Abstract
The chicken anemia virus protein Apoptin selectively induces apoptosis in transformed cells while leaving normal cells intact. This selectivity is thought to be largely due to cell type-specific localization: Apoptin is cytoplasmic in primary cells and nuclear in transformed cells. The basis of Apoptin cell type-specific localization and activity remains to be determined. Here we show that Apoptin is a nucleocytoplasmic shuttling protein whose localization is mediated by an N-terminal nuclear export signal (NES) and a C-terminal nuclear localization signal (NLS). Both signals are required for cell type-specific localization, since Apoptin fragments containing either the NES or the NLS fail to differentially localize in transformed and primary cells. Significantly, cell type-specific localization can be conferred in trans by coexpression of the two separate fragments, which interact through an Apoptin multimerization domain. We have previously shown that Apoptin interacts with the APC1 subunit of the anaphase-promoting complex/cyclosome (APC/C), resulting in G2/M cell cycle arrest and apoptosis in transformed cells. We found that the nucleocytoplasmic shuttling activity is critical for efficient APC1 association and induction of apoptosis in transformed cells. Interestingly, both Apoptin multimerization and APC1 interaction are mediated by domains that overlap with the NES and NLS sequences, respectively. Apoptin expression in transformed cells induces the formation of PML nuclear bodies and recruits APC/C to these subnuclear structures. Our results reveal a mechanism for the selective killing of transformed cells by Apoptin.
Viruses employ diverse strategies to alter host cell functions that, in turn, facilitate the viral life cycle. In this regard, many viral proteins modulate critical cellular processes including transcription, cell signaling, and cell cycle regulation. For example, numerous DNA viruses encode proteins that target key cell cycle regulators in mammalian cells such as p53 and pRB (22, 55). Thus, the study of virus-host interactions has been valuable in understanding a wide variety of cellular regulatory pathways.
Many animal viruses encode proteins that are potent inducers of apoptosis, a common mechanism to facilitate viral egress and promote viral spreading (38). In certain cases the apoptotic activity of the virus is selective to transformed cells. One example is the Apoptin protein encoded by the chicken anemia virus (CAV). CAV is the sole member of the family Circoviridae, genus Gyroviridae, and is the etiologic agent of chicken infectious anemia, which results from large-scale apoptosis of cortical thymocytes and erythroblastoid cells in the bone marrow (18). The CAV genome encodes three proteins; the third protein, VP3 or Apoptin, is responsible for apoptosis induction (32). We have previously shown that Apoptin interacts with the anaphase-promoting complex/cyclosome (APC/C) (42), a major regulator of cell cycle function (reviewed in reference 33). The entry of Apoptin into the nucleus is followed by the inhibition of the APC/C, which leads to cell cycle arrest in G2/M and subsequent induction of apoptosis (42).
Two properties of CAV Apoptin make it of particular interest to study cancer-specific processes. First, as stated above, Apoptin selectively induces apoptosis in transformed cells while leaving primary normal cells intact (7). Transient or long-term expression of Apoptin in primary cells has no deleterious effects. Second, induction of apoptosis in transformed cells occurs irrespective of the status of the p53 tumor suppressor gene (8). The p53 pathway is the major mechanism by which cancer cells are destroyed by chemotherapy and radiotherapy (41). However, because the p53 gene is mutated in approximately half of all human tumors, cancer cells are often refractory to these forms of therapy (40). Therefore, the study of Apoptin represents a system for studying novel p53-independent pathways to apoptosis in cancer cells.
The transformed cell-specific killing effects of Apoptin are largely related to differences in subcellular localization of the protein. In primary cells, Apoptin is localized in the cytoplasm, whereas in transformed cells, Apoptin is nuclear (7). The basis for this cell type-specific localization remains to be determined. We have previously demonstrated that in primary cells Apoptin exists in a more aggregated form in the cytoplasm and may be associated with filament networks (42). This differential localization of Apoptin is highly unique and the study of cellular mechanisms regulating this behavior may provide valuable insights into transformed cell physiology. In the present study we address the basis of Apoptin cell type-specific localization and activity. Our results, in conjunction with previous studies, enable us to propose a model for Apoptin-induced transformed cell-selective killing.
MATERIALS AND METHODS
Cells and adenoviruses.
Primary foreskin fibroblast (PFF), H1299, and HA1-IM (S. Bacchetti, McMaster University, Hamilton, Ontario, Canada) cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum plus 10 μg of streptomycin/ml and 10 U of penicillin/ml (Sigma, St. Louis, MO) at 37°C under 5% CO2 (95% air). Apoptin adenoviruses (42) and the LacZ adenovirus (Ad-LacZ) (3) were prepared as previously described. Cells were infected at ∼80% confluence at a multiplicity of infection of 35. Adenovirus was added to cells in a minimal volume of culture media and gently agitated at 37°C under 5% CO2 for 1 h. After infection, culture medium was added, and cells were incubated for the indicated times.
Plasmid constructions.
The green fluorescent protein-Apoptin construct (GFP-Apwt) and deletion mutants have been described previously (42). GFP-Ap-pmNES, -pmNLS, and -pmNLS2 were generated by PCR site-directed mutagenesis. DsRed-Apoptin fusions were generated by PCR amplification of GFP-Apoptin templates, followed by cloning into pDsRed1-N1 (Clontech, Palo Alto, CA). For the dsRed-Ap(82-121) construct, a start codon was added because the endogenous Apoptin 5′ sequence had been removed. GFP-Ap-RevNES was generated by replacing the wild-type Apoptin nuclear export signal (NES) sequence with an oligonucleotide linker containing the human immunodeficiency virus type 1 (HIV-1) Rev NES. GFP-Ap-SV40NLS was made by PCR amplification of Apoptin amino acids 1 to 88 with simultaneous addition of simian virus 40 (SV40) large T nuclear localization signal (NLS) sequence (PKKKRKV) to the C terminus, followed by cloning into pEGFP-C1. To generate the dsRed-dnRan fusion, a construct expressing dnRan (Lan Xu, University of Massachusetts Medical School, Worcester, MA) was PCR amplified and cloned in frame into pDsRed1-N1. All constructs were confirmed by restriction digest analysis and DNA sequencing.
Immunofluorescence and fluorescence microscopy.
Ad-Apwt-infected PFF and HA1-IM cells were fixed in 4% paraformaldehyde (in phosphate-buffered saline [PBS]), permeabilized in 0.5% Triton X-100 (in PBS), and stained with α-Flag M5 primary monoclonal antibody (MAb; Sigma), followed by α-mouse immunoglobulin G Texas Red-conjugated secondary antibody (Sigma). H1299 cells were transiently transfected by using Effectene reagent and after 16 h, fixed in 4% paraformaldehyde, and stained with either α-Cdc27 MAb (Santa Cruz) or α-PML MAb (Santa Cruz), followed by Cy3-conjugated α-mouse secondary antibody. Cells were mounted on slides and observed by using a Zeiss Axiophot2 fluorescence microscope using Axiovision 4.4 software. Images shown are representative examples.
For fluorescence microscopy, H1299 cells were transiently transfected by using Effectene reagent (QIAGEN, Valencia, CA), and PFF transfections were performed using an Amaxa Nucleofector. After 24 h, cells were fixed in 4% paraformaldehyde and stained with DAPI (4′,6′-diamidino-2-phenylindole). Cells were mounted and visualized as described above.
Quantification of the nuclear/cytoplasmic ratio was performed by densitometric analysis of nuclear and cytoplasmic fluorescence signals for at least three regions of interest (ROI) using Axiovision 4.4 software. Background levels of fluorescence were subtracted from average ROI values, and the ratios of nuclear to cytoplasmic signal were calculated and are represented as histograms.
Nucleocytoplasmic shuttling assays.
A total of ∼5 × 105 PFF cells were transiently transfected with GFP-Apwt and 24 h later were treated with leptomycin B (LMB) at a final concentration of 2.5 ng/ml. A total of ∼5 × 105 H1299 cells were transfected with GFP-Apwt or Rev-GFP and 12 h later were transfected with a construct expressing dsRed-dnRan. Cells were visualized by fluorescence microscopy 8 to 12 h later to minimize effects due to protein synthesis or turnover.
Heterokaryon assays.
A total of ∼5 × 105 H1299 or PFF cells were transiently transfected with dsRed-Apwt or GFP-Apwt, respectively, by Amaxa nucleofection. Immediately after transfection, cells were either mixed or plated separately (for controls) in six-well format on coverslips and left to recover for 12 h. The medium was then removed, and the cells were washed twice in PBS and fused by exposure to a solution of 50% polyethylene glycol 1000 (PEG 1000) in serum-free DMEM for 125 s at room temperature. PEG solution was then removed by a wash with PBS, and fresh medium was added. After overnight recovery, cells were fixed in 4% paraformaldehyde (in PBS) and stained with DAPI, and Apoptin localization was determined by fluorescence microscopy.
Immunoprecipitations and immunoblotting.
For multimerization experiments, ∼107 H1299 cells were transiently transfected with GFP-Apoptin truncation mutants by using Effectene. After 12 h cells were infected with Ad-Apwt and incubated for 24 h. Cells were harvested and lysed in buffer X (50 mM Tris [pH 8.5], 250 mM NaCl, 1 mM EDTA, 1% NP-40, Complete Mini tablet [Roche, Basel, Switzerland]) on ice for 20 min. Cell debris was removed by centrifugation, and supernatants were incubated with 30 μl of Ezview Red α-Flag M2 affinity beads (Sigma) at 4°C for 4 h. Beads were then washed in buffer X, and bound proteins were eluted by boiling in sodium dodecyl sulfate (SDS) sample buffer. For solubility analysis, ∼107 H1299 or PFF cells were infected with Ad-Apwt, followed by incubation for 48 h. Cells were then harvested and immunoprecipitated as described above. For APC1 association studies, ∼107 H1299 cells were transiently transfected with Flag-Apoptin constructs, followed by 48 h of incubation. Cells were then harvested and immunoprecipitated as indicated above. For all experiments, whole-cell extracts were prepared by washing cells once in 1× PBS, resuspension in 1× SDS sample buffer, and incubation at 95°C for 5 min.
For immunoblotting, samples were resolved by SDS-polyacrylamide gel electrophoresis with a 6 or 15% polyacrylamide gel and transferred to nitrocellulose. Blots were blocked with 5% milk in TBS-T and probed with either α-Flag M2 MAb (Sigma), α-GFP MAb (Clontech), or α-APC1 affinity-purified polyclonal rabbit serum, followed by appropriate horseradish peroxidase-conjugated α-immunoglobulin G secondary antibody (Amersham Biosciences). Protein bands were visualized by chemiluminescence using SuperSignal substrate (Pierce, Rockford, IL).
Cell viability and apoptosis assays.
For cell viability assays, H1299 and PFF cells infected with Ad-Apwt, Ad-pmNES, or Ad-LacZ were harvested, washed in PBS, and stained with ViaCount reagent (Guava Technologies, Inc., Burlingame, CA), and viability was quantitated by using a Guava Personal flow cytometer. The datum points were collected as percent cell viability per 5,000 events. Apoptosis assays were performed with GFP-Apoptin fusion proteins as previously described (42).
RESULTS
Apoptin is a nucleocytoplasmic shuttling protein.
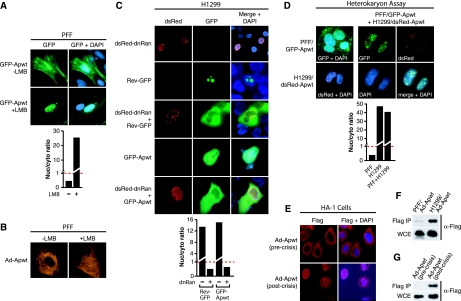
The ability of the CAV protein Apoptin to localize to either the nucleus or the cytoplasm depending on cell type suggested that the protein may undergo nucleocytoplasmic shuttling. To determine whether Apoptin shuttles in primary cells, we transiently transfected PFFs with a plasmid expressing Apoptin fused to the C terminus of GFP (GFP-Apwt) and treated the cells with LMB, a compound that specifically blocks Crm1-mediated nuclear export (25, 52). If Apoptin shuttles in primary cells, then blocking nuclear export should result in the accumulation of Apoptin in the nucleus. Figure 1A shows that LMB treatment resulted in nuclear accumulation of GFP-Apwt in PFFs. To verify this result, PFFs were infected with an adenovirus expressing Flag-tagged Apoptin (42) and treated in the presence or absence of LMB. Figure 1B shows, as expected, that LMB treatment resulted in nuclear accumulation of Flag-Apwt. Consistent with our previous studies (42), Flag-tagged Apoptin exhibited a filamentous staining pattern, which was stable after LMB treatment.
FIG. 1.
Apoptin shuttles between the nucleus and cytoplasm in a Crm1-dependent manner. (A) PFF cells expressing GFP-Apwt were treated in the presence or absence of LMB. (Top) 3 h later, Apoptin localization was visualized by fluorescence microscopy for GFP, and nuclei were visualized by DAPI staining. (Bottom) The ratio of nuclear to cytoplasmic GFP-Apwt signal was quantified for the images presented in the top panel. The dashed red line indicates a ratio of 1 (equal levels of nuclear and cytoplasmic fluorescence). Values greater than 1 indicate predominantly nuclear localization, whereas values less than 1 indicate predominantly cytoplasmic localization. (B) PFF cells were infected with an adenovirus expressing Flag-Apoptin (Ad-Apwt) and treated in the presence or absence of LMB. Cells were stained with an α-Flag antibody, and immunofluorescence analysis was performed 24 h postinfection. (C) H1299 cells were transfected with a construct expressing dsRed-dnRan either alone (top panel) or 12 h after transfection with GFP-Apwt (bottom panel). As a control, H1299 cells were also transfected with either a GFP Rev-GFP fusion alone, Rev-GFP plus dsRed-dnRan, or GFP-Apwt (middle panels). At 12 h after transfection, localization was monitored by fluorescence microscopy for dsRed (left) or GFP (middle) and by DAPI staining (merged, right). Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented is shown. (D) Heterokaryon assay. (Top) PFFs expressing GFP-Apwt and H1299 cells expressing dsRed-Apwt were either mixed or plated separately, incubated for 12 h, and subjected to polyethylene glycol-induced fusion. (Bottom) Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented in the top panel. (E) Immunofluorescence of HA-1 cells infected with Ad-Apwt precrisis (passage 44; upper panels) and postcrisis (passage 94; lower panels). Cells were stained with an α-Flag antibody (left) or DAPI (merged, right). (F) The solubility of Apoptin in PFF and H1299 cell extracts was monitored by immunoprecipitation with an α-Flag affinity resin, and the presence of Apoptin in the immunoprecipitate (top) and WCE (bottom) was monitored by immunoblot analysis. Samples were normalized by equivalent cell number. (G) The solubility of Apoptin in pre- and postcrisis HA-1 cells was monitored by immunoprecipitation as described in panel F.
To test whether Apoptin also shuttles in transformed cells, we developed an assay to monitor the nucleocytoplasmic shuttling of a predominantly nuclear protein. In non-small-cell-lung-carcinoma H1299 cells, we expressed a dominant-negative Ran GTPase mutant (dnRan), which blocks Ran-dependent nuclear import (21). To validate this approach, we performed a control experiment monitoring the effect of dnRan on localization of HIV Rev, a well-characterized nucleocytoplasmic shuttling protein (20, 30). Figure 1C shows, as expected, that a dsRed-dnRan fusion-protein localized to the nuclear periphery (13) and that a Rev-GFP fusion-protein accumulated in the nucleolus (45). Coexpression of dsRed-dnRan and Rev-GFP resulted in the loss of nuclear GFP signal, confirming that Rev-GFP exited the nucleus and was blocked for subsequent reentry. Similarly, expression of dsRed-dnRan resulted in the loss of nuclear GFP-Apwt, indicating that Apoptin shuttled in transformed cells. Collectively, the data of Fig. 1A to C indicate that Apoptin localization is regulated by nucleocytoplasmic shuttling in both primary and transformed cells and that nuclear import and export are mediated by the common nuclear trafficking machinery.
Apoptin nuclear localization is mediated by a dominant transformed cell-specific activity.
To determine whether the transformed or primary cell contained a dominant Apoptin localization activity, we performed a heterokaryon experiment. PFFs were transiently transfected with a plasmid expressing GFP-Apwt, and H1299 cells were transiently transfected with a plasmid expressing Apoptin fused to the C terminus of dsRed (dsRed-Apwt). Twelve hours later, the cells were combined, fused with polyethylene glycol, and allowed to recover overnight. Figure 1D shows that when either PFF/GFP-Apwt or H1299/dsRed-Apwt cells were self-fused, the expected localization patterns were observed. However, fusion of PFF/GFP-Apwt cells with H1299/dsRed-Apwt cells resulted in the translocation of PFF-derived GFP-Apwt to the nucleus where it colocalized with dsRed-Apwt. These results indicate that transformed cells contain a dominant activity that confers Apoptin nuclear localization.
We also monitored localization of Apoptin in HA-1 cells, a clonal population of human embryonic kidney cells transformed with SV40 large T antigen that bypass senescence and enter crisis (6). HA-1 cells were infected with Ad-Apwt and Apoptin localization was monitored both before (passage 44) and after (passage 94) crisis. Figure 1E shows that Apoptin exhibited a filamentous, cytoskeleton-like cytoplasmic staining pattern in precrisis cells, whereas in postcrisis cells Apoptin localized predominantly to the nucleus. Thus, the dominant activity in transformed cells that directs Apoptin to the nucleus appears early during the transformation process.
Biochemical properties of Apoptin differ between primary and transformed cells.
We and others have previously reported that Apoptin forms large cytoplasmic aggregates in primary cells (7, 42). We hypothesized that this aggregation may reflect a fundamental difference in the biochemical state of Apoptin in primary versus transformed cells. The immunoprecipitation experiment of Fig. 1F shows that Apoptin was highly resistant to extraction with a low ionic strength buffer from PFFs compared to H1299 cells. In fact, in primary cells the majority of Apoptin was not solubilized even under high-ionic-strength-conditions (data not shown). Figure 1G shows that Apoptin derived from precrisis HA-1 cells was also highly resistant to extraction compared to postcrisis cells in which Apoptin was readily immunoprecipitated. These results suggest that in transformed cells, nuclear translocation of Apoptin coincides with a large increase in solubility of the protein. The filamentous, cytoplasmic staining pattern observed in primary cells (see Fig. 1B and E and references 7 and 42) suggests that Apoptin may be tightly associated with cellular filament networks, which could explain the difference in solubility between primary and transformed cells.
Functional characterization of Apoptin NES and NLS sequences.
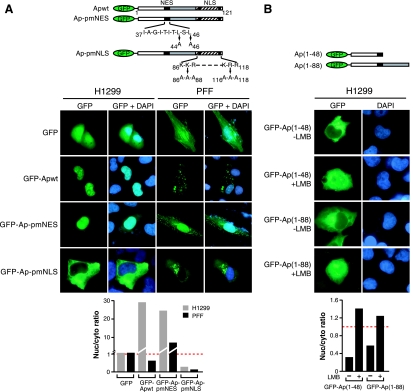
We next sought to characterize the specific sequence elements that contribute to nucleocytoplasmic shuttling. Previous studies have noted a putative canonical NES comprising amino acids 37 to 46 in the N terminus and a putative NLS comprising residues 70 to 121 in the C terminus (shown in Fig. 2A) (9, 47). To determine whether residues 37 to 46 are in fact a functional NES, we constructed a mutant in which the core residues leucine-44 and leucine-46 were mutated to alanine (GFP-Ap-pmNES). Figure 2A shows that in PFFs GFP-Ap-pmNES mislocalized to the nucleus, indicating that the putative NES is functional and that NES-dependent transport of Apoptin is required for cytoplasmic accumulation in primary cells. The putative bipartite NLS contains two domains highly enriched in lysine and arginine (residues 86 to 88 and residues 116 to 118). We constructed an NLS mutant in which both trios of basic amino acids were mutated to alanine (Ap-pmNLS). This mutant mislocalized to the cytoplasm in H1299 cells, indicating the NLS is required for nuclear localization. Thus, Apoptin contains both a functional NES and NLS, a finding consistent with the nucleocytoplasmic shuttling activity described above.
FIG. 2.
Apoptin contains a functional NES and NLS. (A) (Top) Schematic diagrams of N-terminal GFP-tagged Apoptin NES and NLS mutants. (Middle) H1299 and PFF cells expressing GFP-Apoptin mutants were monitored by fluorescence microscopy for GFP (left) and DAPI staining (merged, right). (Bottom) Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented in the middle panel. (B) (Top) Schematic diagrams. (Middle) H1299 cells expressing GFP-Ap(1-48) or GFP-Ap(1-88) were treated in the presence of LMB. After 3 h, Apoptin localization was visualized by fluorescence microscopy for GFP, and nuclei were visualized by DAPI staining. Nuclear entry is presumably facilitated by passive diffusion of the small GFP-Apoptin truncated proteins. (Bottom) Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented in the middle panel.
A recent study has suggested that the Apoptin N-terminal region lacks an NES and that nuclear export is mediated by a noncanonical, Crm1-dependent NES located near the C terminus of the protein (34). To confirm our conclusion that the Apoptin N-terminal region harbors a functional NES, we sought to determine whether the N terminus was sufficient to mediate nuclear export in a Crm1-dependent manner. H1299 cells were transfected with two GFP-fused Apoptin fragments, GFP-Ap(1-48) or GFP-Ap(1-88), both of which contain the N-terminal NES but lack the C-terminal NLS (see Fig. 2A) and putative C-terminal NES (34). Figure 2B shows that both derivatives displayed a predominantly cytoplasmic localization pattern in H1299 cells, indicating the N-terminal NES is sufficient to confer nuclear export. Moreover, treatment with LMB resulted in an increased GFP signal in the nucleus, indicating nuclear export was Crm1 dependent. These results confirm our conclusion that the Apoptin N-terminal region contains a functional, Crm1-dependent NES.
Apoptin fragments containing either the NES or NLS fail to undergo cell type-specific localization.
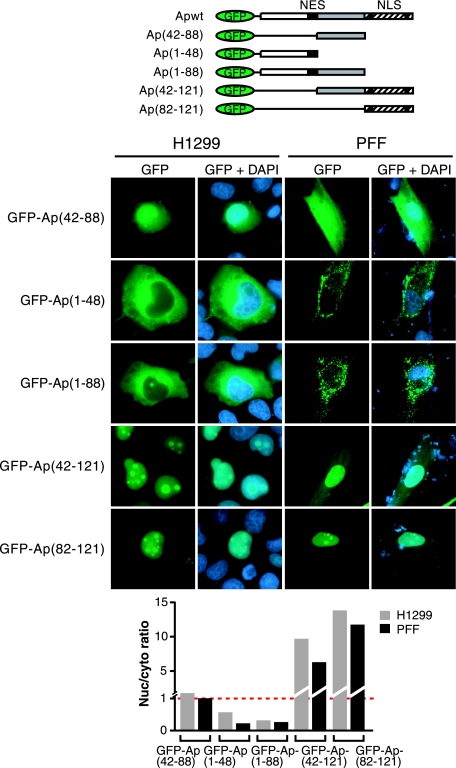
The simplest explanation for the differential localization of Apoptin in primary and transformed cells is that one of the localization signals is subject to cell type-specific regulation. For example, the NLS might be active in transformed cells and inactive in primary cells. To test this model, we analyzed the localization of GFP-fused Apoptin fragments containing either the NLS or NES in primary and transformed cells. Figure 3 shows that GFP-Ap(42-88), which lacks both the NLS and the NES, displayed a diffuse homogeneous localization pattern in both H1299 and PFF cells similar to that of GFP alone. As expected, the GFP-Ap(1-48) and GFP-Ap(1-88) mutants, which contain the N-terminal NES but lack the C-terminal NLS, displayed a predominantly cytoplasmic localization pattern in PFFs and, as shown above, in H1299 cells, indicating the NES is active in transformed cells. Conversely, the GFP-Ap(42-121) and GFP-Ap(82-121) mutants, which contain the C-terminal NLS but lack the N-terminal NES, localized to the nucleus in H1299 cells, as well as in PFFs, indicating the NLS is active in primary cells. Thus, the NLS and NES are active in both primary and transformed cells and, when uncoupled, both localization signals function constitutively.
FIG. 3.
Apoptin fragments containing either the NES or the NLS fail to undergo cell type-specific localization. (Top) Schematic diagrams of GFP-Apoptin deletion mutant derivatives. (Middle) Fluorescence microscopy of H1299 and PFFs expressing GFP-Apoptin derivatives. (Bottom) Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented in the middle panel.
trans restoration of Apoptin cell type-specific localization through protein multimerization.
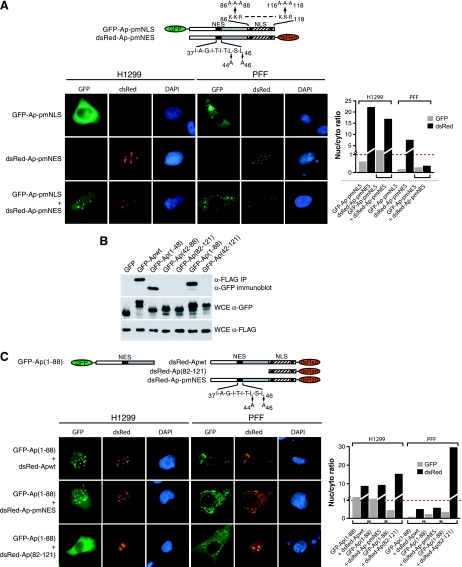
Previous studies have shown that purified, recombinant Apoptin aggregates in vitro (28), raising the possibility that Apoptin functions as a multimer in vivo. As a first test of this possibility, we sought to determine whether cell type-specific localization could be restored in trans by coexpression of two Apoptin fragments, one containing the NLS and the other containing the NES. Figure 4A shows, as expected, that GFP-Ap-pmNLS, which lacks a functional NLS, localized to the cytoplasm of both H1299 cells and PFFs, whereas dsRed-Ap-pmNES, which lacks a functional NES, localized to the nucleus in both H1299 cells and PFFs. However, coexpression of GFP-Ap-pmNLS and dsRed-Ap-pmNES resulted in the localization of both proteins to the nucleus of H1299 cells and the cytoplasm of PFFs. These results suggest that Apoptin is a multimer in vivo and confirm that both localization signals are required for proper cell type-specific localization.
FIG. 4.
Apoptin localization signals are modular and can cooperate in trans to confer cell type-specific localization. (A) H1299 (left panels) and PFF (right panels) cells expressing GFP-Ap-pmNLS, dsRed-Ap-pmNES, or both were monitored by fluorescence microscopy. (Far right) Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented in the panels on the left. (B) H1299 cells were transfected with GFP-Apoptin mutants and 12 h later infected with Ad-Apwt. Apoptin multimerization was monitored by immunoprecipitation of Flag-tagged Apoptin using an α-Flag affinity resin, followed by analysis of the immunoprecipitate (IP) for GFP-Apoptin by immunoblotting with an α-GFP antibody. Expression of the GFP-Apoptin mutants and Flag-Apoptin were also monitored in whole-cell extracts (WCE). Samples were normalized by equivalent cell number. (C) H1299 (left panels) and PFF (right panels) cells were cotransfected with GFP-Ap(1-88) and either dsRed-Apwt (top), dsRed-Ap-pmNES (middle), or dsRed-Ap(82-121) (bottom) and monitored by fluorescence microscopy. (Far right) Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented in panels on the left.
To verify that Apoptin is a multimer in vivo and to map the multimerization domain, we performed a series of coimmunoprecipitation experiments. H1299 cells were transfected with a series of GFP-fused Apoptin derivatives and 12 h later infected with Flag-tagged Ad-Apwt. At 24 h after infection, Flag-Apoptin was immunoprecipitated, and the immunoprecipitate was analyzed by immunoblotting with an α-GFP antibody. Figure 4B shows that only derivatives containing the N-terminal third of Apoptin [GFP-Apwt, GFP-Ap(1-48), GFP-Ap(1-88)] coimmunoprecipitated with the Flag-Apoptin, indicating that this region mediates protein multimerization.
To confirm the results of the coimmunoprecipitation experiments and to verify that multimerization is the basis for restoration of cell type-specific localization in trans, we analyzed three additional Apoptin derivatives in the trans-expression assay. Figure 4C shows that expression of dsRed-Apwt or dsRed-Ap-pmNES restored nuclear localization to the constitutively cytoplasmic GFP-Ap(1-88) mutant in transformed cells and maintained cytoplasmic localization in primary cells. In contrast, dsRed-Ap(82-121) failed to alter cytoplasmic localization of GFP-Ap(1-88) in transformed cells, indicating that the N-terminal multimerization domain is required for trans-association of the localization signals. These results show that the activity of a constitutively cytoplasmic mutant can be restored by expressing an NLS-containing C-terminal fragment in trans.
The specific Apoptin NES is required for cell type-specific localization.
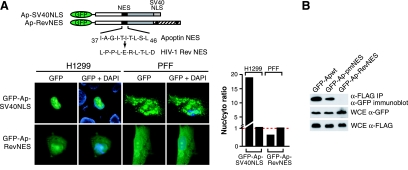
We next sought to determine whether the specific sequence of the Apoptin NES and NLS were required for cell type-specific localization by determining whether they could be functionally substituted with heterologous localization signals. Figure 5A shows that replacement of the Apoptin NLS with that of SV40 large T antigen (Ap-SV40NLS) resulted in nuclear localization in H1299 cells and predominantly cytoplasmic localization in PFFs. Thus, the SV40 large T antigen NLS can functionally substitute for the Apoptin NLS. In contrast, replacement of the Apoptin NES with that of Rev (Ap-RevNES), a well-established, prototypical NES (15, 31, 50), resulted in a similar diffuse localization pattern in both H1299 and PFFs, indicating that the specific sequence of the Apoptin NES is critical for proper cell type-specific localization.
FIG. 5.
The specific Apoptin NES is required for cell type-specific localization and overlaps with the multimerization domain. (A) Fluorescence microscopy of H1299 and PFFs expressing GFP-Apoptin derivatives in which either the NLS was replaced by the SV40 NLS (top) or the NES was replaced by the HIV-1 Rev NES (bottom). (Right) Quantification of the nuclear/cytoplasmic GFP signal ratio for the images presented in panels on the left. (B) Apoptin multimerization was monitored as described for Fig. 4B.
Apoptin NES and multimerization domains overlap.
One explanation for the failure of the Rev NES to functionally substitute for the Apoptin NES is that the Apoptin NES provides an activity in addition to nuclear export. Because both the NES and the multimerization domain map to the N terminus, we reasoned that the NES might be an essential part of the multimerization domain. To test this possibility, we analyzed the two NES derivatives, Ap-pmNES and Ap-RevNES, for their ability to multimerize in the coimmunoprecipitation assay. Figure 5B shows that, compared to GFP-Apwt, the GFP-Ap-pmNES mutant showed a somewhat reduced ability to interact with Flag-Apoptin. Significantly, GFP-Ap-RevNES, which contains a functional NES that differs at multiple residues from the Apoptin NES, failed to detectably coimmunoprecipitate with Flag-Apoptin. Interestingly, the intermediate level of multimerization observed with the Ap-pmNES mutant was sufficient to confer function in the trans assay (see Fig. 4); however, quantification of Fig. 4C revealed that Ap-pmNES was less effective than wild-type Apoptin in mediating trans localization (the nuclear/cytoplasmic ratios were 1.57 and 2.50, respectively), which presumably reflected the decreased multimerization efficiency. Together, these results suggest that the Apoptin multimerization domain overlaps with the NES.
The NLS and nucleocytoplasmic shuttling are required for APC1 interaction and Apoptin-induced cell death.
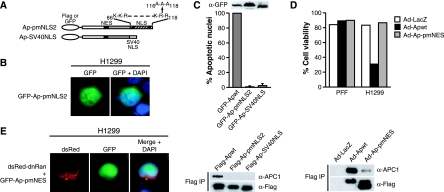
The experiments described above address the contribution of the NES and NLS to protein localization but do not indicate whether these sequences are required for the ability of Apoptin to induce apoptosis. To investigate the role of the NLS, we constructed a second NLS mutant in which lysine-116, arginine-117, and arginine-118 were mutated to alanine (Ap-pmNLS2; Fig. 6A). This mutant protein retained partial nuclear localization in H1299 cells (Fig. 6B). We also analyzed the Ap-SV40NLS mutant, which localized exclusively in the nucleus in H1299 cells (see Fig. 5A). To determine the ability of these two mutants to induce cell death in transformed cells, GFP-Apwt, GFP-Ap-pmNLS2, and GFP-Ap-SV40NLS were transiently expressed in H1299 cells and, 3 days after transfection, the cells were fixed, stained with DAPI, and analyzed by microscopy for apoptotic morphology. As expected, cells expressing GFP-Apwt underwent pronounced apoptosis, whereas the ability of the Ap-pmNLS2 and Ap-SV40NLS mutants to induce apoptosis was severely reduced (Fig. 6C, top).
FIG. 6.
Functionality of both localization signals is critical for APC1 interaction and Apoptin-induced cell death. (A) Schematic diagrams of Apoptin NLS mutations. (B) Fluorescence microscopy of H1299 cells expressing GFP-Ap-pmNLS2. (C) (Top) Apoptosis assays. H1299 cells were transfected with GFP-Apwt, GFP-Ap-pmNLS2, or GFP-Ap-SV40NLS and, after 72 h, fixed, stained with DAPI and analyzed by fluorescence microscopy for GFP expression associated with apoptotic morphology. The percent apoptosis in cells expressing GFP alone was taken as the background and subtracted from all samples. All mutant samples are shown as the percent apoptosis of GFP-Apwt. (Inset) Expression of GFP-Apwt, GFP-Ap-pmNLS2, and GFP-Ap-SV40NLS 24 h after transfection. (Bottom) APC1 association. H1299 cells expressing Flag-tagged versions of Apoptin mutants were immunoprecipitated with an α-Flag antibody, and the immunoprecipitate was analyzed for APC1 and Apoptin by immunoblotting. Sample loading was normalized by equivalent cell number. (D) (Top) PFF or H1299 cells were infected with Ad-LacZ, Ad-Apwt, or Ad-Ap-pmNES, and 72 h later the cell viability was quantified by flow cytometry. (Bottom) APC1 association. Samples were normalized by equivalent cell number. (E) DnRan shuttling assay, performed as described for Fig. 1C.
To understand why the Ap-pmNLS2 and Ap-SV40NLS mutants failed to induce apoptosis in transformed cells even though they localized to the nucleus, we next monitored their ability to interact with the APC1 subunit of the APC/C, which we have previously shown interacts with the C terminus of Apoptin (42). A triple Flag-tagged version of each mutant was transiently transfected into H1299 cells and immunoprecipitated with an α-Flag antibody, and the immunoprecipitate was analyzed for APC1 by immunoblotting. As expected, APC1 was present in the immunoprecipitate from wild-type Apoptin but not in that of the Ap-pmNLS2 or Ap-SV40NLS mutants (Fig. 6C, bottom) despite the presence of both of these mutants in the nucleus. These observations are consistent with our previous results (42) and suggest that the NLS sequence overlaps with the domain required for association with APC1. Furthermore, these results indicate that nuclear localization in the absence of APC1 association is not sufficient to induce apoptosis.
To assess the involvement of the NES in Apoptin-induced cell death, we next investigated the ability of the Ap-pmNES mutant, which contains a wild-type C-terminal domain and localizes to the nucleus in transformed and primary cells (see Fig. 2A), to interact with APC1 and induce apoptosis. Figure 6D shows that despite its nuclear localization, this mutant failed to induce apoptosis in PFFs, a finding consistent with previous studies showing that nuclear localization of Apoptin is not sufficient to induce apoptosis in primary cells (9). Surprisingly, however, this mutant also failed to induce apoptosis in transformed cells (Fig. 6D, top) and exhibited greatly reduced ability to interact with APC1 (Fig. 6D, bottom). The dnRan assay of Fig. 6E shows, as expected, that the Ap-pmNES mutant failed to undergo nucleocytoplasmic shuttling. Thus, the shuttling activity of Apoptin is required for effective interaction with APC1 and induction of apoptosis.
Apoptin recruits APC/C to PML bodies in transformed cells.
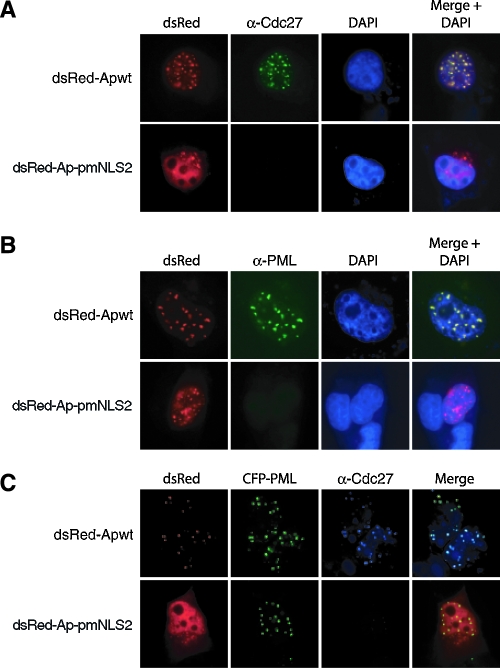
The fact that Apoptin interacts with APC1 (42) suggested that in transformed cells Apoptin would colocalize with APC/C complex. Because the APC1 antibody was not suitable for immunofluorescence (data not shown), to test this prediction we performed immunofluorescence for Cdc27 (also known as APC3), a core APC/C subunit routinely used as a marker for APC/C localization (17, 35). H1299 cells were transiently transfected with a plasmid expressing dsRed-Apwt, incubated for 12 h, fixed, and stained with an α-Cdc27 antibody. Figure 7A shows that dsRed-Apwt colocalized with Cdc27 in the nucleus of transformed cells. However, we failed to detect Cdc27 immunofluorescence in the presence of the Ap-pmNLS2 mutant, which, unlike wild-type Apoptin (42), does not associate with the APC/C and fails to induce apoptosis (Fig. 6C) and G2/M arrest (data not shown). This result is consistent with previous reports showing that several APC/C subunits, including Cdc27, are detected only in mitotic and not in interphase cells (16, 19).
FIG. 7.
Apoptin colocalizes with APC/C within PML bodies in the nuclei of transformed cells. (A and B) H1299 cells were transiently transfected with a plasmid expressing either dsRed-Apwt or dsRed-Ap-pmNLS2 and, after a 12-h incubation, were fixed and stained with an α-Cdc27 antibody (A) or an α-PML antibody (B). (C) H1299 cells were cotransfected with plasmids expressing either dsRed-Apwt or dsRed-Ap-NLS2 and a plasmid expressing CFP-PML. At 12 h after transfection, cells were fixed and stained with an α-Cdc27 antibody.
It has recently been reported that Apoptin associates with PML nuclear bodies (34). To verify this result, cells were transfected with either dsRed-Apwt or, as a control, dsRed-Ap-pmNLS2 and stained with an α-PML antibody. Figure 7B confirms that wild-type Apoptin colocalized with PML bodies within the nucleus of transformed cells. Significantly, in cells transfected with dsRed-Ap-pmNLS2, PML immunofluorescence was undetectable, a finding consistent with previous studies showing that in H1299 cells PML nuclear body formation occurs only under conditions of apoptotic stimuli (11). Thus, Apoptin induces the formation of PML nuclear bodies in transformed cells in a manner that requires APC/C interaction.
The results described above indicate that Apoptin colocalizes with the APC/C (Fig. 7A) and that Apoptin is present in PML bodies (Fig. 7B). These results strongly suggested that in cells expressing Apoptin, APC/C would be present in PML bodies. To provide direct evidence for this supposition, plasmids expressing either dsRed-Apwt or dsRed-Ap-pmNLS2 were cotransfected into H1299 cells with a plasmid constitutively expressing PML fused to the C terminus of cyan fluorescent protein (CFP-PML). At 12 h after transfection, cells were fixed and stained with an α-Cdc27 antibody. Figure 7C shows that, as expected, dsRed-Apwt, Cdc27 and CFP-PML colocalized in the nucleus of transformed cells. Thus, in the presence of Apoptin, the APC/C becomes sequestered in PML bodies.
DISCUSSION
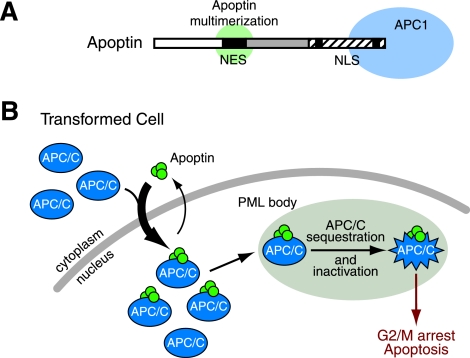
We show here for the first time that Apoptin is a nucleocytoplasmic shuttling protein with a modular structure organized into overlapping functional domains. Whereas previous studies have implicated the existence of putative localization sequences, here we conclusively delineate functional NES and NLS sequences and their respective activities in both primary and transformed cells. A Crm1-dependent NES at the N terminus overlaps with a domain required for Apoptin multimerization, whereas the NLS at the C terminus overlaps with the APC1 interaction domain (Fig. 8A). Overlapping functional domains is a common theme among viruses, which have compact genomes. In particular, the existence of domains that overlap with NES and NLS sequences has been implicated in the regulation of at least one other viral protein, HIV-1 Rev (4, 46). Together the NES, NLS, multimerization, and APC1 interaction domains impart Apoptin with the differential localization, biochemical behavior, and protein interactions that provide the basis for selective killing of transformed cells. The results in the present study allow us to propose a model for how Apoptin selectively kills transformed cells (presented in Fig. 8B and discussed below).
FIG. 8.
Multifunctional localization signals mediate the cell type-specific localization of Apoptin. (A) Schematic diagram of Apoptin's localization signals and overlapping protein interaction domains. (B) A model for the transformed cell-specific killing activity of Apoptin. In transformed cells, Apoptin shuttling is shifted toward nuclear accumulation. The localization of APC/C subunits is not clear; Apoptin may either transport APC/C subunits from the cytoplasm to the nucleus or associate with APC/C subunits already present in the nucleus. Apoptin-mediated recruitment of APC/C into PML nuclear bodies in transformed cells leads to inactivation of APC/C function and subsequent induction of G2/M arrest and apoptosis.
Role of nucleocytoplasmic shuttling.
Unexpectedly, we found that nucleocytoplasmic shuttling is required for both cell type-specific localization and the induction of apoptosis. The equilibrium dynamics of Apoptin nucleocytoplasmic shuttling provides a powerful mechanism for inducing rapid changes in protein localization with minimal changes in cellular activities. Such rapid changes in protein localization and apoptosis induction may be important for the timing of the CAV life cycle and viral egress.
The results presented here and in our previous study (42) indicate that APC1 is a critical target of Apoptin. The APC/C complex has essential nuclear functions such as chromosome segregation and cyclin turnover (33). However, recent studies suggest that many of the APC/C subunits, and perhaps specific APC/C subcomplexes, reside predominantly in the cytoplasm and only transiently associate with nuclear components during mitosis (16, 17, 44). Thus, the nucleocytoplasmic shuttling activity of Apoptin may function to transport APC/C from the cytoplasm to the nucleus, where it is deposited into PML nuclear bodies. Consistent with this proposal, Apoptin mutants that are defective in shuttling show a greatly reduced ability to interact with APC1 and fail to induce apoptosis (Fig. 6D and E).
Role of multimerization.
The multimerization state of Apoptin appears to vary in degree between cell types and affects its localization and apoptotic activities. Although the physical basis of this phenomenon remains to be explained, a higher state of multimerization in primary cells could explain the aggregation, insolubility, and retention of Apoptin in the cytoplasm. Multimerization is also required to establish a high-affinity interaction with APC1, since Apoptin mutants that are decreased in multimerization display a reduced affinity for APC1. Although previous studies have suggested that Apoptin multimerization occurs through direct Apoptin-Apoptin interaction (28), it remains possible that Apoptin molecules associate indirectly through one or more cellular bridging proteins.
Regulation of Apoptin localization and activity.
In the present study we have shown that Apoptin nuclear localization is mediated by a dominant transformed cell-specific activity. However, the nature of the cell type-specific modification that controls Apoptin localization change remains unclear. Some studies (34, 37) have reported that Apoptin regulated localization is controlled by cell-specific phosphorylation on threonine-108 at the C terminus (47). In contrast, we have found that Apoptin derivatives in which the C-terminal region is substituted by heterologous sequences localize normally (see Fig. 5A), indicating that Apoptin sequences at the C terminus are dispensable for regulated localization. Instead, our results indicate that the N-terminal region of Apoptin, containing the NES and multimerization domain, when coupled to a generic NLS activity, mediates regulated localization (see Fig. 5A). This result raises the possibility that the dominant transformed cell-specific activity may negatively regulate the NES, which, in turn, favors NLS function and nuclear localization. In addition, our results are consistent with studies that have shown that the NLS region is not sufficient to confer transformed cell-specific nuclear localization (9, 47). In contrast to that study (47), however, we (42) and others (36) found no evidence that the cellular concentration of Apoptin has any effect on localization.
Role of PML body induction and colocalization with APC/C.
We have shown that introduction of Apoptin into transformed cells results in the formation of PML nuclear bodies and the recruitment of Apoptin-APC/C complexes into these subnuclear structures. The localization of APC/C subunits to PML bodies is a direct result of recruitment by Apoptin and not an indirect consequence of the G2/M arrest: in the absence of Apoptin, PML bodies are not detectable (reference 11 and data not shown), and PML itself is present at very low levels in G2/M (53).
Several studies have shown that cells respond to various types of stimuli by increasing both the size and the number of PML nuclear bodies (11, 23, 43). In addition to established roles in tumor suppression and senescence (reviewed in reference 10), PML nuclear bodies play an integral role in apoptotic processes and have been proposed to serve as a “hub” for nuclear apoptotic reinforcement (5). Cells derived from PML−/− knockout mice are virtually resistant to apoptosis stimulated by intrinsic as well as extrinsic stimuli, indicating a strong connection between PML body formation and induction or execution of programmed cell death. Interestingly, PML nuclear bodies have been implicated in the execution of p53-independent (as well as p53-dependent) apoptotic programs (48, 49, 54), an important aspect of Apoptin-induced cell death. PML nuclear bodies are potential centers for the control and regulation of proteins by posttranslational modification (e.g., sumoylation, acetylation, and ubiquitination) and by degradation (2, 26, 27). For example, many cellular and viral sumoylated proteins are targeted to PML nuclear bodies (39). In many cases, however, the functional significance between protein modification and PML localization remains unclear. Interestingly, studies in yeast have shown that the mitotic functions of the APC/C are modulated by sumoylation and ubiquitination (12). Thus, the Apoptin-mediated recruitment of APC/C components (or subcomplexes) into PML nuclear bodies suggests a functional connection between Apoptin-APC/C interaction, cyclosome inhibition and induction of apoptosis. Abrogation of APC/C activity through targeting to PML nuclear bodies provides a method by which APC/C could be sequestered and inhibited, and apoptotic programming of nuclear origin could ensue.
Inhibition of the APC/C may represent a major tumor suppressor mechanism (reviewed in reference 14). For example, the endogenous APC/C inhibitor RASSF1 is mutated in some human cancers (1). In addition, dysfunction of APC/C and mitotic checkpoint function has been suggested to contribute to transformation and could lead to properties in cancer cells such as unscheduled proliferation and aneuploidy. Several viral proteins, including the adenovirus E4orf4, human T lymphotrophic virus type 1 Tax, and an as-yet-unidentified protein from cytomegalovirus, interact with the APC/C and deregulate its activity (24, 29, 51). These independently evolved viral mechanisms of targeting APC/C also suggest an underlying role for the APC/C as a nodal point in cell cycle control and tumor suppression.
Acknowledgments
We thank Silvia Bacchetti, Lan Xu, and Maria Zapp for reagents and Sara Evans for editorial assistance.
This study was supported in part by an NIH grant to M.R.G. and postdoctoral fellowships from the NCI of Canada and the Medical Foundation Charles A. King Trust to J.G.T. M.R.G. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agathanggelou, A., W. N. Cooper, and F. Latif. 2005. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 65:3497-3508. [DOI] [PubMed] [Google Scholar]
- 2.Anton, L. C., U. Schubert, I. Bacik, M. F. Princiotta, P. A. Wearsch, J. Gibbs, P. M. Day, C. Realini, M. C. Rechsteiner, J. R. Bennink, and J. W. Yewdell. 1999. Intracellular localization of proteasomal degradation of a viral antigen. J. Cell Biol. 146:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacchetti, S., and F. L. Graham. 1993. Inhibition of cell proliferation by an adenovirus vector expressing the human wild-type p53 protein. Int. J. Oncol. 3:781-788. [DOI] [PubMed] [Google Scholar]
- 4.Berger, J., C. Aepinus, M. Dobrovnik, B. Fleckenstein, J. Hauber, and E. Bohnlein. 1991. Mutational analysis of functional domains in the HIV-1 Rev trans-regulatory protein. Virology 183:630-635. [DOI] [PubMed] [Google Scholar]
- 5.Bernardi, R., and P. P. Pandolfi. 2003. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 22:9048-9057. [DOI] [PubMed] [Google Scholar]
- 6.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danen-Van Oorschot, A. A., D. F. Fischer, J. M. Grimbergen, B. Klein, S. Zhuang, J. H. Falkenburg, C. Backendorf, P. H. Quax, A. J. Van der Eb, and M. H. Noteborn. 1997. Apoptin induces apoptosis in human transformed and malignant cells but not in normal cells. Proc. Natl. Acad. Sci. USA 94:5843-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danen-Van Oorschot, A. A., A. J. van der Eb, and M. H. Noteborn. 1999. BCL-2 stimulates Apoptin-induced apoptosis. Adv. Exp. Med. Biol. 457:245-249. [DOI] [PubMed] [Google Scholar]
- 9.Danen-Van Oorschot, A. A., Y. H. Zhang, S. R. Leliveld, J. L. Rohn, M. C. Seelen, M. W. Bolk, A. Van Zon, S. J. Erkeland, J. P. Abrahams, D. Mumberg, and M. H. Noteborn. 2003. Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J. Biol. Chem. 278:27729-27736. [DOI] [PubMed] [Google Scholar]
- 10.Dellaire, G., and D. P. Bazett-Jones. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26:963-977. [DOI] [PubMed] [Google Scholar]
- 11.de Stanchina, E., E. Querido, M. Narita, R. V. Davuluri, P. P. Pandolfi, G. Ferbeyre, and S. W. Lowe. 2004. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 13:523-535. [DOI] [PubMed] [Google Scholar]
- 12.Dieckhoff, P., M. Bolte, Y. Sancak, G. H. Braus, and S. Irniger. 2004. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol. Microbiol. 51:1375-1387. [DOI] [PubMed] [Google Scholar]
- 13.Gorlich, D., N. Pante, U. Kutay, U. Aebi, and F. R. Bischoff. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584-5594. [PMC free article] [PubMed] [Google Scholar]
- 14.Heilman, D. W., M. R. Green, and J. G. Teodoro. 2005. The anaphase promoting complex: a critical target for viral proteins and anti-cancer drugs. Cell Cycle 4:560-563. [PubMed] [Google Scholar]
- 15.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 16.Huang, J., and J. W. Raff. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18:2184-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, J. Y., and J. W. Raff. 2002. The dynamic localization of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localized. J. Cell Sci. 115:2847-2856. [DOI] [PubMed] [Google Scholar]
- 18.Jeurissen, S. H., F. Wagenaar, J. M. Pol, A. J. van der Eb, and M. H. Noteborn. 1992. Chicken anemia virus causes apoptosis of thymocytes after in vivo infection and of cell lines after in vitro infection. J. Virol. 66:7383-7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen, P. M., E. Brundell, M. Starborg, and C. Hoog. 1998. A subunit of the anaphase-promoting complex is a centromere-associated protein in mammalian cells. Mol. Cell. Biol. 18:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalland, K. H., A. M. Szilvay, K. A. Brokstad, W. Saetrevik, and G. Haukenes. 1994. The human immunodeficiency virus type 1 Rev. protein shuttles between the cytoplasm and nuclear compartments. Mol. Cell. Biol. 14:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klebe, C., F. R. Bischoff, H. Ponstingl, and A. Wittinghofer. 1995. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34:639-647. [DOI] [PubMed] [Google Scholar]
- 22.Klein, G. 2002. Perspectives in studies of human tumor viruses. Front. Biosci. 7:d268-d274. [DOI] [PubMed] [Google Scholar]
- 23.Koken, M. H., G. Linares-Cruz, F. Quignon, A. Viron, M. K. Chelbi-Alix, J. Sobczak-Thepot, L. Juhlin, L. Degos, F. Calvo, and H. de The. 1995. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene 10:1315-1324. [PubMed] [Google Scholar]
- 24.Kornitzer, D., R. Sharf, and T. Kleinberger. 2001. Adenovirus E4orf4 protein induces PP2A-dependent growth arrest in Saccharomyces cerevisiae and interacts with the anaphase-promoting complex/cyclosome. J. Cell Biol. 154:331-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 26.Lafarga, M., M. T. Berciano, E. Pena, I. Mayo, J. G. Castano, D. Bohmann, J. P. Rodrigues, J. P. Tavanez, and M. Carmo-Fonseca. 2002. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13:2771-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P. P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leliveld, S. R., Y. H. Zhang, J. L. Rohn, M. H. Noteborn, and J. P. Abrahams. 2003. Apoptin induces tumor-specific apoptosis as a globular multimer. J. Biol. Chem. 278:9042-9051. [DOI] [PubMed] [Google Scholar]
- 29.Liu, B., S. Hong, Z. Tang, H. Yu, and C. Z. Giam. 2005. HTLV-1 Tax directly binds the Cdc20-associated anaphase-promoting complex and activates it ahead of schedule. Proc. Natl. Acad. Sci. USA 102:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev. transactivator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, B. E., J. L. Meinkoth, and M. H. Malim. 1996. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev. proteins: identification of a family of transferable nuclear export signals. J. Virol. 70:2350-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noteborn, M. H., D. Todd, C. A. Verschueren, H. W. de Gauw, W. L. Curran, S. Veldkamp, A. J. Douglas, M. S. McNulty, E. A. van der, and G. Koch. 1994. A single chicken anemia virus protein induces apoptosis. J. Virol. 68:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters, J. M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9:931-943. [DOI] [PubMed] [Google Scholar]
- 34.Poon, I. K., C. Oro, M. M. Dias, J. Zhang, and D. A. Jans. 2005. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res. 65:7059-7064. [DOI] [PubMed] [Google Scholar]
- 35.Rape, M., and M. W. Kirschner. 2004. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 432:588-595. [DOI] [PubMed] [Google Scholar]
- 36.Rohn, J. L., Y.-H. Zhang, S. R. Leliveld, A. A. A. M. Danen-Van Oorschot, N. V. Henriquez, J. P. Abrahams, and M. H. M. Noteborn. 2005. Relevance of Apoptin's integrity for its functional behavior. J. Virol. 79:1337-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohn, J. L., Y. H. Zhang, R. I. Aalbers, N. Otto, J. Den Hertog, N. V. Henriquez, C. J. Van De Velde, P. J. Kuppen, D. Mumberg, P. Donner, and M. H. Noteborn. 2002. A tumor-specific kinase activity regulates the viral death protein Apoptin. J. Biol. Chem. 277:50820-50827. [DOI] [PubMed] [Google Scholar]
- 38.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 39.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 40.Sherr, C. J. 2004. Principles of tumor suppression. Cell 116:235-246. [DOI] [PubMed] [Google Scholar]
- 41.Soussi, T., and G. Lozano. 2005. p53 mutation heterogeneity in cancer. Biochem. Biophys. Res. Commun. 331:834-842. [DOI] [PubMed] [Google Scholar]
- 42.Teodoro, J. G., D. W. Heilman, A. E. Parker, and M. R. Green. 2004. The viral protein Apoptin associates with the anaphase-promoting complex to induce G2/M arrest and apoptosis in the absence of p53. Genes Dev. 18:1952-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terris, B., V. Baldin, S. Dubois, C. Degott, J. F. Flejou, D. Henin, and A. Dejean. 1995. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 55:1590-1597. [PubMed] [Google Scholar]
- 44.van Roessel, P., D. A. Elliott, I. M. Robinson, A. Prokop, and A. H. Brand. 2004. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119:707-718. [DOI] [PubMed] [Google Scholar]
- 45.Venkatesh, L. K., and G. Chinnadurai. 1990. Mutants in a conserved region near the carboxy terminus of HIV-1 Rev. identify functionally important residues and exhibit a dominant negative phenotype. Virology 178:327-330. [DOI] [PubMed] [Google Scholar]
- 46.Venkatesh, L. K., T. Gettemeier, and G. Chinnadurai. 2003. A nuclear kinesin-like protein interacts with and stimulates the activity of the leucine-rich nuclear export signal of the human immunodeficiency virus type 1 rev protein. J. Virol. 77:7236-7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wadia, J. S., M. V. Wagner, S. A. Ezhevsky, and S. F. Dowdy. 2004. Apoptin/VP3 contains a concentration-dependent nuclear localization signal (NLS), not a tumorigenic selective NLS. J. Virol. 78:6077-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Z. G., D. Ruggero, S. Ronchetti, S. Zhong, M. Gaboli, R. Rivi, and P. P. Pandolfi. 1998. PML is essential for multiple apoptotic pathways. Nat. Genet. 20:266-272. [DOI] [PubMed] [Google Scholar]
- 50.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 51.Wiebusch, L., M. Bach, R. Uecker, and C. Hagemeier. 2005. Human cytomegalovirus inactivates the G(0)/G(1)-APC/C ubiquitin ligase by cdh1 dissociation. Cell Cycle 4:1435-1439. [DOI] [PubMed] [Google Scholar]
- 52.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 53.Xu, Z. X., R. X. Zhao, T. Ding, T. T. Tran, W. Zhang, P. P. Pandolfi, and K. S. Chang. 2004. Promyelocytic leukemia protein 4 induces apoptosis by inhibition of survivin expression. J. Biol. Chem. 279:1838-1844. [DOI] [PubMed] [Google Scholar]
- 54.Zhong, S., P. Salomoni, S. Ronchetti, A. Guo, D. Ruggero, and P. P. Pandolfi. 2000. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med. 191:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.zur Hausen, H. 2001. Oncogenic DNA viruses. Oncogene 20:7820-7823. [DOI] [PubMed] [Google Scholar]