Abstract
Recently, substantial evidence has emerged that revealed a very close association between the formation of nitrotyrosine and the presence of activated granulocytes containing peroxidases, such as myeloperoxidase. Peroxidases share heme-containing homology and can use H2O2 to oxidize substrates. Heme is a complex of iron with protoporphyrin IX, and the iron-containing structure of heme has been shown to be an oxidant in several model systems where the prooxidant effects of free iron, heme, and hemoproteins may be attributed to the formation of hypervalent states of the heme iron. In the current study, we have tested the hypothesis that free heme and iron play a crucial role in NO2-Tyr formation. The data from our study indicate that: (i) heme/iron catalyzes nitration of tyrosine residues by using hydrogen peroxide and nitrite, a reaction that revealed the mechanism underlying the protein nitration by peroxidase, H2O2, and NO ; (ii) H2O2 plays a key role in the protein oxidation that forms the basis for the protein nitration, whereas nitrite is an essential element that facilitates nitration by the heme(Fe), H2O2, and the NO
; (ii) H2O2 plays a key role in the protein oxidation that forms the basis for the protein nitration, whereas nitrite is an essential element that facilitates nitration by the heme(Fe), H2O2, and the NO system; (iii) the formation of a Fe(IV) hypervalent compound may be essential for heme(Fe)-catalyzed nitration, whereas O
system; (iii) the formation of a Fe(IV) hypervalent compound may be essential for heme(Fe)-catalyzed nitration, whereas O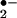 (ONOO− formation), •OH (Fenton reaction), and compound III are unlikely to contribute to the reaction; and (iv) hemoprotein-rich tissues such as cardiac muscle are vulnerable to protein nitration in pathological conditions characterized by the overproduction of H2O2 and NO
(ONOO− formation), •OH (Fenton reaction), and compound III are unlikely to contribute to the reaction; and (iv) hemoprotein-rich tissues such as cardiac muscle are vulnerable to protein nitration in pathological conditions characterized by the overproduction of H2O2 and NO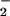 , or nitric oxide.
, or nitric oxide.
Keywords: nitric oxide‖hydrogen peroxide‖nitrite‖peroxidase
As a consequence of aerobic metabolism, vertebrates constantly generate reactive oxygen species (ROS), which include superoxide anion (O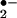 ), hydroxyl radicals (•OH) and hydrogen peroxide (H2O2). The overproduction of ROS has detrimental effects on cellular function in large part through the oxidation of proteins. The discovery of nitric oxide (1, 2) focused attention on reactive nitrogen species, which includes •NO (nitrogen monoxide) that can under go interconversion to form NO+ (nitrosonium), and NO− (nitroxyl anion). •NO reacts with O
), hydroxyl radicals (•OH) and hydrogen peroxide (H2O2). The overproduction of ROS has detrimental effects on cellular function in large part through the oxidation of proteins. The discovery of nitric oxide (1, 2) focused attention on reactive nitrogen species, which includes •NO (nitrogen monoxide) that can under go interconversion to form NO+ (nitrosonium), and NO− (nitroxyl anion). •NO reacts with O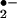 to form peroxynitrite (ONOO−) that can further form peroxynitrous acid (ONOOH), a very unstable and reactive oxidizing species. Involvement of ONOO− is the most widely studied mechanism of protein nitration (3, 4), and the formation of nitrotyrosine (NO2-Tyr) has been detected in various pathological conditions (5–8). However, over the past several years, substantial evidence has revealed a very close association between the formation of NO2-Tyr and the presence of activated granulocytes, which contain significant amounts of peroxidases, such as myeloperoxidase (MPO) (10–12).‡ Two radical intermediates have been proposed to play a key role in peroxidase catalyzed nitration of phenolic rings (13). First, on addition of H2O2, MPO (ground state with ferric form) can be oxidized to form compound I (ferryl π cation radical) that is readily reduced to form hypochlorous acid (HOCl) in the presence of physiological concentrations of chloride (Scheme S1). The reaction could then proceed by Cl− transfer from HOCl to nitrite (NO
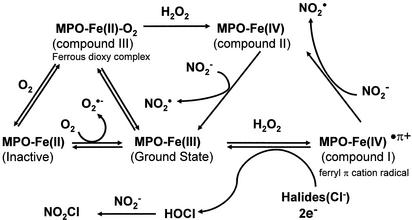
to form peroxynitrite (ONOO−) that can further form peroxynitrous acid (ONOOH), a very unstable and reactive oxidizing species. Involvement of ONOO− is the most widely studied mechanism of protein nitration (3, 4), and the formation of nitrotyrosine (NO2-Tyr) has been detected in various pathological conditions (5–8). However, over the past several years, substantial evidence has revealed a very close association between the formation of NO2-Tyr and the presence of activated granulocytes, which contain significant amounts of peroxidases, such as myeloperoxidase (MPO) (10–12).‡ Two radical intermediates have been proposed to play a key role in peroxidase catalyzed nitration of phenolic rings (13). First, on addition of H2O2, MPO (ground state with ferric form) can be oxidized to form compound I (ferryl π cation radical) that is readily reduced to form hypochlorous acid (HOCl) in the presence of physiological concentrations of chloride (Scheme S1). The reaction could then proceed by Cl− transfer from HOCl to nitrite (NO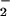 ) to generate nitryl chloride (NO2Cl) as a reaction intermediate (14, 15). The second radical intermediate is nitrogen dioxide (NO
) to generate nitryl chloride (NO2Cl) as a reaction intermediate (14, 15). The second radical intermediate is nitrogen dioxide (NO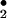 ), which can be generated via either compound I- or II-catalyzed oxidation of NO
), which can be generated via either compound I- or II-catalyzed oxidation of NO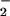 (Scheme S1; ref. 16). In addition, it has been suggested that physiological concentrations of NO
(Scheme S1; ref. 16). In addition, it has been suggested that physiological concentrations of NO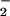 can be oxidized to •NO2 by the Fenton reaction (17), a Fe2+-dependent reaction during which decomposition of H2O2 forms hydroxyl radical (•OH) that oxidizes NO
can be oxidized to •NO2 by the Fenton reaction (17), a Fe2+-dependent reaction during which decomposition of H2O2 forms hydroxyl radical (•OH) that oxidizes NO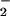 to NO
to NO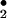 (Fig. 3A; ref. 18).
(Fig. 3A; ref. 18).
Scheme 1.
Figure 3.
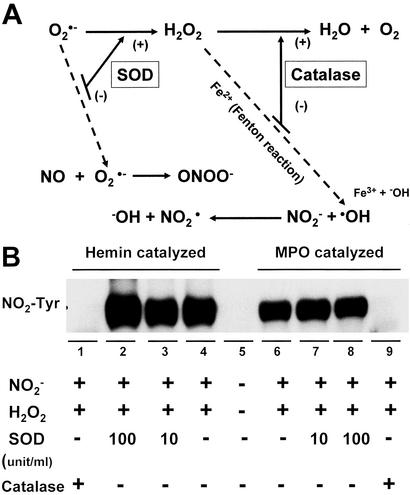
Effects of SODs and catalase on heme and MPO-catalyzed nitration. (A) The scheme shows the effects of SOD and catalase on the reactive oxygen species and reactive nitrogen species generating system. (B) Immunoblotting for NO2-Tyr that was catalyzed by 25 μM heme or 2 units/ml MPO while incubating with NO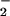 (1 mM) and H2O2 (1 mM) in 0.1 M sodium phosphate buffer (pH 7.4) at 37°C for 30 min. Treatment of SOD (10 and 100 units/ml) had no inhibitory effect on BSA nitration. In contrast, the application of catalase (500 units/ml) abolished both heme and MPO-induced protein tyrosine nitration.
(1 mM) and H2O2 (1 mM) in 0.1 M sodium phosphate buffer (pH 7.4) at 37°C for 30 min. Treatment of SOD (10 and 100 units/ml) had no inhibitory effect on BSA nitration. In contrast, the application of catalase (500 units/ml) abolished both heme and MPO-induced protein tyrosine nitration.
Peroxidases are enzymes that use H2O2 to oxidize substrates. In addition to MPO, the family of human peroxidases includes eosinophil peroxidase, uterine peroxidase, lactoperoxidase, salivary peroxidase, thyroid peroxidase, and intestinal peroxidase (19), all of which share heme-containing homology and can catalyze oxidative reactions. In addition, due to structural similarities, several hemoproteins such as cytochrome P450 isoenzymes, prostaglandin endoperoxide synthase (prostaglandin H synthase), cytochrome c, and hemoglobin are expected to act more or less as an oxidant (17). The biological role of heme has been reviewed extensively (20–22), and the heme molecule has been shown to promote most biological oxidation processes involved in oxygen transport, mitochondrial respiration, drug metabolism, steroid biosynthesis, cellular antioxidant defenses, and signal transduction processes (23, 24). Heme is a complex of protoporphyrin IX with iron that is pentacoordinated and in the high-spin Fe(III) oxidation state. Iron has the capacity to accept and donate electrons readily and to interconvert between ferric (Fe3+) and ferrous (Fe2+), which makes it a useful component of hemoproteins or metalloproteins. However, as a transition metal, iron is able to reduce (active) molecular oxygen and to catalyze the conversion of superoxide and hydrogen peroxide to free radical species. Thus, the iron-containing structure of heme is an oxidant in several model systems where the prooxidant effects of free iron, heme, and hemoprotein may be attributed to the formation of free radical species and hypervalent states of the heme iron that attack cellular components. Therefore, we decided to test the hypothesis that both free heme and iron play a key role in NO2-Tyr formation. To our knowledge, it is the first time that a systematic analysis of NO2-Tyr formation by either heme or free iron was performed and the possible mechanism that is responsible for the heme (iron)-mediated process was explored.
Materials and Methods
Reagents and Antibodies.
BSA, ferriprotoporphyrin IX (heme), sodium nitrite (NaNO2), hydrogen peroxide (H2O2), glucose oxidase, 3-nitro-l-tyrosine, 2,4-dinitrophenylhydrazine, ferrous sulfate (FeSO4), ferric chloride (FeCl3), EDTA, potassium cyanide, and mannitol were purchased from Sigma. All solvents and other reagents were the highest purity commercially available. A rabbit polyclonal antibody against 3-NO2-Tyr was purchased from Upstate Biotechnology (Lake Placid, NY).
Animal Tissue Processing and Sample Preparation.
Male CF-1 mice (body weight, 25–30 g) were obtained from Harlan Breeders (Indianapolis) and were treated in accordance with the NIH Institutional Animal Care and Use Committee Guidebook (2002) as approved by the University of Texas Medical School Animal Care and Use Committee. At death, the brain, heart, liver, kidney, and skeletal muscle from the back were quickly isolated, then immediately frozen in liquid nitrogen and stored at −135°C until further processing. Frozen tissues were pulverized with a pestle and mortar that contained liquid nitrogen. For protein extraction, the tissues were homogenized at 4°C in 20 mM Tris⋅HCl buffer (pH 7.4) containing protease inhibitors (final concentration: 10 μg/ml soybean trypsin inhibitor/10 μg/ml benzamadine/0.005 trypsin inhibitor units/ml aprotinin/10 μg/ml leupeptin/10 μg/ml pepstatin A/5 μg/ml antipain/0.2 mM PMSF/0.1 mM ethylene diamine tetraacetic acid). Each sample was homogenized by using a polytron at 0°C, then sonicated on ice by using a cell disrupter with five pulses at duty cycle of 40% and output of 3. The homogenates were centrifuged at 10,000 × g for 15 min at 4°C, and supernatant fractions were used for further experiments.
Detection of NO2-Tyr Formation with Western Blot Analysis.
BSA (2 mg/ml) in 0.1 M phosphate buffer (pH 7.4) was incubated at 37°C with heme (25 μM), NO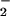 (1 mM), and H2O2 (1 mM) or d-glucose/glucose oxidase (10 mM/100 milliunits) for 30 min. The reaction conditions for iron-catalyzed nitration were similar, except the incubation time was increased to 16 h. After incubation, 75-μl reaction aliquots were mixed with 25 μl of sample loading buffer. Equal amounts of proteins were loaded onto the gel for each experimental sample. Separated proteins were transferred to nitrocellulose membranes that were probed with the anti-3-NO2-Tyr antibody. The nitrated BSAs were visualized with horseradish peroxidase-conjugated secondary antibody, and chemiluminesence was used to identify nitrotyrosine according to the enhanced chemiluminescent Western blotting detection system (Amersham Pharmacia Life Sciences). To confirm that the immunoreactivity to anti-NO2-Tyr antibody is due to the specific reaction with the motif of nitrated tyrosine in BSA, we preincubated the antibody with free 3-nitro-l-tyrosine (10 mM) for 4 h. The preabsorbed anti-NO2-Tyr antibody failed to detect NO2-Tyr that was formed during the reaction. Some gels or transferred membranes were stained with Coomassie brilliant blue or 1× Ponceau S to determine the amount of BSA after the reaction.
(1 mM), and H2O2 (1 mM) or d-glucose/glucose oxidase (10 mM/100 milliunits) for 30 min. The reaction conditions for iron-catalyzed nitration were similar, except the incubation time was increased to 16 h. After incubation, 75-μl reaction aliquots were mixed with 25 μl of sample loading buffer. Equal amounts of proteins were loaded onto the gel for each experimental sample. Separated proteins were transferred to nitrocellulose membranes that were probed with the anti-3-NO2-Tyr antibody. The nitrated BSAs were visualized with horseradish peroxidase-conjugated secondary antibody, and chemiluminesence was used to identify nitrotyrosine according to the enhanced chemiluminescent Western blotting detection system (Amersham Pharmacia Life Sciences). To confirm that the immunoreactivity to anti-NO2-Tyr antibody is due to the specific reaction with the motif of nitrated tyrosine in BSA, we preincubated the antibody with free 3-nitro-l-tyrosine (10 mM) for 4 h. The preabsorbed anti-NO2-Tyr antibody failed to detect NO2-Tyr that was formed during the reaction. Some gels or transferred membranes were stained with Coomassie brilliant blue or 1× Ponceau S to determine the amount of BSA after the reaction.
Detection of NO2-Tyr Formation with Spectrophotometer.
NO2-Tyr formation in BSA was also detected by spectrophotometer. pH-dependent NO2-Tyr absorbance was monitored at 430 nm [ɛ430 = 4,400 M−1⋅cm−1 in alkaline, pH 11.5 (25)] by using a spectrophotometer (CE9500). Briefly, a reaction mixture (in 0.1 M phosphate buffer) containing 2 mg/ml BSA, 25 μM heme, and different concentrations of NO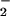 and H2O2 (to observe the concentration dependence of the responses) was incubated at 37°C for 30 min. Then, 900 μl of mixture was mixed with 100 μl of 1 N NaOH, and the absorbance was read immediately. Because heme itself also absorbs at 430 nm, the experimental results were obtained by subtracting the absorbance of the reaction mixture that contained heme, but no added sodium nitrite.
and H2O2 (to observe the concentration dependence of the responses) was incubated at 37°C for 30 min. Then, 900 μl of mixture was mixed with 100 μl of 1 N NaOH, and the absorbance was read immediately. Because heme itself also absorbs at 430 nm, the experimental results were obtained by subtracting the absorbance of the reaction mixture that contained heme, but no added sodium nitrite.
Detection of Carbonyl Formation with Spectrophotometer.
Oxidative damage of protein is accompanied by the formation of protein carbonyl groups that has been used widely as an index of protein oxidation. Protein carbonyls were determined by the spectrophotometric measurement of the formation of 2,4-dinitrophenylhydrazone derivatives (ɛ370 nm = 22,000 M−1⋅cm−1) as described by Levine et al. (26). To subtract the background signal that was generated by NO2-Tyr and heme (both substances absorb at 370 nm), reaction mixtures without treatment of 2,4-dintrophenylhydrazine were used and their readings subtracted from samples treated with 2,4-dinitrophenylhydrazine.
Statistical Analysis.
Data are expressed as mean ± SEM. Results were analyzed by using one-way ANOVA, and statistical significance assumed when P values were <0.05.
Results
Heme-Catalyzed Protein Nitration.
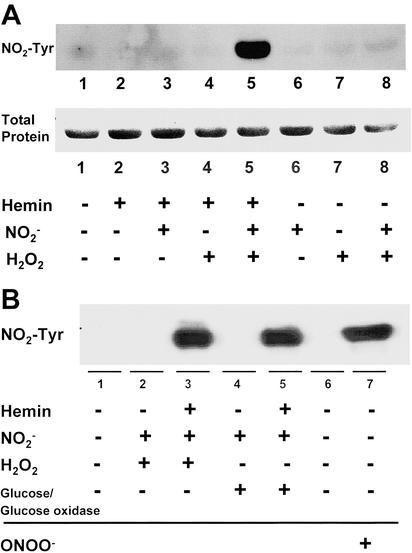
In the presence of hydrogen peroxide, peroxidases are able to oxidize nitrite, which results in protein nitration. To test whether heme by itself has a similar catalytic effect, we incubated BSA with various combinations of heme, nitrite and H2O2 in phosphate buffer at physiological pH for 30 min. As demonstrated in Fig. 1A, Western blot analysis revealed that BSA nitration was elicited, but only by the combination of heme, nitrite, and H2O2. Thus, all components of the heme/NO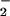 /H2O2 system are critical for NO2-Tyr formation. To further confirm the requirements of the reaction, we used glucose/glucose oxidase (10 mM/100 units) to generate the H2O2 (27). Again, the combination of the H2O2 generator with heme and nitrite induced BSA NO2-Tyr formation (Fig. 1B). Our experiment also showed that the heme (25 μM) catalyzed BSA tyrosine nitration is comparable with the nitration induced by 5 mM peroxynitrite (Fig. 1B). To confirm the specificity of our Western blot analysis, we treated SDS/PAGE transferred membrane with anti NO2-Tyr antibody that was preincubated with 10 mM free 3-nitro-l-tyrosine or with 10 mM free l-tyrosine. Preincubation of the antibody with free 3-nitro-l-tyrosine, but not free l-tyrosine blocked its ability to detect the NO2-Tyr signal that was identified by the original anti NO2-Tyr antibody (data not shown).
/H2O2 system are critical for NO2-Tyr formation. To further confirm the requirements of the reaction, we used glucose/glucose oxidase (10 mM/100 units) to generate the H2O2 (27). Again, the combination of the H2O2 generator with heme and nitrite induced BSA NO2-Tyr formation (Fig. 1B). Our experiment also showed that the heme (25 μM) catalyzed BSA tyrosine nitration is comparable with the nitration induced by 5 mM peroxynitrite (Fig. 1B). To confirm the specificity of our Western blot analysis, we treated SDS/PAGE transferred membrane with anti NO2-Tyr antibody that was preincubated with 10 mM free 3-nitro-l-tyrosine or with 10 mM free l-tyrosine. Preincubation of the antibody with free 3-nitro-l-tyrosine, but not free l-tyrosine blocked its ability to detect the NO2-Tyr signal that was identified by the original anti NO2-Tyr antibody (data not shown).
Figure 1.
Heme-catalyzed protein nitration. Immunoblotting for NO2-Tyr in heme-catalyzed BSA. (A) BSA (2 mg/ml) nitration was detected while incubating with heme (25 μM), nitrite (1 mM), and H2O2 (1 mM) in 0.1 M sodium phosphate buffer (pH 7.4) at 37°C for 30 min. (B) The combination of a H2O2 generator glucose/glucose oxidase (10 mM/100 units) with the same concentration of heme and nitrite also induced BSA nitration. Tyrosine nitration catalyzed by heme is comparable with that induced by 5 mM ONOO−.
Effect of Nitrite on Protein Nitration in Heme/NO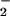 /H2O2 System.
/H2O2 System.
Nitrite has been recognized as one of the key oxidation products of NO. NO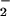 is a small molecule with high affinity for the heme center in peroxidase, which has been reported to enhance the oxidizing capacity of peroxidase/H2O2 (28). To further examine the role of NO
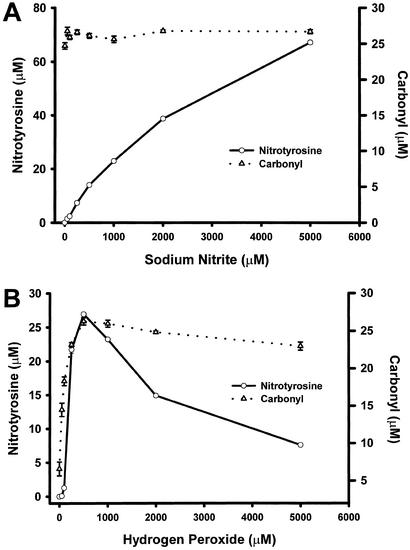
is a small molecule with high affinity for the heme center in peroxidase, which has been reported to enhance the oxidizing capacity of peroxidase/H2O2 (28). To further examine the role of NO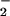 in the formation of NO2-Tyr, we compared BSA protein oxidation and nitration under different concentrations of nitrite (Fig. 2A). In the presence of heme (25 μM) and H2O2 (1 mM), different concentrations of nitrite were added and BSA nitration and oxidation (carbonyl formation) were measured by spectrophotometer. Fig. 2A illustrates a strong nitrite concentration-dependent nitration (r = 0.98). In contrast, carbonyl formation showed no relationship (r ≈ 0) with nitrite concentration. In fact, a high level of BSA oxidation was obtained through the reaction of heme and H2O2 in the absence of nitrite.
in the formation of NO2-Tyr, we compared BSA protein oxidation and nitration under different concentrations of nitrite (Fig. 2A). In the presence of heme (25 μM) and H2O2 (1 mM), different concentrations of nitrite were added and BSA nitration and oxidation (carbonyl formation) were measured by spectrophotometer. Fig. 2A illustrates a strong nitrite concentration-dependent nitration (r = 0.98). In contrast, carbonyl formation showed no relationship (r ≈ 0) with nitrite concentration. In fact, a high level of BSA oxidation was obtained through the reaction of heme and H2O2 in the absence of nitrite.
Figure 2.
Effect of nitrite and hydrogen peroxide on protein nitration and oxidation in the heme/NO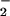 /H2O2 system. Protein nitration (○) was detected spectrophotometrically by monitoring pH-dependent NO2-Tyr absorbance (n = 3). Protein oxidation (carbonyls; ▵) was determined spectrophotometrically by measuring formation of the 2,4-dinitrophenylhydrazone (n = 3). (A) Reaction mixtures (in 0.1 M phosphate buffer) containing 2 mg/ml BSA, 25 μM heme, 1 mM H2O2, and different concentrations of NO
/H2O2 system. Protein nitration (○) was detected spectrophotometrically by monitoring pH-dependent NO2-Tyr absorbance (n = 3). Protein oxidation (carbonyls; ▵) was determined spectrophotometrically by measuring formation of the 2,4-dinitrophenylhydrazone (n = 3). (A) Reaction mixtures (in 0.1 M phosphate buffer) containing 2 mg/ml BSA, 25 μM heme, 1 mM H2O2, and different concentrations of NO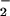 were incubated at 37°C for 30 min. Correlation coefficients (r) were 0.98 for NO2-Tyr formation and ≈0 for carbonyl formation. (B) The reaction mixtures (in 0.1 M phosphate buffer) containing 2 mg/ml BSA, 25 μM heme, 1 mM NO
were incubated at 37°C for 30 min. Correlation coefficients (r) were 0.98 for NO2-Tyr formation and ≈0 for carbonyl formation. (B) The reaction mixtures (in 0.1 M phosphate buffer) containing 2 mg/ml BSA, 25 μM heme, 1 mM NO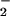 , and different concentrations of H2O2 were incubated at 37°C for 30 min. Both protein nitration and oxidation depend on H2O2 with the optimal concentration of 0.5 mM. Higher concentrations of H2O2 significantly attenuated the level of BSA tyrosine nitration while exhibiting less interference of the carbonyl formation.
, and different concentrations of H2O2 were incubated at 37°C for 30 min. Both protein nitration and oxidation depend on H2O2 with the optimal concentration of 0.5 mM. Higher concentrations of H2O2 significantly attenuated the level of BSA tyrosine nitration while exhibiting less interference of the carbonyl formation.
Effect of Hydrogen Peroxide on Protein Nitration in Heme (Fe)/NO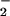 /H2O2 System.
/H2O2 System.
H2O2 is known to be a substrate for peroxidases to catalyze oxidative reaction. To further evaluate the effect of H2O2 on protein nitration and oxidation simultaneously, we measured NO2-Tyr as well as carbonyl formation using spectrophotometer. BSA was exposed to reaction mixtures containing 25 μM heme, 1 mM NO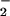 and different concentrations of H2O2 for 30 min at 37°C. The data depicted in Fig. 2B demonstrate that both protein nitration and oxidation rely on the presence of H2O2 with the optimal concentration of 0.5 mM. Interestingly, higher concentrations of H2O2 significantly attenuated the level of BSA tyrosine nitration while exhibiting less interference with carbonyl formation. To obtain additional insight into the mechanism of hydrogen peroxide-dependent protein nitration, we conducted similar experiments but using ferrous iron rather than heme. In contrast to the results using heme, iron-catalyzed BSA nitration displayed only a positive correlation between the concentration of H2O2 and the formation of NO2-Tyr (data not shown). Of note, heme demonstrated a higher efficiency to catalyze nitration in the condition of lower concentration of H2O2.
and different concentrations of H2O2 for 30 min at 37°C. The data depicted in Fig. 2B demonstrate that both protein nitration and oxidation rely on the presence of H2O2 with the optimal concentration of 0.5 mM. Interestingly, higher concentrations of H2O2 significantly attenuated the level of BSA tyrosine nitration while exhibiting less interference with carbonyl formation. To obtain additional insight into the mechanism of hydrogen peroxide-dependent protein nitration, we conducted similar experiments but using ferrous iron rather than heme. In contrast to the results using heme, iron-catalyzed BSA nitration displayed only a positive correlation between the concentration of H2O2 and the formation of NO2-Tyr (data not shown). Of note, heme demonstrated a higher efficiency to catalyze nitration in the condition of lower concentration of H2O2.
Effects of Antioxidant Enzymes on Heme/NO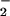 /H2O2 System-Induced Nitration.
/H2O2 System-Induced Nitration.
Heme-containing enzymes are involved in both reduction and activation of oxygen. In the heme molecule, five of the coordination sites of iron are occupied by intrinsic ligands, and the sixth is able to bind O2 or other extrinsic ligands. The strong and symmetrical coordination of the axial ligands enhances the interaction between O2 and iron-porphyrin and results in the formation of O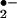 by reduction of O2 (29). As presented in Fig. 3A, superoxide dismutase (SOD) interferes with the reactive oxygen species and reactive nitrogen species generating system by converting O
by reduction of O2 (29). As presented in Fig. 3A, superoxide dismutase (SOD) interferes with the reactive oxygen species and reactive nitrogen species generating system by converting O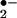 into H2O2. This will reduce the formation of ONOO− but increase the concentration of H2O2, which, in the presence of transition metals such as iron can generate a strong oxidant, hydroxyl radical (•OH), via the Fenton reaction. However, another heme-containing enzyme catalase can degrade H2O2 to water and oxygen thus inhibiting •OH formation. To evaluate the effect of SOD and catalase on protein nitration, we used heme as well as MPO to catalyze BSA NO2-Tyr formation. As shown in Fig. 3B, treatment with SOD had no significant effect on BSA nitration. In contrast, the application of catalase abolished both heme and MPO-induced protein tyrosine nitration. Thus, the above results further confirmed the requirement for H2O2 in protein nitration by the heme/O
into H2O2. This will reduce the formation of ONOO− but increase the concentration of H2O2, which, in the presence of transition metals such as iron can generate a strong oxidant, hydroxyl radical (•OH), via the Fenton reaction. However, another heme-containing enzyme catalase can degrade H2O2 to water and oxygen thus inhibiting •OH formation. To evaluate the effect of SOD and catalase on protein nitration, we used heme as well as MPO to catalyze BSA NO2-Tyr formation. As shown in Fig. 3B, treatment with SOD had no significant effect on BSA nitration. In contrast, the application of catalase abolished both heme and MPO-induced protein tyrosine nitration. Thus, the above results further confirmed the requirement for H2O2 in protein nitration by the heme/O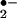 /H2O2 system.
/H2O2 system.
Iron-Catalyzed Protein Nitration.
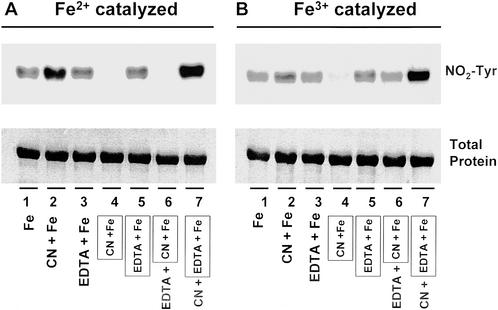
The release of iron in an oxidative manner from heme-containing proteins has received consideration recently (30). As a transition element, iron is able to exist in several oxidative states in response to different liganding environments (31). Therefore, we tested whether iron or chelated iron has a catalytic effect on tyrosine nitration. As shown in Fig. 4, both ferrous and ferric ions are capable of catalyzing the nitration of BSA at tyrosine residues when incubated with BSA in the presence of NO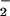 and H2O2. With addition of iron first into the BSA containing mixture, followed by adding of cyanide (CN−) (Fig. 4, lane 2) or EDTA (Fig. 4, lane 3), significant BSA nitration was detected. Nitration also was detected if the iron was mixed with EDTA before adding to the BSA-containing reaction buffer (Fig. 4, lane 5; framed). However, if the iron was first mixed with cyanide (Fig. 4, lane 4; framed) then added to the BSA-containing buffer, no protein nitration was observed. Of interest, addition of EDTA partly reversed the inhibited nitration by the premixture of iron with cyanide (Fig. 4, lane 6), whereas the addition of cyanide into an iron–EDTA mixture markedly enhanced NO2-Tyr formation (Fig. 4, lane 7).
and H2O2. With addition of iron first into the BSA containing mixture, followed by adding of cyanide (CN−) (Fig. 4, lane 2) or EDTA (Fig. 4, lane 3), significant BSA nitration was detected. Nitration also was detected if the iron was mixed with EDTA before adding to the BSA-containing reaction buffer (Fig. 4, lane 5; framed). However, if the iron was first mixed with cyanide (Fig. 4, lane 4; framed) then added to the BSA-containing buffer, no protein nitration was observed. Of interest, addition of EDTA partly reversed the inhibited nitration by the premixture of iron with cyanide (Fig. 4, lane 6), whereas the addition of cyanide into an iron–EDTA mixture markedly enhanced NO2-Tyr formation (Fig. 4, lane 7).
Figure 4.
Iron-catalyzed protein nitration. Immunoblotting for NO2-Tyr in BSA (2 mg/ml) that was incubated with either ferrous (Fe2+; 0.1 mM) or ferric (Fe3+; 0.1 mM) ions plus NO (1 mM) and H2O2 (1 mM) in 0.1 M sodium phosphate buffer (pH 7.4) at 37°C for 16 h. To study the mechanism, cyanide (CN−; 0.8 mM) and EDTA (0.4 mM) were used as chelators. The labeling with the frame indicates the mixture of iron with chelator (CN− or EDTA) before addition to the reaction buffer. The labeling without frame shows that iron and chelator (CN− or EDTA) were added to the BSA-containing reaction buffer in a sequential manner.
(1 mM) and H2O2 (1 mM) in 0.1 M sodium phosphate buffer (pH 7.4) at 37°C for 16 h. To study the mechanism, cyanide (CN−; 0.8 mM) and EDTA (0.4 mM) were used as chelators. The labeling with the frame indicates the mixture of iron with chelator (CN− or EDTA) before addition to the reaction buffer. The labeling without frame shows that iron and chelator (CN− or EDTA) were added to the BSA-containing reaction buffer in a sequential manner.
Effect of Hydroxyl Radical Scavengers on Iron-Catalyzed Protein Nitration.
The redox potential of iron is dramatically increased during its reaction with H2O2 (Fenton reaction) to produce the highly reactive •OH (Fig. 3A), which has been demonstrated to oxidize a wide range of substrates (32). Our observation (Fig. 3B) that catalase treatment abolished heme-catalyzed nitration further supported the key role of H2O2 in heme/iron-induced protein nitration. However, this result could also be considered as a sign of requirement for •OH in the reaction. To further explore the mechanism, we used two hydroxyl radical scavengers, mannitol (0.1 and 1.0 mM) (33) and EtOH (0.1% and 1.0%) (34) to evaluate the role of •OH in iron-catalyzed protein nitration. Treatment with either mannitol or EtOH at different concentrations all failed to affect iron-catalyzed BSA nitration (data not shown).
Protein Nitration in Heme-Rich Organs.
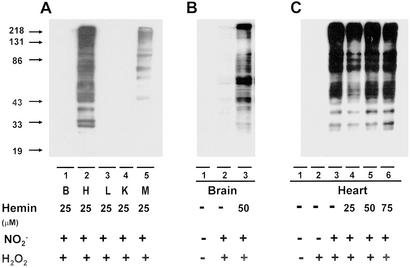
Heme-containing proteins are wildly distributed in the body. Myoglobin, a well-known hemoprotein that contains a protoporphyrin IX iron complex, is mainly concentrated in heart and skeleton muscle. In light of the possibility that a high concentration of heme may facilitate protein nitration, we compared NO2-Tyr formation in tissue homogenates from various organs (Fig. 5A). In the presence of heme, H2O2 and NO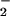 , considerable protein nitration was observed in homogenates of heart and skeleton muscle but not in the brain, liver, and kidney. Increasing the exogenous heme concentration, however, promoted nitration in the brain (Fig. 5B), liver, and kidney (data not shown). To further elucidate the role of endogenous heme in NO2-Tyr formation, we compared protein nitration in homogenates of heart under conditions of with or without exogenous heme. As depicted in Fig. 5C, significant protein nitration could be induced by application of NO
, considerable protein nitration was observed in homogenates of heart and skeleton muscle but not in the brain, liver, and kidney. Increasing the exogenous heme concentration, however, promoted nitration in the brain (Fig. 5B), liver, and kidney (data not shown). To further elucidate the role of endogenous heme in NO2-Tyr formation, we compared protein nitration in homogenates of heart under conditions of with or without exogenous heme. As depicted in Fig. 5C, significant protein nitration could be induced by application of NO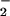 and H2O2 alone (Fig. 5C, lane 3) into the heart homogenate, and the addition of exogenous heme did not further increase the level of NO2-Tyr formation (Fig. 5C, lanes 4–6).
and H2O2 alone (Fig. 5C, lane 3) into the heart homogenate, and the addition of exogenous heme did not further increase the level of NO2-Tyr formation (Fig. 5C, lanes 4–6).
Figure 5.
Protein nitration in heme-rich organs. (A) Immunoblotting for NO2-Tyr in tissue homogenates from brain, heart, liver, kidney, and skeleton muscle was compared. In the presence of heme (25 μM), H2O2 (1 mM), and NO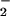 (1 mM), protein nitration was observed in heart and skeleton muscle but not in the brain, liver, and kidney. (B) With increases of exogenous heme concentration to 50 μM, however, nitration in the brain was observed. (C) Immunoblotting for NO2-Tyr in incubations of heart homogenate with or without exogenous heme. A significant nitration could be detected only by application of nitrite (1 mM) and H2O2 (1 mM) into the cardiac muscle homogenate, and addition of exogenous heme did not further increase the level of NO2-Tyr formation.
(1 mM), protein nitration was observed in heart and skeleton muscle but not in the brain, liver, and kidney. (B) With increases of exogenous heme concentration to 50 μM, however, nitration in the brain was observed. (C) Immunoblotting for NO2-Tyr in incubations of heart homogenate with or without exogenous heme. A significant nitration could be detected only by application of nitrite (1 mM) and H2O2 (1 mM) into the cardiac muscle homogenate, and addition of exogenous heme did not further increase the level of NO2-Tyr formation.
Discussion
Our current study has demonstrated that both heme and free iron can effectively catalyze the nitration of tyrosine via a H2O2-dependent mechanism. H2O2 is an essential element that reacts with hemeproteins such as MPO to form the intermediary ferryl species, compounds I and II (Scheme S1). Briefly, the ferric enzyme (ground state) reacts with H2O2 in a two-electron process to generate the ferryl π cation radical known as compound I (O = FeIVP+•). Compound I can undergo further reduction yielding another enzyme intermediate, compound II (O = FeIVP). Thus, the ferryl [Fe(IV)O] form has higher oxidative equivalents then the ferric [Fe(III)] form (35). Both compounds I and II can oxidize NO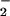 to generate the freely diffusible NO
to generate the freely diffusible NO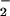 radical, nitrogen-dioxide (NO
radical, nitrogen-dioxide (NO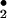 ), which can catalyze protein nitration (36). With remarkable similarity, our study indicated the importance of H2O2 in heme/NO
), which can catalyze protein nitration (36). With remarkable similarity, our study indicated the importance of H2O2 in heme/NO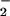 /H2O2 catalyzed NO2-Tyr formation. As illustrated in Fig. 1A, we did not observe nitrotyrosine formation in the presence of heme only (Fig. 1A, lane 2), nitrite only (Fig. 1A, lane 6), or heme plus nitrite only (Fig. 1A, lane 3). However, with the other two reagents present, H2O2 concentration-dependently stimulated both protein nitration and oxidation with an optimal concentration of 0.5 mM (Fig. 2B). Furthermore, treatment with catalase abolished NO2-Tyr formation by the heme/NO
/H2O2 catalyzed NO2-Tyr formation. As illustrated in Fig. 1A, we did not observe nitrotyrosine formation in the presence of heme only (Fig. 1A, lane 2), nitrite only (Fig. 1A, lane 6), or heme plus nitrite only (Fig. 1A, lane 3). However, with the other two reagents present, H2O2 concentration-dependently stimulated both protein nitration and oxidation with an optimal concentration of 0.5 mM (Fig. 2B). Furthermore, treatment with catalase abolished NO2-Tyr formation by the heme/NO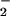 /H2O2 system, which is comparable to the effect of catalase on MPO-induced protein nitration (Fig. 3B). Thus, BSA nitration and oxidation are H2O2-dependent.
/H2O2 system, which is comparable to the effect of catalase on MPO-induced protein nitration (Fig. 3B). Thus, BSA nitration and oxidation are H2O2-dependent.
In addition to involving compounds I and II, ferric (FeIII) peroxidase can be reduced to the ferro (FeII) form. Ferroperoxidase has a high affinity for O2 and can be oxidized to the ferrous dioxy complex (compound III) (Scheme S1; refs. 37 and 38). Compound III is generally considered as enzymatically inactive; however, it can perform its oxidative activities by either undergoing spontaneous decay to ferriperoxidase with the generation of superoxide anion (O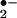 ) or by catalyzing the reduction of endogenous H2O2 to form the ferryl [Fe(IV)O]-peroxidase (Scheme S1; refs. 37 and 39). In the current study, we could not detect protein nitration in the absence of exogenous H2O2 (Fig. 1, lane 3; Fig. 2B); thus, there is no sign of endogenous H2O2 generation. In addition, a low concentration of SOD showed no effect on heme-catalyzed BSA nitration (Fig. 3B; lane 3), and a high concentration of SOD somehow promoted BSA tyrosine nitration (Fig. 3B; lane 2). These results suggest that heme-catalyzed BSA nitration is not a direct result of the formation of superoxide anion. Therefore, there is no evidence to support the formation or involvement of compound III or the possibility of generating ONOO− from O
) or by catalyzing the reduction of endogenous H2O2 to form the ferryl [Fe(IV)O]-peroxidase (Scheme S1; refs. 37 and 39). In the current study, we could not detect protein nitration in the absence of exogenous H2O2 (Fig. 1, lane 3; Fig. 2B); thus, there is no sign of endogenous H2O2 generation. In addition, a low concentration of SOD showed no effect on heme-catalyzed BSA nitration (Fig. 3B; lane 3), and a high concentration of SOD somehow promoted BSA tyrosine nitration (Fig. 3B; lane 2). These results suggest that heme-catalyzed BSA nitration is not a direct result of the formation of superoxide anion. Therefore, there is no evidence to support the formation or involvement of compound III or the possibility of generating ONOO− from O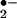 in our current experimental system. Also, although Brennan et al. (40) have demonstrated that a peroxynitrite-like species could be formed while peroxidase catalyze nitration is under acid pH, our neutral (pH 7.4) experimental condition that effectively prevented free ONOO− formation (40) further showed that the possible involvement of ONOO− (Fig. 3A) is negligible.
in our current experimental system. Also, although Brennan et al. (40) have demonstrated that a peroxynitrite-like species could be formed while peroxidase catalyze nitration is under acid pH, our neutral (pH 7.4) experimental condition that effectively prevented free ONOO− formation (40) further showed that the possible involvement of ONOO− (Fig. 3A) is negligible.
Although the inhibitory effect of catalase on heme- (Fig. 3B, lane 1) or Fe- (data not shown) catalyzed nitration may suggest the involvement of the Fenton reaction (Fig. 3A) as suggested by Thomas et al. (41), our data have revealed that a potent hydroxyl radical scavenger, mannitol together with EtOH, had no effect on Fe-catalyzed BSA tyrosine nitration. Thus, heme/iron-mediated protein nitration does not depend on the formation of hydroxyl radical. In addition, Fenton reaction effectively produces hydroxyl radical in several seconds; however, our study showed that a 16-h incubation is needed for iron to induce BSA nitration. Thus, the inefficiency of the iron/H2O2/NO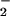 /system in protein nitration casts further doubt on the role of hydroxyl radical in iron-catalyzed tyrosine nitration.
/system in protein nitration casts further doubt on the role of hydroxyl radical in iron-catalyzed tyrosine nitration.
To further elucidate the biochemical basis of protein nitration by the heme/NO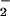 /H2O2 system, we investigated carbonyl and nitrotyrosine formation as markers of protein oxidation and nitration, respectively. The results presented in Fig. 2 offer new facets that allow us to dissect those two reactions. The contrast between the concentration/response curves for BSA nitration and oxidation indicates that they are two distinguishable processes (Fig. 2A), and that there is no competitive mechanism involved in the reactions between BSA oxidation and nitrite oxidation by the oxidant. Despite the fundamental important role of H2O2 in heme- (or Fe-) catalyzed nitration, a high concentration of hydrogen peroxide markedly reduced the level of protein nitration while exerting less effect on BSA oxidation (Fig. 2B). As a reactive oxygen species, H2O2 has been reported to cause the suicide inactivation of peroxidase (42). Excess H2O2 can also oxidize hemoglobin or heme and result in the release of iron (43, 44). Thus, a decreased extent of nitration may reflect the degradation of heme by a higher dose of H2O2. Our current study further supports this suggestion: without the heme structure, iron-catalyzed BSA nitration was positively correlated with the increased concentration of H2O2. It is understandable that due to the lower efficacy of iron in catalyzing NO2-Tyr formation, an attenuated level of nitration was observed when heme structure was damaged by excess H2O2. Another major impression derived from the current data is that the protein oxidation by heme depends more on H2O2 concentration, because the carbonyl formation was not affected by the degradation of heme with higher H2O2 conditions (Fig. 2B).
/H2O2 system, we investigated carbonyl and nitrotyrosine formation as markers of protein oxidation and nitration, respectively. The results presented in Fig. 2 offer new facets that allow us to dissect those two reactions. The contrast between the concentration/response curves for BSA nitration and oxidation indicates that they are two distinguishable processes (Fig. 2A), and that there is no competitive mechanism involved in the reactions between BSA oxidation and nitrite oxidation by the oxidant. Despite the fundamental important role of H2O2 in heme- (or Fe-) catalyzed nitration, a high concentration of hydrogen peroxide markedly reduced the level of protein nitration while exerting less effect on BSA oxidation (Fig. 2B). As a reactive oxygen species, H2O2 has been reported to cause the suicide inactivation of peroxidase (42). Excess H2O2 can also oxidize hemoglobin or heme and result in the release of iron (43, 44). Thus, a decreased extent of nitration may reflect the degradation of heme by a higher dose of H2O2. Our current study further supports this suggestion: without the heme structure, iron-catalyzed BSA nitration was positively correlated with the increased concentration of H2O2. It is understandable that due to the lower efficacy of iron in catalyzing NO2-Tyr formation, an attenuated level of nitration was observed when heme structure was damaged by excess H2O2. Another major impression derived from the current data is that the protein oxidation by heme depends more on H2O2 concentration, because the carbonyl formation was not affected by the degradation of heme with higher H2O2 conditions (Fig. 2B).
In living cells, iron exerts its functions either in the form of hemoproteins or by binding with other molecules (18). Apparently, the most-described iron-mediated oxidative reaction is the iron-catalyzed Haber–Weiss reaction, also called the Fenton reaction. However, as discussed previously, our data show that the formation of superoxide anion (O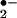 ) and hydroxyl radical (•OH) are not necessary for iron/heme-induced nitration. Recently, considerable effort has been made to determine whether another oxidizing species is formed besides the hydroxyl radical, and the effect of ferryl species has been emphasized (for review, see ref. 45). Thus, the chemical structure of iron and its capacity to drive one-electron reactions makes iron a major player in the production and metabolism of free radicals in response to certain liganding environments. As demonstrated by our experiment (Fig. 4), the reaction of iron with either BSA (lane 1), CN− (lane 2), or EDTA (lane 3) all catalyzed protein nitration. The significance of iron imbalance in its redox potentials is further demonstrated by current study (Fig. 4) that the sequential addition of iron and cyanide (lane 2) and the mixture of iron and cyanide first (lane 4) before application into the BSA-containing buffer created completely opposite results in terms of protein nitration. As a potent iron-chelating agent, CN− can occupy all of the binding sites of iron in a 6:1 ratio and completely block the catalytic effect of iron on BSA nitration (Fig. 4, lane 4). Incomplete binding characteristics of either EDTA or BSA (46) are useful for iron to form its redox status [Fe(IV)] (31) that may further be stabilized by addition of CN− (Fig. 4, lanes 2 and 7). Summarizing current information, the catalytic effect of heme or chelated iron on the BSA tyrosine nitration could be described by following reaction (Eqs. 1–4):
) and hydroxyl radical (•OH) are not necessary for iron/heme-induced nitration. Recently, considerable effort has been made to determine whether another oxidizing species is formed besides the hydroxyl radical, and the effect of ferryl species has been emphasized (for review, see ref. 45). Thus, the chemical structure of iron and its capacity to drive one-electron reactions makes iron a major player in the production and metabolism of free radicals in response to certain liganding environments. As demonstrated by our experiment (Fig. 4), the reaction of iron with either BSA (lane 1), CN− (lane 2), or EDTA (lane 3) all catalyzed protein nitration. The significance of iron imbalance in its redox potentials is further demonstrated by current study (Fig. 4) that the sequential addition of iron and cyanide (lane 2) and the mixture of iron and cyanide first (lane 4) before application into the BSA-containing buffer created completely opposite results in terms of protein nitration. As a potent iron-chelating agent, CN− can occupy all of the binding sites of iron in a 6:1 ratio and completely block the catalytic effect of iron on BSA nitration (Fig. 4, lane 4). Incomplete binding characteristics of either EDTA or BSA (46) are useful for iron to form its redox status [Fe(IV)] (31) that may further be stabilized by addition of CN− (Fig. 4, lanes 2 and 7). Summarizing current information, the catalytic effect of heme or chelated iron on the BSA tyrosine nitration could be described by following reaction (Eqs. 1–4):
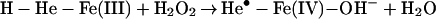 |
1 |
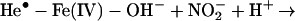 |
2 |
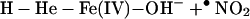 |
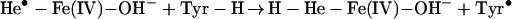 |
3 |
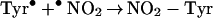 |
4 |
Briefly, hemin or chelated iron reacts with H2O2 to form the ferryl π-cation radical complex, which then oxidizes both nitrite and tyrosine to form nitric dioxide radical (•NO2) and tyrosine radical (Tyr•), respectively. These nitrating species nitrate tyrosyl residues in proteins to form NO2-Tyr.
The average adult human body contains ≈3–4 g of iron, and 65% of it is bound to hemoglobin. Ten percent is a constituent of myoglobin, cytochromes, and iron-containing enzymes, and the rest is bound to the iron storage proteins (47). The higher contents of iron (heme) in certain tissues such as heart could serve as a biological base for iron toxicity on free radical-mediated tissue damage, including formation of nitrotyrosine (Fig. 5). The first evidence (9) that muscle contraction can be altered by nitration of key proteins is suggested by the study of skeletal muscle sarcoplasmic-reticulum (SR) Ca2+-ATPase isoforms 2 (SERCA2a), which suggests that tyrosine nitration may affect Ca2+-ATPase activity. The data obtained in our simple experimental systems regarding heme/iron-induced nitration are surprisingly compatible with empirical results from our study with heme-rich tissues. The elucidation of the behavior of the heme (iron) on reaction with NO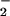 /H2O2 system is essential to the development of therapeutic methods that will focus on the prevention of oxidation.
/H2O2 system is essential to the development of therapeutic methods that will focus on the prevention of oxidation.
Acknowledgments
Z.G. was supported in part by the China Scholarship Council. Financial support of this research was provided by the John S. Dunn, Harold and Leila Mathers, and Robert A. Welch Foundations, as well as the University of Texas and the National Institutes of Health (Grants HL-64221, GM61731, and GMP50 38529).
Abbreviations
- MPO
myeloperoxidase
- SOD
superoxide dismutase
Footnotes
Bian, K., Harari, Y., Zhong, M., Weisbrodt, N. & Murad, F. (2001) FASEB J. 15, A199 (abstr.).
References
- 1.Murad F. Recent Prog Horm Res. 1998;53:43–59. [PubMed] [Google Scholar]
- 2.Murad F. Biosci Rep. 1999;19:133–154. doi: 10.1023/a:1020265417394. [DOI] [PubMed] [Google Scholar]
- 3.Beckman J S, Koppenol W H. Am J Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 4.Beckman J S. FASEB J. 2002;16:1144. doi: 10.1096/fj.02-0133lte. [DOI] [PubMed] [Google Scholar]
- 5.Bian K, Davis K, Kuret J, Binder L, Murad F. Am J Physiol. 1999;277:F33–F40. doi: 10.1152/ajprenal.1999.277.1.F33. [DOI] [PubMed] [Google Scholar]
- 6.Bian K, Harari Y, Zhong M, Lai M, Castro G, Weisbrodt N, Murad F. Mol Pharmacol. 2001;59:939–947. doi: 10.1124/mol.59.4.939. [DOI] [PubMed] [Google Scholar]
- 7.Davis K L, Martin E, Turko I V, Murad F. Annu Rev Pharmacol Toxicol. 2001;41:203–236. doi: 10.1146/annurev.pharmtox.41.1.203. [DOI] [PubMed] [Google Scholar]
- 8.Hanafy K A, Krumenacker J S, Murad F. Med Sci Monit. 2001;7:801–819. [PubMed] [Google Scholar]
- 9.Viner R I, Ferrington D A, Huhmer A F, Bigelow D J, Schoneich C. FEBS Lett. 1996;379:286–290. doi: 10.1016/0014-5793(95)01530-2. [DOI] [PubMed] [Google Scholar]
- 10.Baldus S, Eiserich J P, Mani A, Castro L, Figueroa M, Chumley P, Ma W, Tousson A, White C R, Bullard D C, et al. J Clin Invest. 2001;108:1759–1770. doi: 10.1172/JCI12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duguet A, Iijima H, Eum S Y, Hamid Q, Eidelman D H. Am J Respir Crit Care Med. 2001;164:1119–1126. doi: 10.1164/ajrccm.164.7.2010085. [DOI] [PubMed] [Google Scholar]
- 12.Kenyon N J, van der Vliet A, Schock B C, Okamoto T, McGrew G M, Last J A. Am J Physiol. 2002;282:L540–L555. doi: 10.1152/ajplung.00297.2001. [DOI] [PubMed] [Google Scholar]
- 13.Bian K, Ferid M. Front Biosci. 2003;8:d264–d278. doi: 10.2741/997. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Q, Hurst J K. J Biol Chem. 1997;272:32767–32772. doi: 10.1074/jbc.272.52.32767. [DOI] [PubMed] [Google Scholar]
- 15.Johnson D W, Margeyum D W. Inorg Chem. 1991;30:4845–4851. [Google Scholar]
- 16.van der Vliet A, Eiserich J P, Halliwell B, Cross C E. J Biol Chem. 1997;272:7617–7625. doi: 10.1074/jbc.272.12.7617. [DOI] [PubMed] [Google Scholar]
- 17.Augusto O, Bonini M G, Amanso A M, Linares E, Santos C C, De Menezes S L. Free Radical Biol Med. 2002;32:841–859. doi: 10.1016/s0891-5849(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 18.Halliwell B, Gutteridge J M. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 19.Dunford H B. Xenobiotica. 1995;25:725–733. doi: 10.3109/00498259509061888. [DOI] [PubMed] [Google Scholar]
- 20.Paoli M, Marles-Wright J, Smith A. DNA Cell Biol. 2002;21:271–280. doi: 10.1089/104454902753759690. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima H, Honma Y, Tawara T, Kato T, Park S Y, Miyatake H, Shiro Y, Aono S. J Biol Chem. 2001;276:7055–7061. doi: 10.1074/jbc.M003972200. [DOI] [PubMed] [Google Scholar]
- 22.Nelson M J, Batt D G, Thompson J S, Wright S W. J Biol Chem. 1991;266:8225–8229. [PubMed] [Google Scholar]
- 23.Poulos T L, Li H, Raman C S. Curr Opin Chem Biol. 1999;3:131–137. doi: 10.1016/s1367-5931(99)80024-6. [DOI] [PubMed] [Google Scholar]
- 24.Ponka P. Am J Med Sci. 1999;318:241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Gow A, Duran D, Thom S R, Ischiropoulos H. Arch Biochem Biophys. 1996;333:42–48. doi: 10.1006/abbi.1996.0362. [DOI] [PubMed] [Google Scholar]
- 26.Levine R L, Garland D, Oliver C N, Amici A, Climent I, Lenz A G, Ahn B W, Shaltiel S, Stadtman E R. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 27.Mueller S, Weber A, Fritz R, Mutze S, Rost D, Walczak H, Volkl A, Stremmel W. Biochem J. 2002;363:483–491. doi: 10.1042/0264-6021:3630483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reszka K J, McCormick M L, Britigan B E. Biochemistry. 2001;40:15349–15361. doi: 10.1021/bi011869c. [DOI] [PubMed] [Google Scholar]
- 29.Isogai Y, Iizuka T, Shiro Y. J Biol Chem. 1995;270:7853–7857. doi: 10.1074/jbc.270.14.7853. [DOI] [PubMed] [Google Scholar]
- 30.Comporti M. Free Radical Biol Med. 2002;32:565–567. doi: 10.1016/s0891-5849(02)00758-x. [DOI] [PubMed] [Google Scholar]
- 31.Welch K D, Davis T Z, Aust S D. Arch Biochem Biophys. 2002;397:360–369. doi: 10.1006/abbi.2001.2694. [DOI] [PubMed] [Google Scholar]
- 32.Winterbourn C C. Toxicol Lett. 1995;82–83:969–974. doi: 10.1016/0378-4274(95)03532-x. [DOI] [PubMed] [Google Scholar]
- 33.Aft R L, Mueller G C. J Biol Chem. 1984;259:301–305. [PubMed] [Google Scholar]
- 34.Sankarapandi S, Zweier J L. J Biol Chem. 1999;274:34576–34583. doi: 10.1074/jbc.274.49.34576. [DOI] [PubMed] [Google Scholar]
- 35.Furtmuller P G, Burner U, Jantschko W, Regelsberger G, Obinger C. FEBS Lett. 2000;484:139–143. doi: 10.1016/s0014-5793(00)02143-8. [DOI] [PubMed] [Google Scholar]
- 36.Burner U, Furtmuller P G, Kettle A J, Koppenol W H, Obinger C. J Biol Chem. 2000;275:20597–20601. doi: 10.1074/jbc.M000181200. [DOI] [PubMed] [Google Scholar]
- 37.Chen S X, Schopfer P. Eur J Biochem. 1999;260:726–735. doi: 10.1046/j.1432-1327.1999.00199.x. [DOI] [PubMed] [Google Scholar]
- 38.Ximenes V F, Catalani L H, Campa A. Biochem Biophys Res Commun. 2001;287:130–134. doi: 10.1006/bbrc.2001.5557. [DOI] [PubMed] [Google Scholar]
- 39.Halliwell B, Gutteridge J M. Methods Enzymol. 1984;105:47–56. doi: 10.1016/s0076-6879(84)05007-2. [DOI] [PubMed] [Google Scholar]
- 40.Brennan M L, Wu W, Fu X, Shen Z, Song W, Frost H, Vadseth C, Narine L, Lenkiewicz E, Borchers M T, et al. J Biol Chem. 2002;277:17415–17127. doi: 10.1074/jbc.M112400200. [DOI] [PubMed] [Google Scholar]
- 41.Thomas D D, Espey M G, Vitek M P, Miranda K M, Wink D A. Proc Natl Acad Sci USA. 2002;99:12691–12696. doi: 10.1073/pnas.202312699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnao M B, Acosta M, del Rio J A, Varon R, Garcia-Canovas F. Biochim Biophys Acta. 1990;1041:43–47. doi: 10.1016/0167-4838(90)90120-5. [DOI] [PubMed] [Google Scholar]
- 43.Balla J, Jacob H S, Balla G, Nath K, Eaton J W, Vercellotti G M. Proc Natl Acad Sci USA. 1993;90:9285–9289. doi: 10.1073/pnas.90.20.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagababu E, Rifkind J M. Biochemistry. 2000;39:12503–12511. doi: 10.1021/bi992170y. [DOI] [PubMed] [Google Scholar]
- 45.Welch K D, Davis T Z, Van Eden M E, Aust S D. Free Radical Biol Med. 2002;32:577–583. doi: 10.1016/s0891-5849(02)00760-8. [DOI] [PubMed] [Google Scholar]
- 46.Lovstad R A. Biometals. 1995;8:328–331. doi: 10.1007/BF00141606. [DOI] [PubMed] [Google Scholar]
- 47.Fraga C G, Oteiza P I. Toxicology. 2002;180:23–32. doi: 10.1016/s0300-483x(02)00379-7. [DOI] [PubMed] [Google Scholar]