Abstract
Strong species specificity and similar tropisms suggest mouse cytomegalovirus (mCMV) as a potential vector for transgenes into human cells. We reexamined the dogma that mouse cytomegalovirus cannot productively replicate in human cells and found that mouse cytomegalovirus can produce infectious particles albeit at a level that does not sustain an infection. This finding demonstrates that mouse cytomegalovirus can undergo all processes of its life cycle in human cells but may not be well adapted to circumvent the human cell's intrinsic defenses. The suppression of mCMV production in human cells is affected at several levels, which additively or synergistically result in the appearance of species specificity. Hydrolysis of most newly replicated viral DNA and very low capsid protein transcription reduced the potential particle production to insignificant levels. These effects can be ameliorated by adding human cytomegalovirus tegument proteins and immediate-early protein 1. They function synergistically to produce significant amounts of mCMV in human cells. While the possibility that mouse cytomegalovirus might replicate in human cells raises caution in the use of this virus as a transgene vector, manipulation of the mouse cytomegalovirus genome to allow limited spread to other human cells might also provide an advantage for the distribution of certain transgenic products.
Cytomegaloviruses (CMV) are a widespread subgroup of Herpesviridae. These large double-stranded DNA viruses with genomes of approximately 230 kb are well adapted to their respective hosts and cause only mild symptoms in the immunocompetent host. Strict species specificity has been recognized in cross-infections between mouse CMV (mCMV) and human CMV (hCMV), the two most extensively investigated viruses in this family (23, 24, 34). This apparently absolute species specificity is not due to incompatibilities during infection since adsorption and fusion with the cell membrane and viral DNA penetration of the nuclear compartment take place, with production of hCMV immediate-early protein 1 (IE1) and IE2 and the mCMV equivalents IE1 and IE3 in mouse and human cells, respectively (24). Thus, any block in the progression of the transcription cascade or viral DNA replication must occur at early or late times in the progression to viral particle formation. In vivo, no successful cross-species infection is recognized.
The host has evolved various defenses against invading viruses, most prominently the adaptive and innate immune responses. Viruses have developed counterdefenses, and the larger DNA viruses contain a formidable array of countermeasures that subvert antigen presentation or recognition (14, 29, 34). Once inside the nucleus, the foreign DNA may be recognized and destroyed by nucleases or may become chromatinized. Chromatinization can lead to silencing of the viral genome by enzymatic complexes containing histone deacetylases (HDACs) (30, 31, 39). We have identified some of these intrinsic defense mechanisms and hypothesize that the virus has developed immediate-early-mediated countermeasures against these functions by segregating specific proteins such as HDACs and the repressor protein Daxx (31, 39). For CMV, a standoff has evolved wherein the normal host suffers no serious effects while the virus resides in a latent state in various cell types, periodically reactivating and spreading.
Intrinsic defenses and counterdefenses may function in direct recognition of foreign or damaged DNA and its destruction and in the viral use of cell DNA repair mechanisms (26, 44). Silencing of the viral genome by HDACs and a viral segregation of the HDACs by IE1 may represent another dyad that has evolved into a balance between virus and host. Adaptations to numerous sets of host defenses have made CMV infection successful. These adaptations include molecular optimization for enhancers (22), use of tegument proteins to modify the cellular environment by repression of antiviral protein transcription (interferons), specific signaling and cell cycle regulation (6, 42), and proteosome inhibition (11). In cross-species infections, some viral and host protein interactions may not take place so that the unimpaired human intrinsic defenses can totally repress mCMV infection. Alternatively, the adaptation of viral counterdefenses or host cell modification by viral proteins may be less than optimal at different levels, with only their additive effects producing apparent incompatibility. Proof of the latter possibility would come from the finding that a decreased level of defense or modification of the cellular environment results in mCMV production in human cells. Tropism or permissiveness in the appropriate host may represent a subset of similar components where necessary cellular components are abundant or strong defenses absent or where essential transcription factors are at suboptimal levels in specific cell types. We tested these possibilities since some of the mCMV properties relative to human cells make this virus a desirable vector. Use of any vector in humans must be based on a thorough understanding of its biological consequences with respect to its utility for long-term production of an effector protein or for short-term but repeated production of a desired antigen and most importantly with respect to risk.
Several viruses have been used as vehicles for immunization or gene therapy. In most cases, the viral vector must be disabled to prevent replication. Moreover, previous immunity to a human vector, such as adenovirus type 5 (Ad5), can be bypassed by using the same type of virus from another species, as has been accomplished successfully with the simian Ad5 vector (32). Additional viral vectors are desirable since immunity that develops prevents reuse of the original ones. mCMV has recently been suggested as a viral vehicle for human immunodeficiency virus immunization (43). An mCMV vector encoding the human immunodeficiency virus type 1 gp120 envelope glycoprotein has been used to infect cultured human dendritic cells, which are not easily infected by Ad5 due to lack of the primary receptor for Ad5 (33). Whereas hCMV downregulates the response of dendritic cells of the human host (17), mCMV-infected human dendritic cells are capable of stimulating the expansion of autologous, gp120-specific, class I-restricted T lymphocytes (43). mCMV has the added advantage of strong species specificity for productive infection. mCMV can also abortively infect any human cell type tested (25, 41), can express transgenes (40), and can potentially carry rather large DNA cargo since its genome is large and only about 80 genes appear to be necessary for its replication (L. Cicin-Sain et al., 10th International CMV/Betaherpesvirus Workshop, Williamsburg, Va., 24 to 28 April 2005). These properties make mCMV a desirable vector for transgene delivery.
Here, we provide evidence that there are no fundamental blocks for mCMV transcription, translation, DNA replication, or virus production in human cells. Instead, the results indicate that the lack of mCMV IE1 adaptation to counter certain human host defenses as well as lack of tegument-based changes of the human cell environment decreases the probability of mCMV production to insignificant levels.
MATERIALS AND METHODS
Cells and viruses.
The following cell lines were used: NIH 3T3 (ATCC), Daxx−/− 227 (21), HeLa (ATCC), human foreskin fibroblast, U373MG (ATCC), hCMV IE1-producing U373MG (U373-IE1) (10), hCMV IE1-producing human fibroblast ihfie1.3, provided by E. S. Mocarski (13), and Ad5 E1A-transformed human epithelial kidney HEK293 (ATCC). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% penicillin-streptomycin. For immunohistochemical staining, cells were grown on round coverslips (Corning Glass Inc., Corning, NY) in 24-well plates (Falcon; Becton Dickinson Labware, Lincoln Park, NJ). The mCMV Smith strain was obtained from M. Messerle (3), and green fluorescent protein (GFP)-tagged mCMV RVG102 was a generous gift from J. D. Hamilton (16). hCMV Towne strain (ATCC) inactivation by UV was carried out by 15 min of exposure of plates containing 107 PFU/ml to a 30-W UV light source at a distance of 6 cm. The MG-IE1 recombinant mCMV was generously provided by Wolfgang Brun, Berlin, Germany.
Antibodies.
The ND10-associated protein PML was identified by rabbit serum R14 produced against the N-terminal half of PML and monoclonal antibodies (MAb) against PML (S. Lowe, Cold Spring Harbor, NY). Rabbit antibody against HDAC2 was purchased from Zymed Laboratory, Inc. (San Francisco, CA), and MAb against tubulin was from Sigma Co. (St. Louis, MO). MAb against mCMV IE1 and M112/113 (E1) were generously provided by S. Jonjic (Croatia). Rabbit antibody against E1 (EVMS 55B) was obtained from J. Kerry (9), and rabbit antibody against M141 was generously supplied by A. Campbell (15).
Plaque formation assay.
Viral titers were determined by plaque assay as described previously (39), with slight modifications. Supernatants of cells infected for various times were serially diluted and added to confluent mouse 227 cell monolayers in six-well plates. After absorption for 2 h, medium was removed and cells were washed twice with serum-free DMEM and overlaid with phenol-free DMEM containing 5% FCS, 0.5% low-melting-point agarose (GIBCO), and 1% penicillin-streptomycin. Mean PFU were determined after averaging from different dilutions. In separate experiments, mixed cell cultures were used to detect production of mCMV from human cells. 3T3 cells and human cell lines were infected with wild-type mCMV or RVG102 at a multiplicity of infection (MOI) of 1 PFU/cell in six-well plates. At 24 h postinfection (p.i.), infected cells were washed twice with DMEM (no serum), treated with trypsin-EDTA (GIBCO), and resuspended in 100 μl DMEM (no serum). Infected cells (∼104 cells in 10 μl) were mixed with ∼2 × 106 uninfected 227 cells (1:200 ratio) and cultured in duplicate six-well plates in DMEM with 10% FCS for 48 h. The remaining 80 μl of infected cells was freeze-thawed three times, and the centrifuged supernatants were used to infect 227 cells as a control for removing all input virus. Plaques formed on the mouse cells were then quantitated.
Microarray analysis.
Slides were printed with mCMV-derived 60-mer oligonucleotides at the Microarray Facility of the University of Pennsylvania (Philadelphia, PA) (39a). Total RNAs were isolated from infected cells by use of TRI reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was reverse transcribed, purified, and labeled with Cy3 by use of a SuperScript indirect cDNA labeling system (Invitrogen, Carlsbad, CA). cDNAs were hybridized with the oligoarrays. Hybridization and washing were accomplished using a Tecan HS4800 hybridization station (Tecan Austria GmbH) according to the manufacturer's protocol. Slides were scanned with a microarray-equipped GenePix Pro600 microarray scanner (Union City, CA), and data were analyzed using GenePix software (Union City, CA). Array analysis was carried out twice on slides that carried six sets of all oligonucleotides, and the data were averaged.
Reverse transcription-PCR (RT-PCR).
Total RNAs were isolated using TRI reagent (Invitrogen, Carlsbad, CA). Reverse transcription was carried out using a kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. PCR was performed for M86, M83, and M112/113 by use of the following primers: M86 forward, 5′-GCT CTT CTC GCT GTA CTT GGA CA-3′; M86 reverse, 5′-TTC AAC AAC TTT CGC CTG TAC TAC-3′; M83 forward, 5′-CAG AAA CCC AGG TAC TTG TTG GA-3′; M83 reverse, 5′-TAT CAA GAT CCT CCT GAA ACG ACC-3′; M112/113 forward, 5′-ATG GCC GCG CCA GAT CGA CG-3′; and M112/113 reverse, 5′-CCG ATC CGG ACG ACG AGC CG-3′.
Nick translation.
Double-strand DNA probes for both in situ hybridization and Southern blotting were labeled by nick translation as described previously (36). Briefly, 1 μg plasmid DNA, pSM3fr (38), 10× nick translation buffer, 0.05 mM deoxynucleoside triphosphates (dATP, dCTP, and dGTP), 0.01 mM dTTP, 0.04 mM biotinylated UTP, 1 U of DNA polymerase I, and appropriate concentrations of DNase I (36) were incubated at 15°C for 50 min. Labeled fragments obtained from the protocol were 200 to 500 bases long as determined on 2% agarose gels.
DNA isolation and separation and Southern blotting.
Samples were isolated by Hirt's method with modification for mCMV-infected cells (3). A biotin-labeled probe, pSM3fr, made from the full-length mCMV DNA by nick translation was hybridized to the membrane. After the membrane was washed according to standard protocol, the hybridization signal was detected using an Ambion detection system (Ambion Inc., Austin, TX). For analysis of chromosomal DNA fragmentation by pulsed-field gel electrophoresis, cells grown in six-well plates were infected with wild-type CMV at an MOI of 5 PFU/cell for indicated times. Cells remained untreated or were treated with UV at 10 J/m2 2 h before collection. Cells were then collected and embedded in 1% agarose plugs. The plugs were treated with a buffer containing 100 mM EDTA, 10 mM Tris base, pH 8.0, 1% sodium N-lauroyl-sarcosinate (Sigma), and 0.5 mg/ml proteinase K for 24 h at 50°C and subsequently washed with a buffer containing 20 mM Tris, 50 mM EDTA, pH 8.0. The plugs were loaded on a 1% agarose gel (SeaKem LE agarose; Cambrex Bio Science Rockland, Inc., Rockland, ME), and separation was performed for 24 h with a Bio-Rad contour-clamped homogeneous electric field DR II pulsed-field gel electrophoresis apparatus at 14°C. The separated DNA was stained with ethidium bromide and photographed under UV light.
Immunocytochemistry and fluorescence in situ hybridization.
Immunostaining of nuclear proteins and in situ hybridization to mCMV DNA were performed as described previously (36). The probe for DNA in situ hybridization, pSM3fr, was made from the mCMV DNA genome with a bacterial artificial chromosome system by nick translation as described previously (36).
Immunoblot analysis.
Proteins were separated by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed according to standard procedures. Membranes were stripped with stripping buffer (100 mM β-mercaptoethanol, 2% sodium dodecyl sulfate, 62.5 mM Tris-HCl, pH 6.8), washed with phosphate-buffered saline-0.1% Tween 20, and used to detect additional proteins.
Confocal microscopy.
Cells were examined with a Leica TCS SPII confocal laser scanning system. The red and green channels were recorded simultaneously and/or sequentially and controlled for possible breakthrough between the green and red channels. The images were cropped and adjusted for contrast by Photoshop.
RESULTS
Sequence of immediate-early and early mCMV protein synthesis in human cells.
To determine the point at which the mCMV life cycle is blocked in infected human cells, we tested various cell types at different times postinfection for the presence and abundance of mCMV IE1 and the early proteins encoded by the M112/113 and M141 loci. Western blot analysis (Fig. 1) revealed increasing expression of IE1 and M112/113 over time from the first 24 h to day 5 in control mouse 3T3 cells, with M141 appearing at day 4 in substantial amounts. In contrast, mCMV IE1 diminished after an early increase, M112/113 translation products increased throughout the observation time, and little or no recognizable M141 was produced, as shown, for human fibroblasts (HF) (Fig. 1). Since M112/113 is dependent on the activation of IE3, sufficient IE3 must be produced in the human cell for the vigorous synthesis of these early M112/113 products.
FIG. 1.
Expression of immediate-early and early proteins at different time (hours) of mCMV infection (MOI of 1). The membrane was stripped each time before probing with a specific antibody occurred. Tubulin was used as a loading control. m, mock infected.
Since fibroblasts can suppress a significant number of infecting viral particles (39) and we expected, at most, very-low-level productive infection, we searched for a highly sensitive indicator cell line for plaque production. Based on our previous findings that Daxx segregates the hCMV transcriptional activator pp71 (20), we tested plaque formation in mouse Daxx−/− 227 cells compared with that in mouse 3T3 cells; the 227 cells produced 60 times more plaques. This substantial increase in plaque formation was attributed to the unhindered availability of a potential or functional pp71 equivalent of mCMV or to some other component of Daxx-based inhibition potential. In initial experiments, analysis of supernatants from several human wild-type-mCMV-infected cell lines placed on mouse 227 cells revealed no plaques (HeLa, HEF293, HF, and U373) (data not shown).
State of mCMV DNA replication in human cells.
Although antibodies are not available to test for production of all early mCMV proteins in human cells, the presence of all early proteins necessary for viral DNA replication can be tested based on whether mCMV DNA is replicated in human cells. In 3T3 cells, mCMV DNA is accumulated in replication compartments defined by the M112/113 gene products (38). The combined use of in situ hybridization and immunohistochemistry to identify viral DNA and the location of M112/113 gene products in human and mouse fibroblasts, respectively, at 24 h postinfection (Fig. 2) revealed viral DNA in M112/113 domains (Fig. 2A), present at a high concentration when these domains filled the nuclei of 3T3 cells. A similar distribution of mCMV DNA in human cells (Fig. 2B) indicates that mCMV can replicate in human cells and suggests that all early proteins necessary for viral replication are produced.
FIG. 2.
mCMV DNA replication and IE1 distribution in human fibroblasts. (A) In situ hybridization of mCMV-infected 3T3 cells 24 h p.i., counterstained with antibodies to M112/113 gene products (red). Green dots represent replicated mCMV DNA. (B) Human fibroblasts probed as described for panel A. (C) Human fibroblasts infected with mCMV for 3 h and probed with antibodies against IE1 and PML. (D) Same as panel C, except at 24 h p.i., IE1 (arrows) is segregated beside PML-labeled aggregates. (E) Human fibroblasts infected with mCMV for 24 h and probed with antibodies to M112/113 and HDAC2. Note the accumulation of HDAC2 in the M112/113 domains. (F) Same as panel E but showing only the HDAC2 layer. (G) Human fibroblasts infected for 24 h with mCMV and probed with antibodies to IE1 and HDAC2. (H) Same as panel G but showing only the HDAC2 staining.
A number of ND10-associated proteins, such as the Bloom protein, a RecQ helicase (19), and the heterochromatin and Daxx binding protein ATRX (21), are involved in recombination of DNA. These proteins are dispersed and presumably activated by IE1 in the homologous host. We used Southern blot analysis to determine whether the mCMV DNA produced in human cells is intact and at full size. As shown in Fig. 3A, the appropriate unit-sized DNA band in 3T3 cells and HF is present over time, but degraded viral DNA was dominant in the human cells. In addition, the amount of DNA in HF decreased substantially after day 3, although a faint unit-sized band was detectable throughout the observation period. Thus, the reduction of full-size mCMV DNA in human cells diminished the chance to produce infectious particles. However, the presence of some full-size DNA suggests that encapsidation and viral particle formation might still be possible.
FIG. 3.
(A) Southern blot of mCMV-infected mouse and human cells. 3T3 and human fibroblasts were infected with 1 PFU per cell, and cells were harvested at different times after infection. Coexisting cellular DNA was used as a loading control. (B) Pulsed-field gel electrophoresis-separated DNA from infected and uninfected cells exposed or unexposed to UV. Smears indicate DNA degradation.
One possibility of virus DNA degradation could be that mCMV leads to general DNA degradation in human cells. This possibility was evaluated by testing for host cell DNA degradation after various times of virus infection. As a positive control for degraded host DNA, we used UV-irradiated cells. As shown in Fig. 3B, no general DNA smear is seen in the mock-infected 3T3 cells but after UV irradiation DNA degradation can be observed (Fig. 3B, compare lanes 2 and 5). The same can be observed with human fibroblasts (Fig. 3B, lanes 9 and 12). Viral DNA synthesis is apparent in the mouse cells 24 and 48 h p.i. (Fig. 3B, band between 200 and 300 kb in lanes 3 and 4) but not in human cells in these comparatively loaded gels. The amount of unit-sized viral DNA produced is therefore substantially smaller in human cells than in mouse cells infected with equal amount of virus. Also, no host DNA degradation is evident. All of the DNA degradation can be attributed to the UV irradiation (Fig. 3B, compare lanes 5 and 6 and 12 and 13). From this evidence, we conclude that mCMV DNA degradation is not due to an mCMV-induced general DNA degradation of the human host cell.
mCMV IE1 does not function in human cells as in mouse cells.
IE1 has a strong effect on hCMV propagation in human cells (12, 13) and ND10 destruction and Daxx and HDAC binding in both hCMV and mCMV (31, 39). To determine whether mCMV IE1 would perform the appropriate functions in the human host, we tested the ability of the mCMV IE1 protein to disperse ND10 in infected HF. Immunofluorescence microscopy showed that mCMV IE1, like hCMV IE1, initially accumulated at ND10 (Fig. 2C) but that ND10s, as visualized by anti-PML antibodies, were retained after infection with mCMV even 24 h p.i. (Fig. 2D). Thus, mCMV IE1 cannot disperse the numerous ND10s in HF. Surprisingly, mCMV IE1 (Fig. 2D) and a certain amount of HDAC2 segregated into large compartments beside ND10 at 24 h p.i. These domains are the replication compartments, as indicated by the presence of M112/113 (Fig. 2E and F). Localization of both HDAC2 and IE1 in the same compartment (Fig. 2G and H) shows by implication that they are accumulated in the replication compartments. In addition, we tested whether trichostatin A would substitute for an hCMV IE1. We did not obtain any infectious viral particles (data not shown). These findings suggest that mCMV IE1 functions differently in HF than in mouse cells. Since IE1 of either mCMV or hCMV is not considered essential to viral replicative success, its functional loss should not underlie the failed progression to late stages of virus particle production.
Either IE1 or tegument proteins of hCMV increase mCMV particle production to detectable levels in human cells.
If mCMV IE1 cannot perform certain chromatin or DNA protecting functions in human cells, as suggested by the lack of ND10 dispersal and lack of human HDAC binding (not shown), then providing hCMV IE1 in trans should alleviate these blocks and lead to viral particle production. To take into consideration the possibility of low-level transmission of viral genomes short of plaque formation, human astrocytoma cell line U373 and its constitutively IE1-producing derivative, U373-IE1, as well as human fibroblasts and the derivative producing IE1 (ihfie1.3) were infected with GFP-tagged mCMV at low (0.1) PFU (as determined on 3T3 cells). Comparison of cultures infected at confluence showed that 3T3 cells and HF were equally infected (Fig. 4A and B), indicating equal transmission of viral genomes to the nucleus and equal activation of the major immediate-early promoter (MIEP) in these two fibroblast lines. Surprisingly, the human U373 cells had a much higher apparent infection rate at the same particle input and the number of U373-IE1 cells producing MIEP-driven GFP at 24 h p.i. was even higher than that of the U373 cells (Fig. 4C and D). Assuming the same level of infection, the presence of hCMV IE1 appears to activate the mCMV MIEP in more cells, consistent with Western blot analysis of the actual viral proteins, which also revealed activation of the mCMV MIEP (Fig. 5A). In U373 cells, IE1 expression declined over time and M141 showed the same pattern as for HF, whereas U373-IE1 cells continued to produce IE1, M112/113, and also M141, similarly to 3T3 cells (compare Fig. 5A and Fig. 1). Based on the high infection rate of U373-IE1 cells, we conclude that fibroblasts of both species can totally suppress a considerable amount of infecting virus even at the MIEP level. By use of Southern analysis, the hCMV IE1-expressing cells also produced substantially more mCMV DNA and, importantly, substantially more full-size viral genomes clearly recognized when the time sequence of mCMV DNA production was compared between U373 and U373-IE1 cells, particularly recognizable at late times after infection (Fig. 5B). However, the presence of hCMV IE1 did not prevent mCMV DNA degradation.
FIG. 4.
Plaque formation of GFP-producing mCMV infection in IE1-expressing human cells. Confluent cell lines (indicated at lower left) were infected with equal amounts of mCMV expressing GFP (RVG102), with titers that induced ∼1% GFP-producing 3T3 cells. Much higher numbers of U373 astrocytoma cells produce GFP (C), and more than 50% of the hCMV IE1-expressing U373 cells were labeled with GFP (D). (E to H) Size increase of a plaque over time in U373-IE1 cells; the different days (e.g., D5, day 5) are indicated at the upper right.
FIG. 5.
(A) Expression of immediate-early and early proteins in human U373 and U373-IE1 cells at different times (hours) post-mCMV infection. The membrane was stripped each time before probing with a different specific antibody occurred. Tubulin was used as a loading control. The gel was loaded with less cell lysate for the U373-IE1 sample. (B) mCMV DNA production in mCMV-infected human U373 and U373-IE1 cells, as determined by Southern blotting. Cells were harvested at different times (hours) after infection. m, mock infected.
To address whether these mCMV protein-producing human cells generate virus that can reinfect additional cells and considering the possibility that reinfected cells might not be detected as plaque formation because of low particle production, we photographed a small cluster of cells over several days (Fig. 4E to H). The low-density cluster of GFP-producing U373-IE1 cells at day 5, which is presumed to be a plaque, increased by day 8 to a dense aggregate of GFP-producing cells. By day 12, a few cells appeared rounded and dead. This photographic sequence clearly shows that IE1-producing human cells can produce virus, since the number of divisions necessary to obtain such a cell cluster would have diluted the initial infection even if cells divided with active virus present. We did not observe typical plaques with empty centers indicative of cell lysis in this rapidly dividing cell type.
To determine whether infectious virus was actually produced or whether all the apparent plaques observed were formed from viral DNA produced continuously in cycling cells, we infected U373-IE1 cells with wild-type mCMV and RVG102 at an MOI of 1 (as determined on 3T3 cells), resulting in GFP production in ∼50% of the human cells at 24 h p.i. Cells were then trypsinized, washed repeatedly to remove any input virus, and diluted ∼1:200 with mouse 227 cells, and plaques were quantified (Table 1). HF, HeLa, HEK293, or U373 astrocytoma cells did not produce infectious virus that could start a progressive infection in the highly susceptible mouse cells, indicating that no input virus was transferred to the mouse reporter layer of cells. However, both the astrocytoma cell line and the HF line expressing hCMV IE1 (HF-IE1) produced infectious virus, since plaques formed in the mouse cells. Thus, the presence of hCMV IE1 in human cells alone is sufficient to allow productive mCMV infection, although the plaque-forming number of human cells is ∼10 times smaller in hCMV IE1-producing human fibroblasts and ∼100 times smaller in the hCMV IE1-producing astrocytoma cell line than in mouse 3T3 cells.
TABLE 1.
Plaque formation in mixed cell culturea
| Cell type | No. of plaque-forming cells infected with mCMV strain:
|
|||
|---|---|---|---|---|
| Smith
|
RVG102
|
|||
| Cells | Supernatant | Cells | Supernatant | |
| 3T3 | 403 | 0 | 414 | 0 |
| HeLa | 0 | 0 | 0 | 0 |
| HEK293 | 0 | 0 | 0 | 0 |
| HF | 0 | 0 | 0 | 0 |
| ihfie1.3 | 40 | 0 | 28 | 0 |
| U373 | 0 | 0 | 0 | 0 |
| U373-IE1 | 2 | 0 | 6 | 0 |
Cells were infected with the respective virus and, at 20 h p.i., washed and trypsinized to remove all adherent viruses and then mixed with 227 cells (1:200). As a control for remaining input virus, a large aliquot of infected cells was frozen and thawed three times and the supernatants were used to infect 227 cells (Daxx−/−).
Using the GFP-producing RVG102 strain, we tested whether the mCMV-infected hCMV IE1-producing human cells can produce mCMV particles that productively infect a new set of human cells. Fluorescence microscopy was used to examine this second round of human cells and hCMV IE1-producing human cells infected with supernatant obtained from mCMV-infected human cell lines expressing hCMV IE1. Although numerous green fluorescing cells were observed only 1 day p.i., no plaques formed and no new infectious virus that yielded plaques on the mouse 227 reporter layer was produced (data not shown). Thus, while reinfection of human cells by mCMV produced in human cells is possible, propagation is drastically curtailed in the second round.
Higher input virus overcomes the deficiency of IE1 in hCMV (13). It also substantially increases the tegument proteins from infectious virus and from dense bodies, which might contribute to enhanced levels of proteins such as pp71 above the segregation capability of Daxx and make IE1 unnecessary. Since mCMV tegument proteins might not have been sufficiently adapted to prime human cells for permissive infection, we tested whether hCMV tegument proteins affect mCMV productive infection in human cells. To ensure that UV-irradiated hCMV infected cells properly and discharged tegument proteins into the cells, we tested UV-irradiated hCMV-infected cells by immunohistochemistry. Visualization with antibodies to the tegument protein pp71 showed pp71 as small diffraction-sized dots, presumably single viral particles outside the cell, which accumulated in the nucleus at ND10 3 h p.i. (Fig. 6A). As shown in Fig. 6B, the tegument protein pp65 was seen dispersed in the nucleus and as single particles outside the cell. This shows that the UV-irradiated hCMV tegument proteins enter the nucleus and distribute, as expected from previous analysis (20). Virus production in human cells coinfected with GFP-producing RVG102 mCMV and UV-irradiated hCMV was then assessed by a plaque assay modified to show that free particles were produced after three freeze-thaw cycles and infection of the mouse reporter layer with the clarified cell supernatant (Table 2). At 72 h p.i., some new mCMV was produced even in normal human fibroblasts and this virus production was strongly enhanced by the addition of UV-irradiated hCMV. Such enhancement was more extensive in human fibroblasts than in the astrocytoma cell line, with mCMV levels reaching those detected in the hCMV IE1-producing ihfie1.3 line. Virus production continued, with some decline by day 5. We also tested whether virus production was enhanced by the presence of hCMV IE1 and the tegument proteins of hCMV by coinfecting hCMV IE1-producing human cells with UV-treated hCMV and mCMV. About three times as many plaques were formed on the mouse cell reporter layer when UV-irradiated hCMV was present in the hCMV IE1-producing human cells, indicating an additive or synergistic effect of hCMV IE1 and proteins provided by UV-irradiated hCMV. These results show that human cell permissiveness for mCMV infection is not specifically hCMV IE1 dependent and that the apparently insignificant levels of virus produced in normal cells can be boosted to significant levels by at least two apparently independent means.
FIG. 6.
Immunolocalization of pp71 (A) and pp65 (B) introduced by UV-treated hCMV 3 h p.i., relative to PML. pp71 localization in uninfected (uninf.) HF pp71 (C) relative to PML and (D) relative to Daxx.
TABLE 2.
Plaque formation of mCMV in human cells with the help of UV-inactivated hCMVa
| Cell type and infection | No. of plaque-forming cells at:
|
||
|---|---|---|---|
| 72 h p.i. | 96 h p.i. | 120 h p.i. | |
| HF | 1 | 0 | 0 |
| HF + UV+ hCMV | 9 | 10 | 5 |
| ihfie1.3 | 9 | 6 | 5 |
| ihfie1.3 + UV+ hCMV | 28 | 15 | 9 |
| U373 | 1 | 0 | 0 |
| U373 + UV+ hCMV | 1 | 0 | 0 |
| U373-IE1 | 6 | 7 | 7 |
| U373-IE1 + UV+ hCMV | 17 | 13 | 8 |
The different human cell lines were infected with GFP-tagged mCMV (RVG102) in 24-well plates, with UV-irradiated hCMV (+UV+ hCMV) or without. After overnight incubation, cells were washed two times with DMEM and replenished with fresh medium. After 72, 96, and 120 h p.i., the cells were collected and frozen and thawed three times, and the clarified supernatant (1 ml) was used in a plaque assay with mouse 227 cells (Daxx−/−).
The viral tegument protein pp71 is thought to activate the MIEP in part by interacting with the repressor Daxx (2, 5, 7, 20, 27). We therefore tested whether pp71 alone of the tegument proteins could induce production of significant levels of infectious mCMV in human cells. A telomerase-immortalized human fibroblast line constitutively producing pp71 (4) was used for these experiments. As shown in Fig. 6C, this cell line has pp71 segregated to ND10, as indicated by PML colabeling. The same ND10s also contain Daxx (Fig. 6D), which has previously been identified as the adapter protein that brings pp71 to ND10 (18, 20). Production of infectious GFP-producing mCMV in the pp71-producing cell line relative to production in the control telomerase parent cell was evaluated. The human cells were trypsinized 24 h p.i., washed twice, and mixed 1:200 with the indicator mouse 227 cells. They were scored 72 h later for plaques, which are revealed as large clusters of GFP-producing cells. As a control for potential carryover of input virus, the supernatants of cells frozen/thawed three times were used instead of live cells. The individual supernatants came from eight times as many of the different infected cells as were used in the live-cell mixing. As shown in Table 3, no potentially present input virus produced a plaque from any of the tested infected cell lines, but the live cells cocultured with the mouse indicator cells must have produced some infectious virus, as a few definite plaques formed. However, the pp71-expressing fibroblasts did not produce substantially more cells that could productively infect the mouse indicator cells than the control cells. pp71 alone is therefore not able to affect the cross-species success as much as the tegument proteins introduced into the cells by infection with UV-irradiated hCMV. Another tegument protein or combination of tegument proteins that can have the observed effect remains to be identified.
TABLE 3.
Comparison of telomerase-immortalized human fibroblasts and a pp71-expressing variant in mCMV plaque formationa
| Means of infection | No. of plaque-forming cellsb
|
|||
|---|---|---|---|---|
| wt mCMV-GFP
|
mCMV-GFP-hIE1
|
|||
| HF-tel | HF-tel-pp71 | HF-tel | HF-tel-pp71 | |
| Supernatant | 0 | 0 | 0 | 0 |
| Live cell | 1 | 3 | 14 | 39 |
Cells were infected with the respective virus and, at 24 h p.i., washed and trypsinized to remove all adherent viruses and then mixed with the reporter cells (1:200). As a control for the remaining input virus, a large aliquot of infected cells was frozen and thawed three times and the supernatant was used to infect the reporter 227 cells.
wt mCMV-GFP, wild-type mCMV expressing GFP; mCMV-GFP-hIE1, hCMV IE1- and GFP-producing mCMV; HF-tel, control telomerase-immortalized human fibroblast; HF-tel-pp71, telomerase-immortalized human fibroblast constitutively producing pp71.
Since hCMV IE1 releases Daxx and thus frees pp71 from ND10, we asked whether human IE1 would enhance the activity of pp71 in the cross-species infection to a recognizable level. For this purpose, an hCMV IE1- and GFP-producing mCMV was used. When fibroblasts were infected with this virus, plaques were produced at elevated levels, as expected from the presence of hCMV IE1 (Table 3). When this virus was used to infect pp71-expressing fibroblasts, it produced over three times the number of plaques as the control telomerase-immortalized cell line, suggesting a synergistic effect due to IE1-based release of the Daxx-bound and thus inactive pp71.
mCMV DNA microarray analysis of infected human cells.
To identify potential obstructions to virus particle formation, we used an mCMV DNA microarray system (39a) to compare various cell lines infected equally with mCMV. We detected 153 gene products in 3T3 cells but only 53 in human U373 cells; in the presence of hCMV IE1, these astrocytoma cells expressed 116 mCMV genes. HF expressed 70 genes, and HF-IE1 expressed 103 genes. The genes expressed in the two human cell lines and their hCMV IE1-producing equivalents did not totally overlap, indicating that even for mCMV there is a human-cell-dependent difference in viral proteins expressed. Use of background levels as a cutoff (set at 500) may have led to an underestimation of the number of transcribed genes; some of the sites of oligonucleotide deposition were below background levels and apparently true negatives. This possibility was tested for capsid transcripts (see below).
For comparative purposes, only mCMV genes with an apparent hCMV equivalent and with a kinetic class of the hCMV equivalent (8) are listed in Table S4 in the supplemental material. Comparing the data from the five cell lines, we find that the immediate-early proteins of mCMV in human cells are not regulated by the presence of hCMV IE1. Despite near-background levels of mCMV IE1 transcripts in U373 cells and human fibroblasts in the mCMV DNA microarray analysis, mCMV IE1 expression was clearly evident (Fig. 1 and 5). The essential DNA polymerase accessory protein transcript (M44) was present at low copy number and only mildly induced by IE1; however, the proteins produced must be present in sufficient amounts for mCMV DNA synthesis to occur in all human cell lines tested. Some early proteins, such as M26 and M33, were present below the background level and may not be necessary for early replicative functions in these cells (42).
Structural proteins such as the capsid protein (M86), the large tegument protein (M48), and the minor capsid protein (M46) were all present at substantially lower levels than in the mouse cells and often differentially upregulated either in U373-IE1 cells or in HF-IE1 (see Table S4 in the supplemental material). In U373 cells, the minor capsid protein could be the limiting factor for mCMV particle production since very few transcripts were detected, whereas in HF-IE1, the major capsid protein transcripts appeared limiting. However, in both, levels must have been sufficient for some infectious particle production (see below) (Fig. 7). Of the late kinetic structural proteins, the upper (M82) and lower (M83) matrix proteins were more strongly upregulated in HF-IE1 than in U373-IE1 cells, consistent with the greater production of mCMV by fibroblasts than by U373 cells. The two human cell types producing hCMV IE1 did not always upregulate different proteins equally, possibly reflecting different intracellular interactions with IE1. This may be particularly important for the protease transcripts, whose product cleaves the assembly protein precursor (M80). This protease transcript is nearly as upregulated by IE1 in HF as in 3T3 cells, but IE1 in U373 cells shows no effect.
FIG. 7.
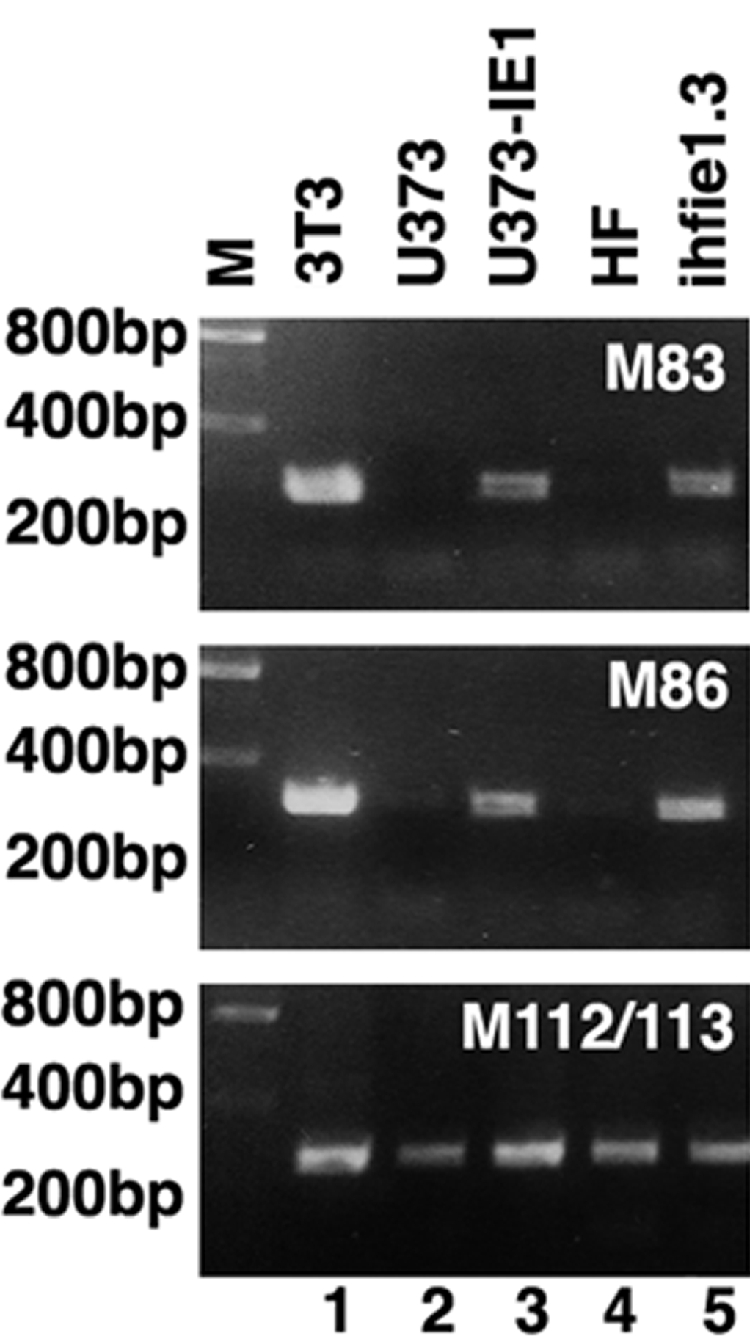
RT-PCR analysis of late transcripts in mCMV-infected normal and hCMV IE1-producing human cells. The early transcript of M112/113 was used as a positive control, as it is transcribed in all mCMV-infected human cells. M, marker.
For the transmembrane glycoproteins, which are equivalent in hCMV and mCMV, there was no recognizable upregulation of gH (M75) by IE1 and in all cell types the signal was below background. By contrast, gM (M100) was highly upregulated in HF-IE1 to a level near the level in mouse cells, although no IE1-dependent increase was seen in U373-IE1 cells. gB (M55) was upregulated by hCMV IE1 in both cell types. These findings underline the variability in transcript upregulation due to a combination of hCMV IE1 and cell-specific factors. In addition, these results show that hCMV IE1 does not equally affect the late class of mCMV proteins in human cells.
Since capsid proteins are essential for infectious particle production and we detected no signal above background levels in the virus particle-producing U373-IE1 cells, we tested the different cell lines for the presence of the M86 transcript by RT-PCR. After 30 cycles, when the amount of transcript tested was in the linear range, we found no signal for the two human control cell lines but a strong signal for the respective hCMV-IE1-expressing cells (Fig. 7). Thus, M86 transcripts were present, although at very low levels. Presumably, the microarray analysis was insufficiently sensitive at lower signal levels to distinguish between relevant transcript expression and their absence, although high positive signals and substantially increased levels in the presence of IE1 were defining for the presence, abundance, and augmentation of mCMV transcripts in human cells by hCMV IE1.
DISCUSSION
Cytomegaloviruses are considered to be highly species specific, and within a species, they show pronounced permissiveness in certain cell types. Since cytomegaloviruses can infect most cells, including those of different species, and produce IE proteins, any block in virus production is not at the receptor level for cell entry and penetration to the nucleus and not due to incompatibilities in promoter recognition or successful activation. Thus, the block in production of infectious progeny may be at different levels of the transcription cascade or in the essential interactions between proteins of the host and the pathogen, specifically those viral proteins that have been optimized to counter cellular defenses. Our analysis of the progression of mCMV through the immediate-early, early, and late stages in the productive cycle shows that insufficient intact progeny DNA and lack of specific late proteins in mCMV-infected human cells likely account for the effective nonpermissiveness. These impediments or barriers were alleviated by selected hCMV components that helped mCMV to produce infectious particles in human cells.
Sequential analysis of the three stages in the viral transcription cascade revealed no intrinsic block at the immediate-early or early stage, at least with respect to DNA synthesis in the heterologous host. However, there was a pronounced degradation of newly replicated mCMV DNA in human cells. This decrease in the normally sized viral DNA represents a recognizable lowered potential for the production of infectious virus. A trivial explanation would be the induction of a general DNA degradation by mCMV in human cells. This is apparently not the case as no general host DNA destruction is recognized in the first 48 h p.i., when viral DNA degradation was obvious. Also, the mCMV 38.5 gene product has antiapoptotic activity in human cells (28). The degraded viral DNA may reflect faulty protection of replicated mCMV DNA due to low levels of specific DNA binding proteins or proper charge neutralization by proteins such as histones or spermidine/spermine. Alternatively, the degradation may be a secondary indirect effect of impaired encapsidation due to inadequate production of the capsid proteins or to DNA entry into available capsids because of faulty processing for entry followed by nucleolytic attack. The presence of hCMV IE1 resulted in a substantial increase in the amount of viral DNA and of full-size viral DNA (Fig. 5). The larger amount of full-size DNA may increase particle formation and increase late protein transcription, including that of the capsid proteins. The minor or major capsid protein may still remain an essential limiting factor for mCMV production in human cells.
Events that reduce the chance of full mCMV permissive infection in human cells may not be a single block but rather occur at different levels. Although the immediate-early protein synthesis takes place and induces early viral synthetic functions, we observed directly that neither dispersion of ND10 nor nuclear distribution of mCMV IE1 occurred in human cells, indicating improper function in the human cell context. Our analysis of whether modifications of cellular properties by hCMV IE1 might elevate mCMV production to recognizable levels indicated that expression of hCMV IE1 in two different human cell lines did indeed lead to mCMV particle formation, which was unexpected, considering that IE1 of neither virus is essential in the homologous context. Thus, some functions are presumably enhanced rather than induced de novo by hCMV IE1 augmentation, and, clearly, increased capsid transcript synthesis is an important effect. Reversal of the putative silencing effect by IE1 (30, 31, 37) does not appear to be the essential function, since trichostatin A did not lead to mCMV replication in human cells. Moreover, IE1 of neither virus substantially augments the MIEP of the other virus in the present infection study. Thus, it seems more likely that some late functions of mCMV must be enhanced by hCMV IE1.
There may be several mechanisms that lead to the enhancements of these late functions. mCMV particle production in human cells may have gone undetected were it not for the use of the mouse cell line 227, in which the ND10-associated Daxx has been genetically ablated (21). In this mouse cell line, mCMV produces 60-fold more plaques than in permissive fibroblasts (3T3). Thus, Daxx in mouse cells is a strong inhibitor of virus production. Normally, the repressor Daxx is bound by mCMV IE1 and presumably inactivated (39). In mouse cells, but not in human cells, Daxx is interferon upregulated (30a). Although extrapolation from human cells, where Daxx binds the hCMV tegument protein pp71 (18, 20, 27), may be premature, hCMV tegument proteins introduced into human cells substantially augmented mCMV production in the absence of hCMV IE1. The mode of introduction, UV-treated hCMV, may however have had complex side effects through the signaling induced by hCMV binding and the resultant activation of the interferon pathway (a negative) and the repression of such a pathway by tegument proteins such as pp65 (a positive).
Daxx inhibits mCMV particle formation, as implied by the 60-fold increase in PFU by the absence of mouse Daxx. If human Daxx (hDaxx) has the same effect in human cells, then its removal should increase mCMV formation. This can be accomplished by hCMV IE1 introduction into the cell and has the anticipated effect by providing it either as a constitutively expressed protein or as a newly expressed protein from recombinant mCMV. The mechanism of hCMV IE1, then, is through inactivation of a host repressor. mCMV IE1 can, like its hCMV counterpart, interact with human HDAC but brings the HDAC into the prereplication domain in human cells, potentially suppressing the mCMV genomes. The introduced hCMV IE1 would treat the respective HDAC as in the hCMV infection and thus prevent the deacetylation in the mCMV genome, allowing for enhanced transcription.
Since pp71 alone did not augment mCMV production, other tegument proteins or combinations of such proteins that have this effect need to be identified. However, the likely mechanism for a synergistic effect between hCMV IE1 and hCMV pp71 on mCMV production in human cells is the release of pp71 from its segregated position at ND10 through the inactivation of Daxx by IE1. In combination with hCMV IE1, either the tegument protein supplementation or pp71 alone synergizes the hCMV IE1 promotion of mCMV production in human cells. Human Daxx may have been neutralized by hCMV pp71 to relieve mCMV from hDaxx-based repression. These results seem analogous to those reported by Schierling et al. (35). Alternatively, the combination of hCMV pp71 and hDaxx may have augmented the IE protein synthesis of mCMV as it does for IE proteins in hCMV (7). In this context, it is worth noting that the very permissive human fibroblasts have very low Daxx content despite a large number of prominent ND10s (Negorev et al., submitted). In our experiments, pp71 and other tegument proteins like ppUL35 involved in activation of IE transcription and ultimately particle formation (35) were added. hCMV tegument also provides pp65, which suppresses the interferon response by modulating the interferon response factor 3 (1) and which might have relieved some of the interferon-based inhibition on mCMV. When combined, hCMV IE1 and hCMV tegument protein more than tripled mCMV infectious virus in human cells as measured by plaque assay, suggesting that IE1 and tegument proteins augment at different levels and add independent shifts in the cellular environment which are positive for mCMV production.
Our observations suggest that the apparent block in cross-species infection rests in the ability of the cell to suppress the non-species-specific virus more efficiently than the more adapted species-specific virus and that this suppression is exerted at many different levels. These barriers can be overcome with proteins including the hCMV IE1 protein and, separately, hCMV tegument proteins produced by the adapted species. The mechanisms likely include tegument protein pp71 binding to the Daxx repressor, pp65 as a repressor of the interferon response, and the antagonistic effect of IE1 on both Daxx and HDAC. Most probably, this includes only some of the factors at the cell level specifically leading to the low production of certain late proteins with the consequence of viral DNA degradation. The additive effect of several such inadequacies will appear as an effective block for a sustainable infection. Under normal conditions, mCMV will remain unable to produce viral progeny in human cells since many selective steps or recombinations with hCMV genes may need to occur simultaneously to produce a new human pathogen. Which hCMV genes introduced into mCMV would make a human pathogen that produces a number of viral particles equivalent to that produced by hCMV should be evaluated to estimate the probability that it could happen by recombination when using this vector for human therapy. On the other hand, in order to make mCMV a helpful vector one may want to grow the virus in human cells to avoid mouse antigen being transmitted. Development of an increasingly hybrid recombinant virus and cells that supplement necessary hCMV proteins should reveal the relevant contributions of hCMV genes and produce the desired antigens against hCMV in a live virus for immunization. Selected recombinants and supplementing cell lines may also allow the differential production of a virus that retains species specificity or allows minor or major replicative success, depending on the task for which it will be designed.
In conclusion, mCMV can produce infectious virus in human cells but the amounts are normally so small that in practice productive reinfection is not sustained. The suppression of mCMV replication in human cells is affected at several levels, which additively or synergistically result in the appearance of species specificity. The practical consequences are that this virus can be modified for a variety of purposes, including those that require a limited spread, the limited spread being safeguarded by the multiple suboptimally adapted viral counterdefenses.
Supplementary Material
Acknowledgments
This study was supported by funds from NIH AI 41136 and NIH GM 57599, the G. Harold and Leila Y. Mathers Charitable Foundation, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health. Core grant CA-10815 is acknowledged for support of the microscopy and sequencing facility.
We are grateful for reagents from J. D. Hamilton, A. Campbell, J. Jonjic, S. Lowe, J. Kerry, W. A. Besnahan, M. Messerle, and Wolfgang Brun.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst, E. M., I. Crnkovic-Mertens, and M. Messerle. 2004. Cloning of beta-herpesvirus genomes as bacterial artificial chromosomes, p. 221-239. In S. Zhao and M. Stodolsky (ed.), Methods in molecular biology, vol. 256. Bacterial artificial chromosomes. Humana Press Inc., Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciocco-Schmitt, G. M., Z. Karabekian, E. W. Godfrey, R. M. Stenberg, A. E. Campbell, and J. A. Kerry. 2002. Identification and characterization of novel murine cytomegalovirus M112-113 (e1) gene products. Virology 294:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Davignon, J. L., D. Clement, J. Alriquet, S. Michelson, and C. Davrinche. 1995. Analysis of the proliferative T cell response to human cytomegalovirus major immediate-early protein (IE1): phenotype, frequency and variability. Scand. J. Immunol. 41:247-255. [DOI] [PubMed] [Google Scholar]
- 11.Fortunato, E. A., V. Sanchez, J. Y. Yen, and D. H. Spector. 2002. Infection of cells with human cytomegalovirus during S phase results in a blockade to immediate-early gene expression that can be overcome by inhibition of the proteasome. J. Virol. 76:5369-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 76:4441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutermann, A., A. Bubeck, M. Wagner, U. Reusch, C. Menard, and U. H. Koszinowski. 2002. Strategies for the identification and analysis of viral immune-evasive genes—cytomegalovirus as an example. Curr. Top. Microbiol. Immunol. 269:1-22. [DOI] [PubMed] [Google Scholar]
- 15.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 75:6292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry, S. C., K. Schmader, T. T. Brown, S. E. Miller, D. N. Howell, G. G. Daley, and J. D. Hamilton. 2000. Enhanced green fluorescent protein as a marker for localizing murine cytomegalovirus in acute and latent infection. J. Virol. Methods 89:61-73. [DOI] [PubMed] [Google Scholar]
- 17.Hertel, L., V. G. Lacaille, H. Strobl, E. D. Mellins, and E. S. Mocarski. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 77:7563-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, H., H. Sindre, and T. Stamminger. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76:5769-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2004. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell Sci. 117:3807-3820. [DOI] [PubMed] [Google Scholar]
- 22.Isomura, H., T. Tsurumi, and M. F. Stinski. 2004. Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J. Virol. 78:12788-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, K. S., and R. I. Carp. 1971. Growth of murine cytomegalovirus in various cell lines. J. Virol. 7:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafemina, R. L., and G. S. Hayward. 1988. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J. Gen. Virol. 69:355-374. [DOI] [PubMed] [Google Scholar]
- 25.Lafemina, R. L., M. C. Pizzorno, J. D. Mosca, and G. S. Hayward. 1989. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology 172:584-600. [DOI] [PubMed] [Google Scholar]
- 26.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 102:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, K. R., K. V. Rowley, A. Rinaldi, I. P. Nicholson, A. M. Ishov, G. G. Maul, and C. M. Preston. 2002. Activity and intracellular localization of the human cytomegalovirus protein pp71. J. Gen. Virol. 83:1601-1612. [DOI] [PubMed] [Google Scholar]
- 28.McCormick, A. L., C. D. Meiering, G. B. Smith, and E. S. Mocarski. 2005. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J. Virol. 79:12205-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mocarski, E. S., Jr. 2004. Immune escape and exploitation strategies of cytomegaloviruses: impact on and imitation of the major histocompatibility system. Cell. Microbiol. 6:707-717. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Negorev, D. G., O. V. Vladimirova, A. Ivanov, F. Rauscher III, and G. G. Maul. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 31.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto, A. R., J. C. Fitzgerald, W. Giles-Davis, G. P. Gao, J. M. Wilson, and H. C. Ertl. 2003. Induction of CD8+ T cells to an HIV-1 antigen through a prime boost regimen with heterologous E1-deleted adenoviral vaccine carriers. J. Immunol. 171:6774-6779. [DOI] [PubMed] [Google Scholar]
- 33.Rea, D., F. H. Schagen, R. C. Hoeben, M. Mehtali, M. J. Havenga, R. E. Toes, C. J. Melief, and R. Offringa. 1999. Adenoviruses activate human dendritic cells without polarization toward a T-helper type 1-inducing subset. J. Virol. 73:10245-10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddehase, M. J., J. Podlech, and N. K. Grzimek. 2002. Mouse models of cytomegalovirus latency: overview. J. Clin. Virol. 25(Suppl. 2):S23-S36. [DOI] [PubMed] [Google Scholar]
- 35.Schierling, K., C. Buser, T. Mertens, and M. Winkler. 2005. Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation. J. Virol. 79:3084-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, Q., L. Li, A. M. Ishov, V. Revol, A. L. Epstein, and G. G. Maul. 2003. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J. Virol. 77:5821-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, Q., L. Li, and G. G. Maul. 2005. Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter. J. Virol. 79:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, Q., and G. G. Maul. 2003. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J. Virol. 77:1357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Tang, Q., E. Murphy, and G. G. Maul. 2006. Experimental confirmation of global murine cytomegalovirus open reading frames by transcriptional detection and partial characterization of newly described gene products. J. Virol. 80:6873-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Den Pol, A. N., E. Mocarski, N. Saederup, J. Vieira, and T. J. Meier. 1999. Cytomegalovirus cell tropism, replication, and gene transfer in brain. J. Neurosci. 19:10948-10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Den Pol, A. N., J. Vieira, D. D. Spencer, and J. G. Santarelli. 2000. Mouse cytomegalovirus in developing brain tissue: analysis of 11 species with GFP-expressing recombinant virus. J. Comp. Neurol. 427:559-580. [DOI] [PubMed] [Google Scholar]
- 42.Waldhoer, M., T. N. Kledal, H. Farrell, and T. W. Schwartz. 2002. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J. Virol. 76:8161-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, R., D. L. Doolan, T. P. Le, R. C. Hedstrom, K. M. Coonan, Y. Charoenvit, T. R. Jones, P. Hobart, M. Margalith, J. Ng, W. R. Weiss, M. Sedegah, C. de Taisne, J. A. Norman, and S. L. Hoffman. 1998. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science 282:476-480. [DOI] [PubMed] [Google Scholar]
- 44.Weitzman, M. D., C. T. Carson, R. A. Schwartz, and C. E. Lilley. 2004. Interactions of viruses with the cellular DNA repair machinery. DNA Repair 3:1165-1173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








