Abstract
The RNase P protein gene (rnpA) completely overlaps the rpmH gene (encoding ribosomal protein L34) out of frame in the thermophilic bacterium Thermus thermophilus. This results in the synthesis of an extended RNase P protein (C5) of 163 aa and, by inference, of 240 aa in the related strain Thermus filiformis. Start codons of rnpA and rpmH, apparently governed by the same ribosome binding site, are separated by only 4 nt, which suggests a regulatory linkage between L34 and C5 translation and, accordingly, between ribosome and RNase P biosynthesis. Within the sequence encoding the N-terminal extensions and downstream of rpmH, several Thermus species exhibit in-frame deletions/insertions, suggesting relaxed constraints for sequence conservation in this region. Roughly the N-terminal third of T. thermophilus C5 was further shown to be dispensable for RNase P function in vitro by using a precursor tRNAGly substrate from the same organism. Taken together, these data reveal a mode of gene expression that is to our knowledge unprecedented in bacteria.
The ubiquitous enzyme RNase P catalyzes endonucleolytic 5′ maturation of tRNA primary transcripts in all three domains of life (Archaea, Bacteria, and Eukarya) and in mitochondria and chloroplasts (1, 2). Bacterial RNase P enzymes are composed of a catalytic RNA subunit, ≈400 nt in length, and a single small protein of typically 120 aa (3). The RNA subunits alone are catalytically active in vitro but require elevated salt concentrations to compensate for the absence of the protein subunit (4). RNase P holoenzymes are highly efficient catalysts, consistent with their generally low cellular abundance (2).
In the majority of bacteria, the rnpA gene encoding the RNase P protein has been identified immediately downstream of the gene for the ribosomal protein L34 (rpmH) and close to the origin of replication oriC (5–7). In Escherichia coli, rpmH and rnpA were shown to be part of the same operon, with two major promoters preceding the rpmH structural gene (7–9). E. coli L34 is produced in excess over the RNase P protein (termed C5), which appears to be regulated at the transcriptional and the translational level. First, three mRNA species derived from the rpmH–rnpA operon were detected, two shorter ones lacking the rnpA cistron and a longer and much less abundant one including it (9). Second, the rnpA codon usage does not correspond to that of highly expressed E. coli genes, such as rpmH (7).
We have investigated rnpA expression in thermophilic bacteria of the genus Thermus. This work led to the discovery of an unprecedented mode of (nonviral) gene expression in bacteria: rnpA completely overlaps rpmH in a different reading frame, resulting in the synthesis of unusually long RNase P proteins (163 aa in T. thermophilus).
Materials and Methods
For a detailed description of bacterial strains, cloning procedures, plasmid constructs, and protein and RNA preparation, see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Preparation of Recombinant Proteins.
Proteins recombinantly expressed in E. coli cells were purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography. C564 was obtained from its purified precursor by tobacco etch virus protease cleavage according to the manufacturer's instructions. For details, see Supporting Text.
Transformation of T. thermophilus HB27 and Isolation of Recombinant C51/C-His.
T. thermophilus HB27 was grown in rich medium at 70°C under aeration and transformed essentially as described (10). Suspensions (400 ml) of HB27 cells transformed with plasmid p16C5His were cooled on ice after overnight culturing, mixed with 1/10 vol of ice-cold 50% trichloroacetic acid, and incubated on ice for 15 min. Precipitated proteins were pelleted at 4°C by centrifugation for 10 min at 5,000 × g. Proteins were washed three times in ice-cold acetone; each washing step involved sonication to redissolve the pellet followed by centrifugation. Air-dried pellets of total cellular protein were resuspended in 14 ml of buffer D (50 mM Tris⋅HCl, pH 7.4/8 M urea/100 mM NaCl/12 mM imidazole), and the pH was adjusted to 7.4 with NaOH. Recombinant C51/C-His was extracted by incubating the 14 ml of total cellular protein with ≈1 ml Ni-NTA agarose (Qiagen, Chatsworth, CA; preequilibrated in buffer D) for 2 h at room temperature. The slurry was transferred to an empty column and washed extensively (15 column bed volumes) with buffer D (gravity flow). C51/C-His was eluted with four times 0.5 ml of buffer D containing 0.5 M imidazole (gravity flow); residual protein was eluted by overnight incubation of the resin with 1 ml of the same buffer. Eluted C51/C-His fractions were concentrated by using Microcon YM-10 centrifugal filter devices (Millipore). Matrix-assisted laser desorption ionization–MS analysis of C51/C-His was performed with a Reflex IV time-of-flight mass spectrometer (Bruker Daltronics, Bremen, Germany) as described (11).
Immunoblotting.
Western blotting and immunodetection were performed essentially as described (12), using Immobilon P (Millipore) membranes and an alkaline phosphatase-conjugated anti-rabbit antibody (Roche Molecular Biochemicals).
Immunoprecipitation.
Immunoprecipitation was essentially performed as described (ref. 13; Supporting Text).
Processing Assays.
Activity of native T. thermophilus RNase P coupled to anti-C587 antibody/protein A-Sepharose beads (0.6 μl in a final reaction volume of 21 μl) was analyzed in buffer F [50 mM Mes/0.1 M NH4OAc/10 mM Mg(OAc)2, pH 7.0; under shaking to keep the beads in suspension] in the presence of <1 nM of 5′ labeled precursor tRNA (ptRNA)Gly at 37°C. Multiple turnover rates (kobs) for RNase P holoenzymes reconstituted with C51, C551, and C564 were measured at 70°C in the presence of 100 nM protein, 10 nM RNase P RNA, and 100 nM ptRNAGly in buffer H [50 mM Hepes/0.1 M NH4OAc/30 mM Mg(OAc)2, pH 7.0]. Substrate and RNase P RNA were preincubated at 60°C in buffer H for 5 and 10 min, respectively. RNase P RNA and protein were then combined and incubated for 2 min at 60°C in the same buffer before starting the reaction by addition of substrate. Aliquots were withdrawn at different time points, and cleavage products were analyzed by denaturing 20% PAGE, visualized, and quantified as described (14). C587 was inactive under the above-mentioned and several other conditions tested (e.g., assay temperature variation between 37°C and 70°C, inclusion of up to 0.5 M urea in assays to increase its solubility, enzyme/substrate excess).
Results
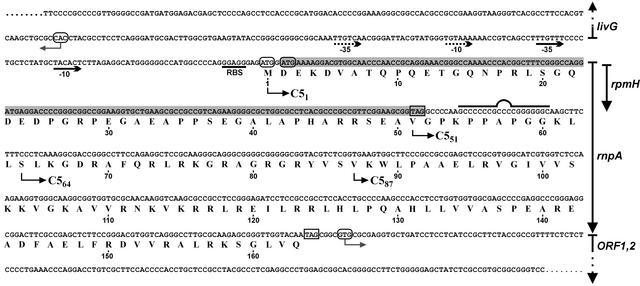
As a first step toward identifying the protein subunit of RNase P in T. thermophilus HB8, we cloned and sequenced the corresponding genomic region encoding rpmH and, by inference, rnpA (Fig. 1). Utilization of the rpmH start codon was verified by determining the amino acid sequence of L34 prepared from T. thermophilus HB8 cells (Theodora Choli, personal communication). Downstream of rpmH, we observed amino acid similarities between the putative rnpA gene product of T. thermophilus and other bacterial RNase P (C5) proteins in the region of amino acids 95–163 (Fig. 1). The only potential rnpA start codon downstream of rpmH was a GUG codon (Val-87, Fig. 1), the most frequently used type of start codon in bacterial rnpA genes (data not shown). This predicted an unusually short C5 protein of 77 aa (C587, amino acids 87–163, Fig. 1) with 45% similarity to the E. coli homolog. A recombinant His-tagged version of C587 was only soluble in the presence of 6 M urea and failed to stimulate the processing reaction catalyzed by T. thermophilus RNase P RNA at low Mg2+ (10 mM, data not shown). Insolubility of C587 is attributable to the lack of conserved structural elements, which became evident only with the publication of RNase P protein structures (see Discussion).
Figure 1.
Genomic context of the rpmH–rnpA genes in T. thermophilus HB8. The rpmH (L34) reading frame is shaded, and the amino acid sequence for the overlapping rnpA reading frame is given and numbered beneath the nucleotide sequence. Start and stop codons are outlined by round and square boxes, respectively. Promoter −35 and −10 elements identified by Maseda and Hoshino (ref. 15; National Center for Biotechnology Information nucleotide gi|971299|) or putative (dotted lines) are marked below the sequence. livG, ORF1,2: adjacent genes. The sequence encoding the potential RNA hairpin 3′ of rpmH is marked by the bulged line. The recombinant versions of C51, C551, and C587 (starting points indicated by arrows) tested for RNase P activity additionally carried an N-terminal His tag.
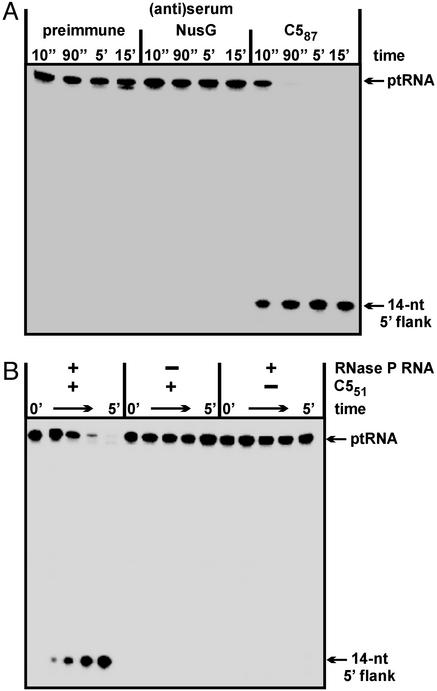
We raised a polyclonal rabbit antiserum against recombinant C587 and analyzed its ability to immunoprecipitate RNase P activity present in T. thermophilus extracts. Immunoprecipitates obtained with the anti-C587 antiserum were rich in RNase P activity, whereas essentially no activity was recovered with the control antisera (Fig. 2A). Thus, epitopes recognized by the C587-specific antiserum were also present in the native T. thermophilus RNase P holoenzyme complex. Attempts to identify the native rnpA gene product in preparative immunoprecipitates by N-terminal sequencing were unsuccessful. The same pertains to extensive efforts to purify the native RNase P holoenzyme by chromatographic methods, which we attribute to the vigorous T. thermophilus protease activities that become immediately unleashed as a consequence of cell disruption (see Supporting Text).
Figure 2.
Endonucleolytic processing of 5′ 32P-labeled ptRNAGly by RNase P holoenzymes at 37°C. (A) RNase P activity test in the presence of protein A Sepharose beads coated with antibodies from three different sera (preimmune serum and antisera specific for T. thermophilus transcription factor NusG or C587) and preincubated with a T. thermophilus DEAE chromatography fraction enriched in RNase P activity. (B) Activation of T. thermophilus RNase P RNA (400 nM) by C551 (80 nM); assay conditions were 37°C, 50 mM Mes, 0.1 M NH4OAc, 10 mM Mg(OAc)2, pH 6.0, < 1 nM ptRNAGly.
Successful immunoprecipitation of RNase P activity with the anti-C587 antiserum together with the observation that C587 cannot function as a cofactor suggested that C587 lacks part of the native T. thermophilus C5 protein. An N-terminal extension seemed possible because the rnpA reading frame can be continued in the upstream direction by another 191 aa until interrupted by a stop codon in the livG gene (not shown). Limited sequence similarity to other bacterial C5 proteins was indeed evident upstream of the initially assumed GUG start codon (data not shown). We therefore expressed an arbitrarily extended C5 variant (C551, Fig. 1). In contrast to C587, C551 was soluble in the absence of urea and formed a functional holoenzyme with the homologous RNA subunit (Fig. 2B).
Genomic Analysis of Other Thermus Species.
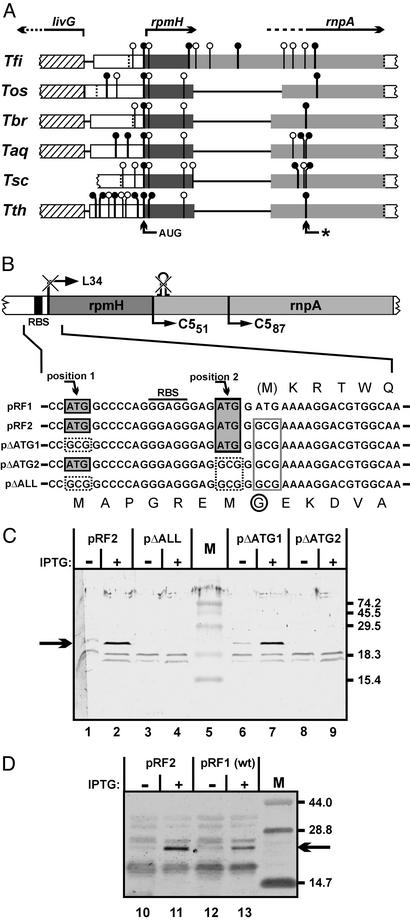
Assuming that the rnpA translational initiation site would be conserved in different species of the genus Thermus, we pursued a phylogenetic approach toward rnpA start codon identification. In addition to T. thermophilus HB8, we cloned and sequenced the rpmH–rnpA genomic regions of several other Thermus species (Fig. 3A and Supporting Text). The analysis for potential start codons in the rnpA reading frame revealed that Thermus oshimai and Thermus filiformis possess a GUC instead of a GUG codon at the initially assumed (Val-87) start position (Fig. 3A, arrow with asterisk), which further argued against this site as a translational start in Thermus. In addition, stop codons interrupting the rnpA reading frame in the upstream direction are found earliest in the livG–rpmH spacer region or in the livG structural gene in all strains analyzed. Compared with Thermus brockianus, Thermus aquaticus, and Thermus scotoductus, the rnpA-coding portion immediately downstream of rpmH is extended by 3 and 231 nt in T. thermophilus and T. filiformis, respectively, and shortened by 33 nt in T. oshimai. These length variations represent multiples of 3 nt, thus preserving the displacement of the rnpA reading frame relative to the rpmH frame. The only potential start codon found in all six species at an identical position is an AUG located 4 nt upstream of rpmH. These findings prompted us to consider the possibility that translation of rnpA is initiated upstream of rpmH, implying that rnpA completely overlaps rpmH. This was also consistent with results from RT-PCR analysis (see Fig. 5, which is published as supporting information on the PNAS web site), which indeed demonstrated the presence of such a full-length rnpA mRNA in T. thermophilus.
Figure 3.
(A) Alignment of rpmH–rnpA coding regions from different Thermus species. Start codons [vertical lines with circles; ●, commonly used codons (AUG and GUG); ○, rarely used codons (U/CUG, AUC/U)], and stop codons (dotted vertical lines) downstream of livG in the rnpA reading frames of six different members of the genus Thermus are indicated. Genes livG, rpmH, and rnpA and their direction of transcription are indicated at the top. Lines connecting the boxes represent sequence gaps introduced by the alignment program (OMIGA 2.0, Accelrys, Cambridge, U.K.). The AUG codon found in all six species and the initially assumed rnpA start codon from T. thermophilus (*) are marked by arrows at the bottom. Tfi, T. filiformis; Tos, T. oshimai; Tbr, T. brockianus; Taq, T. aquaticus; Tsc, T. scotoductus. (B) Expression plasmids carrying the rpmH–rnpA genomic region of T. thermophilus under control of a T7 promoter; pRF1 represents the WT situation. (Upper) Schematic representation: the rpmH start codon and the hairpin structure eliminated and disrupted, respectively, in all shown constructs except for pRF1; also shown are the native RBS and the N termini of the truncated RNase P proteins C551 and C587. (Lower) Sequences surrounding the inactivated start codon of rpmH are shown in detail. Gray-shaded boxes: potential rnpA start codons in the WT sequence; dotted boxes: mutation of potential rnpA start codons to GCG; box outlined in gray: mutated rpmH start codon. The amino acid sequences corresponding to the L34 and C5 reading frames are shown above and below the nucleotide sequences, respectively. M indicates the methionine start codon of L34 mutated in all constructs except for pRF1; the encircled G indicates the substitution of Gly for Asp resulting from mutation of the rpmH start codon. (C) Detection of a polypeptide with an apparent size of 22 kDa (arrow on the left) by Western blotting using the C587-specific antiserum before (−) and after (+) induction of transcription by T7 RNA polymerase from the four plasmids in E. coli BL21(DE3) host cells. M, marker proteins with sizes (in kDa) indicated on the right. IPTG, isopropyl β-d-thiogalactoside. (D) Attenuated expression of C51 in E. coli cells harboring plasmid pRF1 versus pRF2; for details, see legend to C.
Mutational Analysis of Potential rnpA Start Codons.
To precisely explore translational initiation of rnpA, we put the complete rpmH–rnpA region of T. thermophilus under control of a T7 promoter for inducible expression in E. coli BL21(DE3) (plasmid pRF1). To improve rnpA expression, we constructed pRF2, a derivative of pRF1 in which we inactivated the start codon of rpmH and disrupted a potential transcription terminator immediately downstream of rpmH (Figs. 1 and 3B). Isopropyl β-d-thiogalactoside induction of E. coli cells harboring pRF2 resulted in expression of a single protein with an apparent size of ≈22 kDa that reacted with the C587 antiserum (Fig. 3C, lane 2). This size was consistent with usage of the AUG start codon 4 nt upstream of the rpmH start site. However, an AUG codon 22 nt upstream of the rpmH initiation site represented an additional candidate for the rnpA start site (Fig. 3B). Therefore, both AUG codons were inactivated simultaneously (pΔALL) or individually (pΔATG1 and pΔATG2, Fig. 3B). Expression of the “22-kDa” protein was observed only with an intact AUG codon 4 nt upstream of the (mutated) rpmH start codon (Fig. 3C, plasmids pRF2 and pΔATG1), indicating that this AUG used in E. coli represents the genuine translational start point of rnpA in T. thermophilus, and by inference, in other members of the genus Thermus. Recombinant versions of the 22-kDa protein (C51, Fig. 1) and C551, equipped with N-terminal His tags, were then tested for activity. T. thermophilus RNase P reconstituted with C51 and C551 showed very similar turnover numbers at 70°C (kobs = 40 ± 8 and 38 ± 10 min−1 for C51 and C551, respectively; for assay conditions, see Materials and Methods), which corresponded to an ≈2,000-fold stimulation over the RNA-alone reaction (kobs of 0.02 min−1). Cleavage occurred at the canonical −1/+1 site, as inferred from coelectrophoresis of processing reactions catalyzed by E. coli M1 RNA (data not shown). In contrast, C587 completely failed to stimulate the RNA-alone reaction under these and several other conditions (see Materials and Methods). These results suggest that the N-terminal extension in C51 has no significant effect on RNase P protein function, although we cannot exclude that the processing of other natural ptRNA substrates may be influenced by its presence.
Expression of C51 was also seen with plasmid pRF1 (Fig. 3D, lane 13) representing the WT situation, although at ≈5-fold lower levels than for E. coli cells harboring pRF2 (Fig. 3D, lane 11). This result demonstrated that the better positioning of the rpmH start codon with respect to the ribosome binding site (RBS) in pRF1 did not prevent a fraction of E. coli ribosomes from initiating translation in the rnpA frame beginning 4 nt upstream of rpmH.
Recombinant Expression of C5 in T. thermophilus.
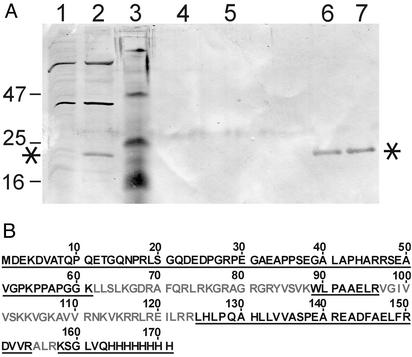
Because recombinant C551 and C51 had very similar activities, we considered the possibility that T. thermophilus C5 is synthesized as a precursor protein that is converted to its mature form by proteolytic processing. In T. thermophilus lysates, purified recombinant C551 and C51 were indeed rapidly degraded to a relatively stable intermediate C564 (see Fig. 6, which is published as supporting information on the PNAS web site). A recombinant version of C564, starting with Ser-64 (Fig. 1), resulted in a holoenzyme with a kobs of 26 ± 4 min−1, thus within 1.5-fold of the activities obtained with C551 and C51 under the same conditions (see above). To analyze whether proteolytic truncation might also occur in vivo, we expressed rnpA in T. thermophilus HB27 cells from an E. coli–Thermus shuttle plasmid. The recombinant plasmid, p16C5His, contained T. thermophilus rnpA under control of the T. thermophilus 16S rRNA promoter (16). Cells grown overnight were treated with trichloroacetic acid to instantly denature and precipitate cellular proteins. The recombinant C5 protein, equipped with a C-terminal His tag (C51/C-His, Fig. 4), was then purified by Ni-NTA affinity chromatography. The eluted protein could be immunostained with the C587 antiserum (Fig. 4A, lanes 6 and 7) and migrated as a band of ≈22 kDa, comparable in size to C51 expressed from pRF1 and pRF2 in E. coli (Fig. 3 C and D). The protein was already detectable in the total protein fraction (Fig. 4A, lane 2), but not in a total protein extract from the parental T. thermophilus strain lacking p16C5His (Fig. 4A, lane 1). Further analysis by MS (see Fig. 4B and Fig. 7, which is published as supporting information on the PNAS web site) revealed that the genuine rnpA translation product is indeed initiated at the AUG start codon 4 nt upstream of rpmH in T. thermophilus. However, in contrast to the in vitro situation, no significant steady-state levels of shorter fragments derived from proteolysis were seen in vivo (Fig. 4A, lanes 6 and 7).
Figure 4.
C51/C-His expression in T. thermophilus HB27. (A) Western blot analysis. Total cellular protein was obtained by treatment of cell suspensions with trichloroacetic acid. Total protein from T. thermophilus HB27 without plasmid (lane 1) and harboring the C5 expression plasmid p16C5His (lane 2). Lane 3, prestained marker proteins with sizes (in kDa) indicated on the left. Lanes 4 and 5, the two main elution fractions from the Ni-NTA column that had been loaded with total protein derived from the plasmid-free parental HB27 strain. Lanes 6 and 7, corresponding elution fractions from the Ni-NTA column loaded with total protein derived from the HB27 strain harboring plasmid p16C5His. For details, see Materials and Methods. Asterisks mark the recombinant C51/C-His protein (encoded by plasmid p16C5His), which was further analyzed by matrix-assisted laser desorption ionization (MALDI)–time-of-flight MS (see Fig. 7). (B) Sequence of C51/C-His; black underlined letters indicate regions (positions 1–61, 89–96, and 125–171) identified by MALDI mass fingerprinting. Positions 62–88, 97–124, and 155–157 (gray letters) could not be identified. These regions are rich in Lys and Arg residues, and the tryptic fragments derived from this section of the protein would be too small to be detected by MALDI-MS under our standard conditions. The total sequence coverage is 68%.
In conclusion, our results indicate that the RNase P holoenzyme in T. thermophilus includes an unusually large protein subunit of 163 aa, whose N-terminal third seems to be functionally dispensable and whose length is only exceeded by that of T. filiformis (Fig. 3A), predicted to be the largest bacterial RNase P protein known so far (240 aa).
Discussion
Regulation of rnpA Expression.
The genes for the small ribosomal protein L34 and the RNase P protein are colocalized in the majority of bacterial genomes (see Introduction), suggesting cotranscription as shown for E. coli (9). Our study has revealed an even more intimate relation of rpmH and rnpA in Thermus, such that the rnpA cistron completely overlaps, out of frame, that of rpmH. The genetic coupling of rpmH and rnpA in most bacteria as well as the mode of rnpA expression revealed for Thermus make it likely that the two genes are coregulated, possibly linking RNase P to ribosome synthesis.
E. coli cells contain a 60- to 100-fold molar excess of ribosomes over RNase P RNA (17). Expression of ribosomal protein L34 is expected to exceed that of the RNase P protein by a similar factor. In the case of T. thermophilus, different mechanisms may contribute to unequal expression of rpmH and rnpA, as discussed in the following.
A potential transcription terminator was identified downstream of rpmH (Fig. 1). Similar hairpin structures (with or without a stretch of U residues on the 3′ side of transcripts) are encoded in several bacterial rpmH–rnpA operons, either in the intergenic region (Bacillus subtilis) or in the 5′ portion of the rnpA cistron (e.g., E. coli). Hairpin structures immediately downstream of the rpmH stop codon are also encoded in the genomes of T. brockianus, T. scotoductus, and T. aquaticus, whereas T. oshimai has a deletion of exactly this structural element. T. filiformis, deviating from the other strains by an insertion in this region (Fig. 3A), encodes four consecutive stable hairpins downstream of rpmH (data not shown). However, at least for T. thermophilus, RT-PCR analysis (see Fig. 5) did not suggest that the hairpin structure causes substantial premature termination of transcription after RNA polymerase has traversed rpmH.
The codon usage of rnpA is expected to be constrained in the region overlapping rpmH. Indeed, several codons rarely used in highly expressed genes in Thermus, such as CCA, GAU, UCA, or GAA (18), are present in this region of T. thermophilus rnpA, suggesting a slower rate of translation and possibly a higher frequency of abortive translation for ribosomes translating in the rnpA relative to those translating in the rpmH frame.
A purine-rich RBS precedes the rnpA and rpmH start codons in Thermus. The distance of the RBS is 3 nt to the rnpA and 7 nt to the rpmH start codon (Figs. 1 and 3B) in all Thermus strains sequenced (Fig. 3A, data not shown). In highly expressed Thermus genes, such as those of the spc ribosomal protein operon of T. aquaticus (18), a distance of 4–7 nt was found. Thus, the distance between the RBS and the rnpA start codon seems to be suboptimal, which may cause less abundant translational initiation in the rnpA versus the rpmH reading frame. Increased C51 expression in E. coli after inactivation of the rpmH start codon in plasmid pRF2 versus pRF1 (Fig. 3D) is consistent with the notion that the the rpmH and rnpA start codons compete for initiating ribosomes, at least in this heterologous context.
Role of the N-Terminal Extension.
The N-terminal part of native T. thermophilus C5, including 8 gly and 11 Pro residues, were neither detrimental nor necessary for RNase P function in our in vitro assay, although we cannot exclude at this point an unknown critical in vivo function of the N-terminal addition. However, the N-terminal length variation in different Thermus strains (Fig. 3A) may argue against such a critical function. Also, a homology search did not reveal any convincing similarity of the T. thermophilus N-terminal region to other bacterial or archaeal proteins. It therefore appears that cells may simply tolerate the N-terminal extension of C5, which is dispensable for RNase P activity. This situation is reminiscent of bacteriophage MS2 where the frame for the lysis protein (75 aa) partially overlaps out of phase with the cistron encoding the coat protein (19). The N-terminal 40 aa of the lysis protein, which have little structural order compared with the essential C-terminal domain, were found to be dispensable for function. Likewise, most amino acid exchanges between MS2 and related phages fr and R17 occur within these N-terminal 40 aa. It was concluded that the overlap with the coat protein gene is not required for extra coding capacity but serves to couple synthesis of the lysis protein to that of the coat protein (19). Thus, acquisition of nonfunctional N-terminal extensions for the sake of translational coregulation seems to emerge as a principle not only applying to phages, but also to chromosomal genes in bacteria.
N-Terminal Truncation of T. thermophilus C5.
Alignment of T. thermophilus C5 variants with the RNase P proteins from B. subtilis and S. aureus (see Fig. 6) suggests that not only C51 and C551 but also C564 includes all conserved structural elements of bacterial C5 proteins. Nevertheless, RNase P reconstituted with C564 (100 aa) showed a somewhat lower turnover number (26 ± 4 min−1) than holoenzymes containing C51 (40 ± 8 min−1) or C551 (38 ± 10 min−1). Because the N terminus (Ser-64) of C564 is only 4 aa apart from the first conserved element (helix α1, see Supporting Text), the additional 13 aa in C551 might somehow stabilize the protein, consistent with the finding that bacterial RNase P proteins have an average size of 120 aa (3).
C587 lacks α-helix 1 and β-strand 1 and carries a partial deletion of β-strand 2. The failure of C587 to form a functional holoenzyme with its cognate RNA is therefore probably caused by a lack of essential structure elements or defective folding as suggested by its insolubility. Our findings are not consistent with those of Kim et al. (20), who analyzed, among several other deletion constructs, N-terminal deletion variants of E. coli C5 (MC5ΔN39 and MC5ΔN45) lacking four and nine more residues than C587 (alignment not shown), resulting in complete deletion of α1, β1, and β2. Both variants were reported to retain some activity in vitro, moderately complement the phenotype of a thermosensitive rnpA mutant strain (E. coli A49), and bind to E. coli RNase P RNA in vitro, although with reduced affinity.
Evolutionary Aspects.
How might this mechanism of rnpA gene expression have evolved in the genus Thermus? One clue could be the high genomic G+C content (69%) of T. thermophilus HB8 (21). Other thermophilic bacteria have G+C contents <50% (22, 23). Thus, the correlation between thermostability and development of G/C-rich genomes is a peculiarity of the genus Thermus and other members of the Deinococcus group (24). The pressure to avoid A-T base pairs has certainly minimized the number of fortuitous stop codons in the genome. We surmise that a successful switch to the rnpA start codon upstream of rpmH occurred in the progenitor of extant Thermus species, and a primordial rnpA start codon at a position similar to that in most bacteria was abandoned simultaneously or subsequently by mutational drift. This scenario requires the resulting N-terminal extension not to be detrimental to protein function, a prerequisite fulfilled because C51 was found to be active. Because the extension is not essential for RNase P function, there was relatively little pressure to conserve it as long as stop codons and frameshifts between the start codon and the functional C-terminal RNase P protein domain were avoided. Indeed, this conforms to our findings. The Thermus species analyzed here exhibit length and sequence variation in their N-terminal extensions, but have preserved the start codon upstream of rpmH (Fig. 3A) and avoided shifts in the reading frame between the N terminus and the C-terminal RNase P protein domain. However, because other bacteria survive well without it, the special mode of rpmH/rnpA expression in Thermus may not necessarily have brought about a selective advantage.
Evolutionary events leading to substantial reading frame extensions are less likely for bacteria with A/T-rich genomes because of the higher probability that the extension is interrupted by a fortuitous stop codon. Indeed, the largest (predicted) rpmH/rnpA overlap known apart from Thermus is 11 codons in Mycoplasma genitalium (data not shown). Thus, the rnpA reading frame extension in Thermus seems to be unique among extant bacteria. Even in the related Deinococcus radiodurans (67% G+C content; ref. 25), the start codon of the putative rnpA gene was assigned downstream of and in the same frame as rpmH. The putative gene product is also extended (166 aa), but caused by internal and C-terminal expansions (25). In the thermophile T. maritima, thought to represent a deeper phylogentic branch than Thermus (24), the putative start codon of rnpA (encoding an average-sized RNase P protein of 117 aa) directly overlaps with the stop codon of rpmH (ref. 23; unpublished data); a theoretical N-terminal extension of rnpA is unfeasible because of a stop codon occurring in the rnpA reading frame within rpmH.
Supplementary Material
Acknowledgments
We are grateful to Theodora Choli for making the T. thermophilus L34 protein sequence available to us, Judith Schlegl for cloning of T. thermophilus rpmH-rnpA, Horacio Alberto Avila Ulloa and Francois Franceschi for N-terminal protein sequencing, Ralph A. D. Williams for providing cells and genomic DNA from several Thermus species, José Berenguer for providing strain HB27, and Sybille Siedler and Barbara Wegscheid for kinetic measurements. This work was supported by Deutsche Forschungsgemeinschaft Grant HA 1672/4-2/4-3/13-1 and Bundesministerium für Bildung und Forschung Grant 031U215B (to Reinhard Lührmann).
Abbreviations
- RBS
ribosome binding site
- NTA
nitrilotriacetic acid
- ptRNA
precursor tRNA
Footnotes
References
- 1.Altman S, Kirsebom L A. In: The RNA World. 2nd Ed. Gesteland R F, Cech T, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 351–380. [Google Scholar]
- 2.Frank D N, Pace N R. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 3.Brown J W. Nucleic Acids Res. 1998;26:351–352. doi: 10.1093/nar/26.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 5.Ogasawara N, Yoshikawa H. Mol Microbiol. 1992;6:629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- 6.Salazar L, Fsihi H, de Rossi E, Riccardi G, Rios C, Cole S T, Takiff H E. Mol Microbiol. 1996;20:283–293. doi: 10.1111/j.1365-2958.1996.tb02617.x. [DOI] [PubMed] [Google Scholar]
- 7.Hansen F G, Hansen E B, Atlung T. Gene. 1985;38:85–93. doi: 10.1016/0378-1119(85)90206-9. [DOI] [PubMed] [Google Scholar]
- 8.Hansen F G, Hansen E B, Atlung T. EMBO J. 1982;1:1043–1048. doi: 10.1002/j.1460-2075.1982.tb01294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panagiotidis C A, Drainas D, Huang S-C. Int J Biochem. 1992;24:1625–1631. doi: 10.1016/0020-711x(92)90180-9. [DOI] [PubMed] [Google Scholar]
- 10.de Grado M, Castán P, Berenguer J. Plasmid. 1999;42:241–245. doi: 10.1006/plas.1999.1427. [DOI] [PubMed] [Google Scholar]
- 11.Hartmuth K, Urlaub H, Vornlocher H P, Will C L, Gentzel M, Wilm M, Lührmann R. Proc Natl Acad Sci USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 18.60–18.75. [Google Scholar]
- 13.Lygerou Z, Pluk H, van Venrooij W J, Séraphin B. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- 14.Warnecke J M, Held R, Busch S, Hartmann R K. J Mol Biol. 1999;290:433–445. doi: 10.1006/jmbi.1999.2890. [DOI] [PubMed] [Google Scholar]
- 15.Maseda H, Hoshino T. FEMS Microbiol Lett. 1995;128:127–134. doi: 10.1111/j.1574-6968.1995.tb07511.x. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann R K, Erdmann V A. J Bacteriol. 1989;171:2933–2941. doi: 10.1128/jb.171.6.2933-2941.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong H, Kirsebom L A, Nilsson L. J Mol Biol. 1996;261:303–308. doi: 10.1006/jmbi.1996.0461. [DOI] [PubMed] [Google Scholar]
- 18.Jahn O, Hartmann R K, Erdmann V A. Eur J Biochem. 1991;197:733–740. doi: 10.1111/j.1432-1033.1991.tb15965.x. [DOI] [PubMed] [Google Scholar]
- 19.Berkhout B, de Smit M H, Spanjaard R A, Blom T, van Duin J. EMBO J. 1985;4:3315–3320. doi: 10.1002/j.1460-2075.1985.tb04082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim M, Hyun Park B, Lee Y. Biochem Biophys Res Commun. 2000;268:118–123. doi: 10.1006/bbrc.2000.2084. [DOI] [PubMed] [Google Scholar]
- 21.Oshima T, Imahori K. Int J Syst Bacteriol. 1974;24:102–112. [Google Scholar]
- 22.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, et al. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 23.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, et al. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 24.Reysenbach A-L, Wickham G S, Pace N R. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, et al. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.