklenar. 1988. The definition of generalized helicoidal parameters and of axis of curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 6:63–91. [DOI] [PubMed] [Google Scholar]
- 49.Kan, L.-S., and A. Ono. 1994. Triplex formation as functions of variation of sequence and chain length of deoxyoligonucleotides at varied concentrations of NaCl and MgCl2. J. Chin. Chem. Soc. 41:865–869. [Google Scholar]
- 50.Tsay, L. M., S. B. Lin, H. T. Tsay, H. H. Chen, and L.-S. Kan. 1995. A study of 5′-d-T-(C-T-)2C-(T-)4C-(T-C-)2T with 5′-d-A-(G-A-)2G. A hairpin triplex with three T.A.T and three C+.G.C base triads. J. Biomol. Struct. Dyn. 12:1235–1245. [DOI] [PubMed] [Google Scholar]
- 51.Radhakrishnan, I., and D. J. Patel. 1992. Three-dimensional homonuclear NOESY-TOCSY of an intramolecular pyrimidine·purine·pyrimidine DNA triplex containing a central G·TA triple: nonexchangeable proton assignments and structural implications. Biochemistry. 31:2514–2523. [DOI] [PubMed] [Google Scholar]
- 52.Bornet, O., and G. Lancelot. 1995. Solution structure of a selectively 13C-labeled intramolecular DNA triplex. J. Biomol. Struct. Dyn. 12:803–814. [DOI] [PubMed] [Google Scholar]
- 53.Van Duynhoven, J. P. M., J. Goudriaan, C. W. Hilbers, and S. S. Sijmenga. 1992. Quantitative evaluation of TOCSY data. application to sugar ring conformational analysis. J. Am. Chem. Soc. 114:10055–10056. [Google Scholar]
- 54.Saenger, W. 1984. Principles of Nucleic Acid Structure. Springer-Verlag, New York.
- 55.Chin, T.-M., C.-M. Chang, H.-W. Huang, and L.-L. Lo. 2004. Bimolecular triplex formation between 5′-d-(AG)nT4(CT)n and 5′-d-(TC)n as functions of helix length and buffer. J. Biomol. Struct. Dyn. 22:35–43. [DOI] [PubMed] [Google Scholar]
- 56.Wang, S., and E. T. Kool. 1994. Recognition of single-stranded nucleic acids by triplex formation: the binding of pyrimidine-rich sequences. J. Am. Chem. Soc. 116:8857–8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kandimalla, E. R., and S. Agrawal. 1995. Single strand targeted triplex formation: parallel-stranded DNA hairpin duplexes for targeting pyrimidine strands. J. Am. Chem. Soc. 117:6416–6417. [Google Scholar]
- 58.Prakash, G., and E. Kool. 1992. Structural effects in the recognition of DNA by circular oligonucleotides. J. Am. Chem. Soc. 114:3523–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]











