Abstract
Although distinct pathological stages of breast cancer have been described, the molecular differences among these stages are largely unknown. Here, through the combined use of laser capture microdissection and DNA microarrays, we have generated in situ gene expression profiles of the premalignant, preinvasive, and invasive stages of human breast cancer. Our data reveal extensive similarities at the transcriptome level among the distinct stages of progression and suggest that gene expression alterations conferring the potential for invasive growth are already present in the preinvasive stages. In contrast to tumor stage, different tumor grades are associated with distinct gene expression signatures. Furthermore, a subset of genes associated with high tumor grade is quantitatively correlated with the transition from preinvasive to invasive growth.
The current hypothesis of tumorigenesis in humans suggests that cancer cells acquire their hallmarks of malignancy through the accumulation of advantageous gene activation and inactivation events over long periods of time (1). For breast cancer development, this multistep process may manifest itself as a sequence of pathologically defined stages. It is widely held that breast cancer initiates as the premalignant stage of atypical ductal hyperplasia (ADH), progresses into the preinvasive stage of ductal carcinoma in situ (DCIS), and culminates in the potentially lethal stage of invasive ductal carcinoma (IDC) (for review, see ref. 2). This linear model of breast cancer progression has been the rationale for the use of detection methods such as mammography in the hope of diagnosing and treating breast cancer at earlier clinical stages (3). However, the stages of DCIS and IDC are heterogeneous with respect to mitotic activity and cellular differentiation both within a tumor and among individual tumors. To further characterize DCIS and IDC with respect to this heterogeneity, several tumor-grading systems have been created. Such systems are used clinically to subtype the stages of DCIS and IDC into three tumor grades in which grade I, II, and III lesions correspond to well, moderately, and poorly differentiated breast tumors, respectively (4, 5). Tumor grade has been a highly valuable prognostic factor for breast cancer, as poorly differentiated, high-grade DCIS or IDC lesions are associated with significantly poorer clinical outcome (4–6).
The molecular basis of breast tumorigenesis remains poorly understood. Although loss of heterozygosity and comparative genomic hybridization analyses have provided compelling evidence that ADH and DCIS are precursors to IDC (2, 7), such approaches have limited utility in identifying the biologically relevant genes that correlate with the different pathological stages. Genome-wide microarray-based gene expression analysis would be expected to provide a new opportunity to discover genes specifically activated or inactivated during the course of breast cancer progression. However, the application of such technology to interrogate the transcriptome in the different stages of breast tumorigenesis has been hampered by the microscopic size of the premalignant and preinvasive stages. The microscopic nature of these lesions precludes the use of traditional tissue RNA extraction techniques, as the contaminating cells that constitute the majority of a clinical sample compromise the resulting gene expression data. Therefore, to circumvent such issues, we and others have successfully combined the use of laser capture microdissection (LCM) and DNA microarray technologies to perform cellular-based, rather than tissue-based, gene expression profile analyses (8). The feasibility of this approach has been demonstrated with a limited number of breast specimens (9, 10). Herein, we applied this technology platform to a significantly larger cohort of breast cancer specimens to explore the gene expression changes that are associated with the various stages of breast cancer progression. Contrary to our initial expectation that the pathologically discrete stages (ADH, DCIS, and IDC) might be associated with unique gene expression signatures, we find that the three distinct stages of breast cancer are highly similar to each other at the level of the transcriptome. This finding supports the idea that the distinct stages of progression are evolutionary products of the same clonal origin, and that genes conferring invasive growth are active in the preinvasive stages. In addition, we provide evidence that different tumor grades are associated with distinct transcriptional signatures and that tumor grade is linked with the DCIS–IDC stage transition.
Materials and Methods
Clinical Specimen.
All breast specimens were obtained from the Massachusetts General Hospital between 1998 and 2001 (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). Thirty-six breast cancer patients were selected, 31 of whom were diagnosed with two or more pathological stages of breast cancer progression, and 5 of whom were diagnosed with preinvasive disease only. Three healthy women who underwent elective mammoplasty reduction were selected as disease-free normal controls. Tissue specimens that demonstrated one or more pathological lesions (ADH, DCIS, and IDC) were selected for the study. ADH cases were selected as proliferative epithelial lesions that possessed some, but not all, of the features of DCIS (11). DCIS was classified according to the European classification scheme (5): low-grade DCIS is characterized by a clinging, cribriform, or micropapillary proliferation of small, monomorphic cells with rare mitoses, whereas high-grade DCIS is characterized by a solid or clinging proliferation of large, pleomorphic cells with frequent mitoses. IDC was classified according to by the Nottingham combined histological grade (4). Estrogen receptor (ER) and progesterone receptor (PR) expression were determined by immunohistochemical staining (negative when none of the tumor cell nuclei showed staining), and Her-2 expression determined by immunohistochemistry or fluorescence in situ hybridization (FISH). This study was approved the Massachusetts General Hospital human research committee in accordance with National Institutes of Health human research study guidelines.
LCM and RNA Isolation and Amplification.
Normal, ADH, DCIS, or IDC was laser capture microdissected in triplicate (from consecutive tissue sections) as described (9) by using a PixCell II LCM system (Arcturus Engineering, Mountain View, CA). Patient-matched normal breast epithelium was microdissected from normal breast tissue that was at a minimum of 0.3 cm from any premalignant or malignant lesion. Malignant epithelial cells were microdissected from areas of tumor in which the tumor grade consisted of one type. Total RNA was extracted from the captured cells by using the Picopure RNA Isolation kit (Arcturus Engineering). T7-based RNA amplification was carried out by using the RiboAmp kit (Arcturus Engineering) according to the manufacturer's instructions. To obtain enough amplified RNA (aRNA) for a microarray experiment, a second round of RNA amplification was performed on all samples. To serve as reference in microarray hybridizations, a human universal reference RNA from Stratagene was amplified identically.
Fabrication of Microarrays.
Sequence-verified human cDNA clones were obtained from Research Genetics (Huntsville, AL). cDNA inserts were amplified by PCR, purified, and spotted onto a 1 × 3-inch SuperAmine (TeleChem International, Sunnyvale, CA) glass microscope slide by using an OmniGrid robotic arrayer (GeneMachines, San Carlos, CA).
Probe Labeling and Hybridization.
cDNA was transcribed from aRNA in the presence of 5-(3-aminoallyl)-2′-deoxyuridine 5′-triphosphate (aminoallyl dUTP) by using Stratagene's FairPlay kit. Cy3 or Cy5 mono-reactive dye (Amersham Pharmacia) was conjugated onto purified cDNA, and the residual dye was removed by using QiaQuick PCR Purification columns (Qiagen, Valencia, CA). Each Cy5-labeled cDNA was hybridized together with the Cy3-labeled reference probe to a microarray in 40 μl of hybridization solution (5× SSC/0.1 μg/μl COT I/0.2% SDS/50% formamide) at a concentration of 25 ng/μl per channel for 17 h at 42°C in >60% relative humidity.
Washing, Scanning, and Image Analysis.
After hybridization, slides were washed as follows: 1× SSC/0.2% SDS at 42°C for 5 min (two times), 1× SSC/0.2% SDS at 55°C for 5 min, 0.1× SSC/0.2% SDS at 55°C for 5 min, and 0.1× SSC at room temperature for 2 min. Washed slides were scanned by using ScanArray 5000 (Perkin–Elmer), and Cy5/Cy3-signals were quantitated by using IMAGENE 4.2 (BioDiscovery, Los Angeles).
Data Processing.
Fluorescent intensities of Cy5 and Cy3 channels on each slide were subjected to spot filtering and normalization. Spots flagged by imagene were excluded from further analysis. Normalization was performed by using a robust nonlinear local regression method (12). The normalized ratios of Cy5/Cy3 were used to represent the relative gene expression levels in the experimental samples. Measurements from replicate samples were averaged after normalization.
Data Analysis.
Hierarchical cluster analysis was performed in genemaths (v1.5, Applied-Maths, Austin, TX) by using correlation coefficient as measure of similarity between two genes or samples and complete linkage for clustering. Linear discriminant analysis with variance was performed within genemaths. The open source statistical environment R (www.r-project.org) and the Bioconductor packages were used for statistical analysis (13).
Quantitative Real-Time PCR (QRT-PCR) Analysis.
For the nonamplified RNA QRT-PCR validation study, we independently laser captured ≈40,000 normal breast epithelial cells from case 215, and ≈40,000 abnormal epithelial cells from DCIS (from cases 89, 178, 179) or IDC (from cases 97, 169, 170). Total RNA was isolated and converted to double-stranded cDNA. For the validation studies using aRNA, 2 μg of aRNA from each microdissected sample was converted into double-stranded cDNA. In all cases (cDNA derived from nonamplified and amplified RNA), the double-stranded cDNA was quantitated with PicoGreen (Molecular Probes) by using a spectrofluorometer (Molecular Devices) and quantitative analysis of gene expression (QRT-PCR) was performed with an ABI 7900HT (Applied Biosystems) as described (9). Each reaction was performed in triplicate by using 2.5 ng of cDNA from each sample as template. The relative standard curve method was used for linear regression analysis of unknown samples (9) and data presented as fold change between samples. The sequences of the PCR primer pairs and fluorogenic probes for each gene are in Table 3, which is published as supporting information on the PNAS web site.
In Situ Hybridization.
Frozen sections of breast carcinoma or normal breast were cut at five microns and fixed in 10% neutral buffered formalin. Digoxigenin (DIG)-labeled sense and antisense CRIP1 RNA probes were applied to the tissue sections, covered with a glass coverslip, and sealed. The slides were heated to 85°C for 5 min and hybridized overnight at 60°C. After stringent wash conditions at 60°C, the DIG-labeled RNA was detected by using rabbit anti-DIG-Alkaline Phosphotase (DAKO, catalog no. 5105). Fast red (DAKO, catalog no. K0597) was used as substrate and the slides were counterstained with hematoxylin.
Results
LCM-Based Approach to Gene Expression Profiling.
To determine the gene expression profiles of the premalignant, preinvasive and invasive stages of breast cancer progression, we integrated the use of LCM and T7-based RNA amplification with DNA microarrays. LCM provides a highly accurate means by which to procure the specific cells that constitute the different stages of progression and avoids contamination by surrounding stromal and inflammatory cells (Fig. 1). This allows the subsequent gene expression profiles to be obtained with cellular-based, rather than tissue-based, resolution. As shown in Fig. 1, all three stages of breast cancer progression plus the adjacent normal epithelium were laser-capture microdissected from the same clinical specimen (patient case 79). Because the numbers of cells that constitute the early stages of breast cancer are often limited in a clinical biopsy, we used T7-based linear RNA amplification as a means to obtain sufficient amounts of RNA for microarray analysis. Using this approach, we routinely generated >40 μg of aRNA from 2,000 to 2,500 captured cells. To address whether linear RNA amplification accurately preserves the differences in mRNA abundance between samples, we conducted QRT-PCR analysis comparing nonamplified total RNA with the corresponding amplified RNA from captured normal breast epithelium (from case 215) and DCIS (from cases 89, 178, and 179) or IDC (from cases 97, 169, and 170). Based on our initial microarray experiments, we selected three genes (CRIP1, IFI-6–16, PNMT) that are up-regulated and two genes (ELF5, NDRG2) that are down-regulated in breast cancer relative to normal breast epithelium for the QRT-PCR validation experiment. We calculated the gene expression ratios of DCIS or IDC vs. normal epithelium for each of these five genes using either the amplified total RNA or nonamplified RNA. Linear regression analysis of the gene expression data derived from nonamplified RNA and amplified RNA across six different clinical specimens demonstrates an R2 value of 0.96 (Fig. 5, which is published as supporting information on the PNAS web site). This result indicates that T7 amplification of RNA is highly accurate in preserving the differential gene expression between samples. Therefore, the combined use of LCM and T7-based RNA amplification represents a reliable approach to gene expression profiling at the level of cellular resolution.
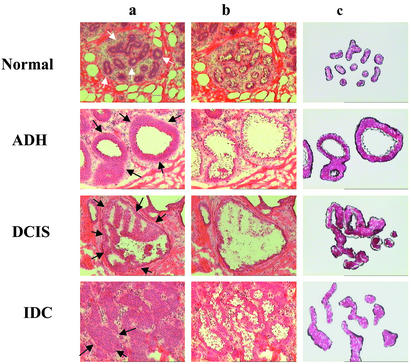
Figure 1.
LCM. Phenotypically normal breast epithelium (white arrows) and phenotypically abnormal epithelium (black arrows) from ADH, DCIS, and IDC from a single breast specimen (case 79) were captured from hematoxylin and eosin-stained sections (8 μm). Images of precapture (lane a), postcapture (lane b), and the captured epithelial compartments (lane c) are shown.
Gene Expression Profiles of Breast Tumor Stages.
We selected 36 different clinical breast cancer specimens, 31 of which contain two or more synchronous pathological stages of breast cancer progression and 32 of which contain normal breast tissue (Table 2). We microdissected from each cancer specimen phenotypically abnormal epithelial cells constituting the different stages of breast cancer progression and phenotypically normal epithelial cells constituting the terminal duct lobular unit (TDLU), the anatomic substructure from which breast cancer arises (14) (see Fig. 1). In addition, we microdissected phenotypically normal TDLU breast epithelial cells from three mammoplasty reduction specimens that serve as noncancerous normal breast controls. Our intent in this study was to discover the most consistently up- or down-regulated genes at each stage of disease progression in all patients.
The distinct components (normal, ADH, DCIS, or IDC) within each clinical specimen were microdissected in triplicate (Table 2), resulting in a total of 300 samples. Each of the independently captured samples was interrogated in duplicate with a 12,000-gene cDNA microarray, generating >7 × 106 data points from 600 microarrays. On the basis of data suggesting that closely adjacent breast cancer and phenotypically normal terminal duct lobular units may share loss of heterozygosity for certain genes (15), we first performed a cluster analysis to determine whether the patient-matched phenotypically normal epithelium from cancer specimens is equivalent to that derived from noncancerous specimens. This analysis demonstrated that all patient-matched “normal” samples are highly similar to those from the mammoplasty reduction specimen normal breast controls (data not shown). Although this does not rule out the presence of subtle differences between mammoplasty reduction specimens and disease-matched “normals,” it suggests that at a broad level, patient-matched normal breast epithelium can serve as an appropriate baseline control for evaluating tumor progression. Next, to focus our analysis on those genes that are consistently expressed differentially between normal and various neoplastic lesions, we carried out linear discriminant analysis of pair-wise comparisons of normal vs. ADH, normal vs. DCIS, and normal vs. IDC by using the software genemaths (v.1.5). Genes with large discriminant function coefficients (>0.5) were selected, resulting in a total of 1,940 genes for further exploration (Table 4, which is published as supporting information on the PNAS web site).
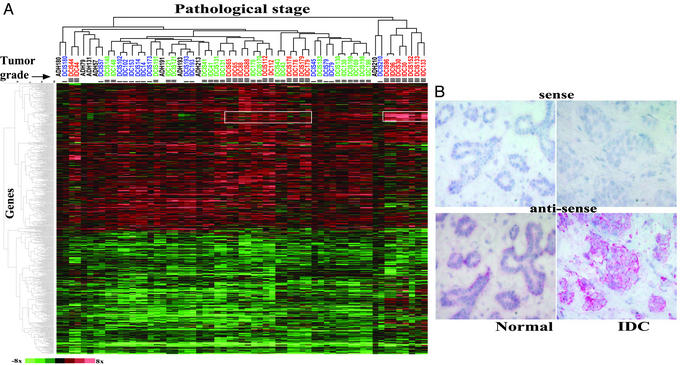
One important advantage of our LCM-based approach is the ability to procure both normal and diseased cell populations from the same biopsy. Therefore, we represented the expression level of each gene in a disease state as the ratio to the patient-matched normal. Hierarchical clustering of the 61 distinct tumor stage samples and the 1,940 genes reveals two main clusters (Fig. 2A). One cluster demonstrates increased expression in a majority of the diseased samples, and another cluster shows a relatively uniform decrease in expression across all samples. Most of these alterations (both increases and decreases) occur at the earliest pathologically defined stage, ADH, and such alterations generally persist throughout the later stages of DCIS and IDC. However, on hierarchical clustering of all samples, different stages did not form distinct groups. Instead, the different synchronous stages of progression within an individual patient cluster more closely to one another than to their respective stage from different patients (Fig. 2A, e.g., cases 193, 44, 210, and 179). In addition, this analysis reveals a pattern of gene expression that correlates with high tumor grade (Fig. 2A, white outlined rectangles).
Figure 2.
Expression profiles of breast cancer progression. (A) Two-dimensional hierarchical clustering of the data matrix consisting of 1,940 genes by 61 samples of different pathological stages (Table 4). Rows represent genes and columns represent samples, which are color-coded by tumor grade (blue, green, and red correspond to grades I, II, and III, respectively). Color scale is shown at bottom left. (B) In situ hybridization of CRIP1 mRNA. DIG-labeled RNA probes from both the antisense and sense (negative control) strands of CRIP1 transcript were hybridized to sections of normal and IDC components of case 179. Hybridization signals were visualized by alkaline phosphatase-conjugated anti-DIG antibody using fast red as substrate.
To confirm our microarray findings, we performed QRT-PCR analysis of several differentially expressed genes. The QRT-PCR data correlate well with the patterns of expression revealed by microarray analysis, and two examples include those for CRIP1 and ELF5. In agreement with the microarray results, QRT-PCR demonstrated over-expression of CRIP1 (>2-fold) in seven of eight ADH, 27 of 30 DCIS, and 23 of 25 IDC cases, and under-expression of ELF5 (>2-fold) in seven of eight ADH, 28 of 30 DCIS, and 25 of 25 IDC cases (see Fig. 6, which is published as supporting information on the PNAS web site). In addition, we performed in situ hybridization for CRIP1 to confirm its cellular specificity. As expected from the use of LCM, CRIP1 signal localized to the epithelial cells, and its intensity was markedly increased in the IDC compartment of the same biopsy (Fig. 2B), thus verifying the microarray-derived results at the level of cellular resolution.
Gene Expression Profiles of Breast Tumor Grades.
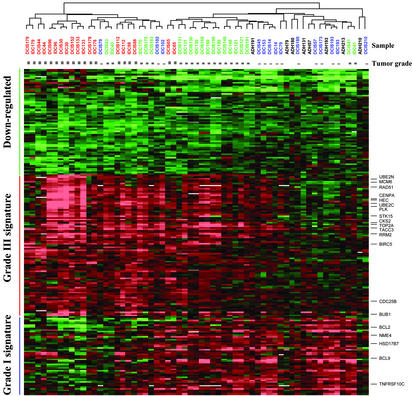
Although stage-specific gene expression patterns are not readily discerned in Fig. 2, a gene cluster characterized by elevated levels of expression in grade III tumors as compared with grade I tumors is apparent (Fig. 2, see white rectangles). To examine this observation more closely, we performed discriminant analysis and extracted two sets of genes, each comprising the top 100 genes correlating with grade I and grade III samples, respectively, from the 1,940 gene set (Table 5, which is published as supporting information on the PNAS web site). To test for statistical significance of these genes, we calculated t statistic for each gene comparing grade I and grade III samples. P values were estimated based on 6,000 permutations of the original data set. To correct for multiple hypothesis testing, we obtained adjusted P values by using the Benjamini and Hochberg false discovery rate procedure (16). This analysis indicates that these 200 genes are all significant at the level of P < 0.01 (Table 5). Gene expression values were expressed as ratios of ADH, DCIS, or IDC to the patient-matched normal and two-dimensional clustering analysis was performed revealing three major gene clusters (Fig. 3). One cluster of genes demonstrates decreased expression in all samples with subtle quantitative differences between grade I and grade III (green bar). A second cluster of genes (denoted as the grade III signature) shows markedly increased expression in grade III samples (red bar), whereas a third cluster (grade I signature) demonstrates increased expression primarily in grade I samples (blue bar). Most striking is the existence of reciprocal gradients in the intensities of the grade I and grade III signatures (Fig. 3). Notably, most grade II lesions exhibit a hybrid of grade I and grade III signatures (e.g., cases 130, 169, and 198). Some grade II lesions, however, show an expression pattern that is most similar to either grade I or grade III lesions (cases 41 and 43, respectively), and a few grade III samples demonstrate coexpression of some genes that are characteristic of the grade I signature (cases 65, 88, and 112). All ADH samples demonstrate a grade I gene expression signature and cluster with the low-grade DCIS and IDC samples.
Figure 3.
Two-dimensional clustering of 61 samples and the top 200 genes correlating with tumor grade. Genes (rows) and samples (columns) were clustered independently by hierarchical clustering. Three main clusters are highlighted by color bars. See Fig. 2A for color scale and designations. The original data are in Table 5.
The Relationship of the DCIS–IDC Stage Transition to Tumor Grades.
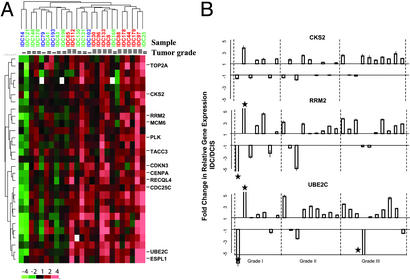
Thus far, we have demonstrated that the greatest alterations in gene expression are seen among the different histological grades of breast cancer, and that no consistent gene expression alterations unique to each of the three different pathological stages are readily apparent (Figs. 2 and 3). This finding is consistent with a previous study indicating that expression of several prominent tumor markers (p53, ERBB2, Ki-67, ER, PR, and bcl2) correlate with tumor grade but not with the distinction between DCIS and IDC (17). Nonetheless, it is of great interest to understand the transcriptional program that drives invasive growth due to its clinical importance. Therefore, we tested the possibility that the DCIS–IDC transition may be associated with subtle quantitative differences in gene expression. Relative gene expression of the 1,940 gene set between IDC and DCIS were calculated for the 25 patient-matched pairs. Two-dimensional clustering of the resulting data set identified a cluster of 29 genes consistently over-expressed in IDC relative to its matched DCIS, especially in grade III samples (Fig. 4A, and Table 6, which is published as supporting information on the PNAS web site). To examine this observation more closely, we applied a paired t test to the 11 grade III DCIS-IDC pairs and identified 85 genes with increased expression in IDC (P < 0.01, one-sided; Table 1. Adjusted P values using the Benjamini and Hochberg false discovery rate procedure (16) detected 15 or 50 of these genes at the significance level of 0.05 or 0.1, respectively (Table 7, which is published as supporting information on the PNAS web site). The lack of statistical significance of many of the 85 genes after correction for multiple testing is likely due to the small sample size (n = 11). Nevertheless, remarkably, 39 of these genes are also found within the 100-gene grade III signature (P value <2.2e-16, χ2 test; Table 2). These include genes involved in the cell cycle (e.g., MCM6, TOP2A, CKS2, CDC25C, and UBE2C), centrosomal function (TACC3, CENPA, and STK15), and DNA repair (RAD51 and RRM2). The higher levels of gene expression in IDC vs. DCIS were verified for three (CKS2, RRM2, and UBE2C) of these genes by QRT-PCR; quantitative increases in gene expression in IDC occurred in a majority of these cases, especially in grade III cases (Fig. 4B). Thus, a significant subset of genes that are expressed at higher levels in grade III DCIS relative to grade I DCIS are further elevated in IDC, revealing an apparent link between tumor grade and stage progression.
Figure 4.
Genes with increased expression in IDC relative to DCIS. (A) Cluster of 29 genes showing consistent up-regulation in IDC. Expression values are expressed as log2-ratios of expression in IDC to that in patient-matched DCIS. Color scheme is shown at bottom left; see Fig. 2A for sample color designations. The original data are in Table 5. (B) Confirmation by QRT-PCR of increased expression in IDC for CKS2, RRM2, and UBE2C. Fold changes from DCIS to IDC and associated standard errors are plotted. Data shown are averages of triplicate QRT-PCR measurements. Values outside the scale in the y axis are marked by a star. The patient case numbers from left to right are: 14, 25, 79, 102, 173, 180, and 193 for grade I; 41, 43, 121, 130, 131, 148, 169, 170, and 198 for grade II; and 8, 30, 44, 65, 88, 96, 112, 133, 178, and 179 for grade III.
Table 1.
Genes with increased expression both in high tumor grade and the DCIS-IDC transition
| Clone ID | t statistic | P value | Adj. P value* | Description |
|---|---|---|---|---|
| 869375 | 6.86 | 2.20E-05 | 0.02683578 | IDH2 | isocitrate dehydrogenase 2 |
| 504308 | 6.46 | 3.60E-05 | 0.02683578 | FLJ10540 | hypothetical protein |
| 825606 | 5.95 | 7.10E-05 | 0.02683578 | KNSL1 | kinesin-like 1 |
| 951241 | 5.92 | 7.40E-05 | 0.02683578 | ANKT | nucleolar protein ANKT |
| 280375 | 5.58 | 1.20E-04 | 0.0304866 | PRO2000 | PRO2000 protein |
| 564981 | 5.58 | 1.20E-04 | 0.0304866 | Similar to RIKEN cDNA 2810433K01 |
| 1476053 | 5.4 | 1.50E-04 | 0.03425014 | RAD51 |
| 769921 | 4.97 | 2.80E-04 | 0.04935153 | UBE2C | ubiquitin-conjugating enzyme E2C |
| 128711 | 4.72 | 4.10E-04 | 0.04935153 | ANLN | anillin, actin binding protein |
| 814270 | 4.61 | 4.80E-04 | 0.05092141 | PMSCL1 | polymyositis/scleroderma autoantigen 1 |
| 209066 | 4.58 | 5.00E-04 | 0.05092141 | STK15 | serine/threonine kinase 15 |
| 2017415 | 4.52 | 5.60E-04 | 0.05338307 | CENPA | centromere protein A |
| 823598 | 4.28 | 8.10E-04 | 0.0703179 | PSMD12 | proteasome 26S subunit |
| 878330 | 4.17 | 9.60E-04 | 0.07962393 | EST |
| 785368 | 4.05 | 1.20E-03 | 0.08461982 | TOPK | PDZ-binding kinase |
| 839682 | 3.96 | 1.30E-03 | 0.08461982 | UBE2N | ubiquitin-conjugating enzyme E2N |
| 756595 | 3.89 | 1.50E-03 | 0.08461982 | S100A10 | S100 calcium-binding protein A10 |
| 347373 | 3.86 | 1.60E-03 | 0.08461982 | TCEB1 | transcription elongation factor B |
| 624627 | 3.84 | 1.60E-03 | 0.08461982 | EST |
| 1517595 | 3.84 | 1.60E-03 | 0.08461982 | RRM2 | ribonucleotide reductase M2 |
| 825470 | 3.83 | 1.70E-03 | 0.08461982 | TOP2A | topoisomerase (DNA) II alpha |
| 259950 | 3.67 | 2.20E-03 | 0.08771114 | CML66 | CML tumor antigen 66 |
| 292936 | 3.67 | 2.20E-03 | 0.08771114 | FLJ10468 | hypothetical protein |
| 1416055 | 3.67 | 2.20E-03 | 0.08771114 | KIAA0165 | homolog of yeast extra spindle poles |
| 744047 | 3.55 | 2.60E-03 | 0.09967783 | PLK | polo-like kinase |
| 705064 | 3.44 | 3.20E-03 | 0.10860726 | TACC3 |
| 2322367 | 3.44 | 3.20E-03 | 0.10860726 | RTN4 | reticulon 4 |
| 66406 | 3.39 | 3.50E-03 | 0.116593 | EST |
| 462926 | 3.36 | 3.60E-03 | 0.11872875 | NEK2 | NIMA-related kinase 2 |
| 815501 | 3.3 | 4.00E-03 | 0.12440908 | MGC2721 | hypothetical protein |
| 1035796 | 3.21 | 4.60E-03 | 0.13817458 | EST |
| 700792 | 3.15 | 5.10E-03 | 0.14848582 | CDKN3 | cyclin-dependent kinase inhibitor 3 |
| 2018131 | 3.09 | 5.80E-03 | 0.16117884 | RACGAP1 | Rac GTPase activating protein 1 |
| 743810 | 3.03 | 6.30E-03 | 0.17362215 | MGC2577 | hypothetical protein |
| 781047 | 2.94 | 7.40E-03 | 0.18297681 | RRM2 | ribonucleotide reductase M2 |
| 1422338 | 2.94 | 7.40E-03 | 0.18297681 | BUB1 |
| 796694 | 2.92 | 7.70E-03 | 0.18720879 | BIRC5 | survivin |
| 725454 | 2.87 | 8.30E-03 | 0.18975804 | CKS2 | CDC28 protein kinase 2 |
Paired t test was performed on 11 patient-matched DCIS–IDC pairs to identify genes with increased expression in IDC. A total of 85 genes were identified at one-sided P < 0.01 (Table 7).
Adjusted P value by the Benjamini and Hochberg procedure. The 39 genes shown here are those also identified in the 100-gene grade III signature. Genes in bold are validated by QRT-PCR (Fig. 4B).
Discussion
A major challenge to human breast cancer research has been the characterization of the molecular events that are associated with breast cancer progression. Progress in achieving this goal has been hindered by the practical aspects of applying advanced molecular techniques to the microscopic premalignant and preinvasive stages of breast cancer. In this study, we have successfully combined laser capture microdissection, RNA amplification, and microarray technologies to generate epithelial-specific, in situ gene expression profiles of the premalignant, preinvasive, and invasive stages of breast cancer. In doing so, we were able to study the interrelationship of the phenotypically distinct stages of breast cancer progression within an individual patient and between different patients with breast cancer.
One surprising result from this study was the remarkable similarity in the expression profiles of the distinct pathological stages (Fig. 2A). As compared with patient-matched normal epithelium, significant global alterations in gene expression occur at ADH, the earliest phenotypically recognized stage of progression, and such alterations are maintained in the later stages of DCIS and IDC. Although unexpected, these findings are consistent with those generated from the analysis of a limited cohort of DCIS and IDC specimens (18). Our observations are also consistent with earlier loss of heterozygosity/comparative genomic hybridization studies in which the many genetic abnormalities associated with DCIS and IDC were also shown to be present in ADH (2, 7, 15, 19). Together, these data suggest a clonal relationship between the distinct pathological stages. Furthermore, these results suggest that the gene expression profile of early stage disease may, in fact, reflect the progressive potential of the pathological lesion. This hypothesis is supported by a recent report in which breast cancer metastatic potential could be reliably predicted from the gene expression profile of the primary tumor (20).
Although our analysis did not identify gene expression differences that are specific to the distinct pathological stages of breast cancer, distinct gene expression alterations were found to be associated with different tumor grades. Grade I and grade III breast tumors exhibit reciprocal gene expression patterns, whereas grade II tumors exhibit a hybrid pattern of grade I and grade III signatures. Together, these unique signatures may underlie the molecular basis of the current pathological grading systems for breast cancer. Such systems rely mainly on histomorphological criteria, which, although highly successful in discriminating grade I from grade III tumors, are inadequate to score grade II tumors consistently (4). This difficulty may be explained by the overlapping nature of grade II signature with those of grade I and grade III. There are at least two possibilities regarding the hybrid nature of the grade II signature. First, the hybrid signature may simply reflect a mixture of grade I and grade III cells in the samples. Second, this hybrid signature may reflect a transition state from the grade I to grade III tumor. Although conceivable, the first possibility is less likely as we specifically dissected cells from areas of morphologically uniform tumor grade. Based on our observation that some grade II tumor samples clustered with either grade I or grade III tumors (Fig. 3, cases 41 and 43), we suggest that a gene expression-based molecular grading system may allow for greater precision in classifying breast cancer.
Our analysis also identified a subset of genes with quantitative expression levels that correlate with advanced tumor grade and with the transition from DCIS to IDC (Fig. 4A and Table 2). This observation is provocative in that it provides an apparent link between tumor stage and tumor grade. Support for this finding is provided by the clinical observation that poorly differentiated (grade III) DCIS is more likely to be associated with occult invasive disease than its well-differentiated (grade I) counterparts (21). Therefore, our data suggest that the transcriptional program that drives cancer cells to an advanced tumor grade may also confer invasiveness. RRM2, a gene we identified correlating with both advanced tumor grade and stage (see Table 2), may serve as an example of such a possibility. RRM2 is the rate-limiting component for the conversion of ribonucleotides to deoxyribonucleotides during DNA synthesis; increased RRM2 expression is thus expected to support the rapid cell division of high grade tumors. Unexpectedly, RRM2 also cooperates with a wide variety of oncogenes (H-ras, rac-1, v-fms, v-src, A-raf, v-fes, and c-myc) in promoting malignancy and metastatic potential (22, 23). Therefore, RRM2 may play a dual role in supporting rapid cell proliferation on the one hand and promoting invasive growth behavior on the other, thus linking higher tumor grade (higher proliferation rate) and the DCIS–IDC transition (invasion). Further elucidation of the signaling pathways that link these genes to the process of tumor grade and stage progression may provide key insights into the molecular mechanism driving breast tumorigenesis.
Supplementary Material
Acknowledgments
We thank D. Haber, D. Louis, F. Koerner, E. Schmidt, and W. Wang for helpful discussions and reading of the manuscript. We also thank Ana Sollberger, Amber Kahle, and Katrina Mesina for technical assistance. This work is supported in part by a grant from the Massachusetts Department of Public Health Breast Cancer Program, a collaborative grant from the Dana–Farber/Partners Cancer Care Women's Cancer Program, and a grant from the Avon Foundation (to D.C.S.).
Abbreviations
- LCM
laser capture microdissection
- CGH
comparative genomic hybridization
- QRT-PCR
quantitative real-time PCR
- ADH
atypical ductal hyperplasia
- DCIS
ductal carcinoma in situ
- IDC
invasive ductal carcinoma
- DIG
digoxigenin
- aRNA
amplified RNA
References
- 1.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Allred D C, Mohsin S K, Fuqua S A. Endocrinol Relat Cancer. 2001;8:47–61. doi: 10.1677/erc.0.0080047. [DOI] [PubMed] [Google Scholar]
- 3.Tabar L, Dean P B, Kaufman C S, Duffy S W, Chen H H. Surg Oncol Clin North Am. 2000;9:233–277. [PubMed] [Google Scholar]
- 4.Dalton L W, Pinder S E, Elston C E, Ellis I O, Page D L, Dupont W D, Blamey R W. Mod Pathol. 2000;13:730–735. doi: 10.1038/modpathol.3880126. [DOI] [PubMed] [Google Scholar]
- 5.Holland R, Peterse J L, Millis R R, Eusebi V, Faverly D, van de Vijver M J, Zafrani B. Semin Diagn Pathol. 1994;11:167–180. [PubMed] [Google Scholar]
- 6.Page D L, Gray R, Allred D C, Dressler L G, Hatfield A K, Martino S, Robert N J, Wood W C. Am J Clin Oncol. 2001;24:10–18. doi: 10.1097/00000421-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell P, Pekkel V, Fuqua S A, Osborne C K, Clark G M, Allred D C. J Natl Cancer Inst. 1998;90:697–703. doi: 10.1093/jnci/90.9.697. [DOI] [PubMed] [Google Scholar]
- 8.Luo L, Salunga R C, Guo H, Bittner A, Joy K C, Galindo J E, Xiao H, Rogers K E, Wan J S, Jackson M R, Erlander M G. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- 9.Sgroi D C, Teng S, Robinson G, LeVangie R, Hudson J R, Jr, Elkahloun A G. Cancer Res. 1999;59:5656–5661. [PubMed] [Google Scholar]
- 10.Luzzi V, Holtschlag V, Watson M A. Am J Pathol. 2001;158:2005–2010. doi: 10.1016/S0002-9440(10)64672-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page D L, Rogers L W. Hum Pathol. 1992;23:1095–1097. doi: 10.1016/0046-8177(92)90026-y. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y H, Dudoit S, Luu P, Lin D M, Peng V, Ngai J, Speed T P. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ihaka R, Gentleman R. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 14.Wellings S R, Jensen H M. J Natl Cancer Inst. 1973;50:1111–1118. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- 15.Deng G, Lu Y, Zlotnikov G, Thor A D, Smith H S. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 16. Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. B, 289–300.
- 17.Warnberg F, Nordgren H, Bergkvist L, Holmberg L. Br J Cancer. 2001;85:869–874. doi: 10.1054/bjoc.2001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porter D A, Krop I E, Nasser S, Sgroi D, Kaelin C M, Marks J R, Riggins G, Polyak K. Cancer Res. 2001;61:5697–5702. [PubMed] [Google Scholar]
- 19.Zhuang Z, Merino M J, Chuaqui R, Liotta L A, Emmert-Buck M R. Cancer Res. 1995;55:467–471. [PubMed] [Google Scholar]
- 20.van 't Veer L J, Dai H, van de Vijver M J, He Y D, Hart A A, Mao M, Peterse H L, van der Kooy K, Marton M J, Witteveen A T, et al. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 21.de Mascarel I, MacGrogan G, Mathoulin-Pelissier S, Soubeyran I, Picot V, Coindre J M. Cancer. 2002;94:2134–2142. doi: 10.1002/cncr.10451. [DOI] [PubMed] [Google Scholar]
- 22.Fan H, Villegas C, Wright J A. Proc Natl Acad Sci USA. 1996;93:14036–14040. doi: 10.1073/pnas.93.24.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan H, Villegas C, Huang A, Wright J A. Cancer Res. 1998;58:1650–1653. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.