Abstract
The UL32 gene of human cytomegalovirus (CMV) encodes a prominent betaherpesvirus-conserved virion tegument protein, called pp150 (basic phosphoprotein/ppUL32), that accumulates within a cytoplasmic inclusion adjacent to the nucleus at late times during infection. Using a UL32 deletion mutant (ΔUL32-BAC) (where BAC is bacterial artificial chromosome), we demonstrate that pp150 is critical for virion maturation in the cytoplasmic compartment. Cotransfection of a pp150 expression plasmid with ΔUL32-BAC DNA led to complementation of the replication defect with focus formation due to secondary spread. Deletion of the amino terminus of pp150 or disruption of the betaherpesvirus conserved regions, CR1 and CR2, revealed these regions to be critical for replication. In contrast, deletion of the carboxyl terminus only partially compromised maturation while disruption of glycosylation sites had no effect. An African green monkey CMV UL32 homolog complemented ΔUL32-BAC replication but murine CMV M32 failed to complement, consistent with evolutionary divergence of rodent and primate cytomegaloviruses. Infection with ΔUL32-BAC showed normal expression of all kinetic classes of viral genes and replication of viral DNA, with accumulation of viral DNA-containing particles in the cytoplasm; however, mutant virus did not spread to adjacent cells. In contrast to this block in virion infectivity, cell-to-cell transfer of pp65-containing particles was observed, suggesting that release of dense bodies continued in the absence of pp150. These observations demonstrate that pp150 is critical for virion egress, possibly at the stage of final envelopment.
The process of herpesvirus virion maturation and egress begins in the nucleus, where preformed capsids incorporate newly replicated viral DNA in a herpesvirus-conserved cleavage/packaging process (15, 37). The acquisition of an initial envelope at the inner nuclear membrane is also conserved and requires budding into the space between the inner and outer nuclear membranes, dependent upon herpesvirus-common viral proteins that modify the nuclear lamella (14, 23, 38, 42, 53). Following this step, maturation may follow one of two directions that remain controversial. Egress from cells was initially believed to require a vesicle derived from the outer nuclear membrane surrounding the newly enveloped virus particle, followed by vesicle transport via the secretory pathway and release of vesicle contents at the plasma membrane (32, 44). However, an alternative process has gained experimental support. The initial envelope obtained at the inner nuclear membrane is lost by a fusion event at the outer nuclear membrane (deenvelopment), releasing free nucleocapsids into the cytoplasm, where egress continues through a second, or final, envelopment step (32). Although electron micrographs have long suggested that this pathway was possible, the study of mutations that affect herpesvirus core genes encoding essential structural proteins has led to accumulated support for this envelopment/deenvelopment/reenvelopment egress pathway (2, 7, 24, 25, 31, 32, 54). Support for this model has come from studies of alphaherpesviruses (12, 31, 32, 54) as well as the betaherpesvirus human cytomegalovirus (HCMV) (9, 19, 21, 22, 47, 52, 55).
Envelopment of HCMV tegument-containing particles lacking capsids or viral DNA, called dense bodies, occurs only within a cytoplasmic inclusion and not in the infected cell nucleus (37). This inclusion becomes a prominent feature of infected cells by late times of infection and is likely to be associated with the final envelopment step based on lipid content of the virion envelope (55), localization studies (17, 19, 47, 50), and recent studies of viral mutants disrupting the gene (UL99) encoding the myristoylated structural protein pp28 (9, 22, 47, 52). pp28 plays a role as a cytoplasmic egress tegument protein that is conserved among mammalian herpesviruses (35, 36). HCMV nucleocapsids probably transit the cytoplasm to final envelopment sites in the endoplasmic reticulum-Golgi intermediate compartment or possibly even the trans-Golgi network that is within the cytoplasmic inclusion.
Eight herpesvirus core (UL48, UL71, UL76, UL77, UL93, UL94, UL95, and UL99) and two betaherpesvirus-conserved (UL32 and UL96) HCMV tegument protein genes play critical roles in viral replication (13, 31, 44, 58). Although the role of pp28 in envelopment has been investigated (9, 22, 52), little is known about the contributions of other tegument proteins to envelopment. The betaherpesvirus-conserved UL32 gene product pp150 (also called basic phosphoprotein or ppUL32) is a major tegument phosphoprotein that localizes to prominent cytoplasmic inclusions where final envelopment of virions and noninfectious enveloped particles likely occurs (17, 19, 47, 50). Tegument proteins (pp65 and pp28 as well as pp150) and envelope glycoproteins (glycoprotein B [gB] and gH) colocalize with nucleocapsids at these inclusions, consistent with their role in virion maturation and/or egress (26, 47). HCMV pp150 has been implicated as a partner interacting with capsids, based on electron cryomicroscopy studies using isolated B capsids purified from either HCMV- or simian CMV (SCMV)-infected cells (10, 56). The amino terminus of HCMV pp150 or the SCMV UL32 homolog has been shown to mediate binding to SCMV or HCMV capsids (5). Studies that preceded recent global mutagenesis of the viral genome (13, 58) first suggested a role for UL32 late in replication (33, 59). pp150 is a major component of mature virions as well as noninfectious enveloped particles (20) but is only a minor component of dense bodies (43). Antisense inhibition of pp150 appeared to block nuclear egress of DNA-containing capsids, even though the approach did not eliminate protein expression (33). Furthermore, the antisense method exhibited potential nonspecific effects, such as the reduced levels of gB that may have contributed to the observed inhibition of replication. Although reports vary, pp150 has been detected within the endoplasmic reticulum-Golgi intermediate compartment (19), in close proximity to the microtubule organizing center (47), and in both the nuclear and cytoplasmic compartments of infected cells (17, 45). Binding and localization studies suggest pp150 may support egress or envelopment of viral particles, possibly contributing to one or more steps, including initial envelopment, tegumentation or trafficking, and final envelopment.
This study investigates the role of pp150 in HCMV replication, employing a multifaceted approach to dissect UL32 function by evaluating viral replication in cells transfected with a mutant, ΔUL32-BAC (where BAC is bacterial artificial chromosome), alone or together with pp150 expression plasmids, and compares the phenotype to those of parental Towne-BAC and rescued-UL32-BAC strains (13). Four approaches were employed: (i) transfection of ΔUL32-BAC alone, (ii) cotransfection of ΔUL32-BAC with pp150 expression plasmids to evaluate the formation of foci arising through secondary spread, (iii) infection with mutant virus collected from cotransfected cells, and (iv) infection with mutant virus propagated on a pp150-expressing cell line. These experiments demonstrate that UL32 is necessary for virion maturation at a time that coincides with accumulation of viral structural proteins at the cytoplasmic inclusion. Although glycosylation sites are not involved in pp150 function and the carboxyl-terminal region may be deleted without a major reduction in function, the amino terminus and conserved regions (CR1 and CR2) are critical in supporting virion maturation. Finally, we show that cell-to-cell transfer of pp65 is independent of UL32 function, suggesting that the release of dense bodies follows a pathway that is distinct from that of virions.
MATERIALS AND METHODS
Cells.
All cells were grown at 37°C in a 5% CO2 atmosphere. Primary human foreskin fibroblasts (HFFs) and the UL32-HFF line were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FetalClone III (HyClone, Logan, UT). 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS).
DNA constructs.
Production of HCMV TownevarATCC strain bacterial artificial chromosome (Towne-BAC), ΔUL32-BAC, and rescued-UL32-BAC was previously described (13, 28). Each has the same BAC elements, including a chloramphenicol resistance gene and a eukaryotic green fluorescent protein (GFP) expression cassette replacing the 9-kbp US1 to US12 region of the viral genome (28), and each was propagated in either DH10B (at 37°C) or DY380 (at 32°C) bacteria grown in Luria-Bertani broth for 15 to 20 h. BAC DNA was purified with a NucleoBond BAC Maxi kit (BD Biosciences), and 2 μg of resultant DNA was digested with HindIII and separated on a 0.8% agarose gel to confirm the cleavage pattern of viral DNAs. A pp150 expression plasmid (pON2780) was constructed using the primers 5′-TCGTCGCGATCGACCATGAGTTTGCAGTTTATCGGTCTAC-3′and 5′-TCGTCGCGATCGTTCCTCCGTGTTCTTAATCTTCTCG-3′to amplify the UL32 open reading frame (ORF) from Towne-BAC. This product was cleaved with PvuI and cloned into the compatible PacI sites of the pLNCX vector (34). The UL32 ORF was engineered in frame with a 3′ c-Myc sequence contained within pLNCX. All of the UL32 expression constructs used in this study included an in-frame c-Myc epitope tag that did not compromise function in any assays based on direct comparison with a nontagged construct. A UL32 N-terminal truncation mutant(pON2781) was constructed using the primers 5′-CGCGGATCCCCACCATGGACCTCTCCTCAGTGCTC-3′and 5′-CTTGCGGCCGCCTACAAGTCTTCTTCAGAAATCAGCTTTTGTTCTTCCTCCGTGTTCTTAATCTT-3′to amplify a 5′ UL32 truncation fragment from pON2780. This fragment was cleaved with BamHI and NotI and cloned into pcDNA3.1zeo+ (Invitrogen). A UL32 C-terminal truncation mutant (pON2782) was constructed using the primers 5′-CGCGGATCCCCACCATGGAACAAAAGCTGATTTCTGAAGAAGACTTGATGAGTTTGCAGTTTATCGGT-3′and 5′-CTTGCGGCCGCCTAGCGTGATTGCAAAGCCGCGCT-3′to amplify a fragment from pON2780. This fragment was cleaved with BamHI and NotI and cloned into pcDNA3.1zeo+. A substitution mutation within CR1 produced the plasmid pON2783, and this mutant was constructed using the primers 5′-GTGCTGTTCAACGAGCTCATCATCGATTTGGGATACTACCGCGAGCTG-3′and 5′-CAGCTCGCGGTAGTATCCCAAATCGATCATGAGCTCGTTGAACAGCAC-3′. A UL32 CR2 substitution mutant (pON2784) was constructed with the primers 5′-CTGGTCAACGCCGTCGCAATCGATGCAGCAACGGGCCGTCTCATC-3′and 5′-GATGACACGGCCCGTTGCTGCATCGATTGCGACGGCGTTGACCAG-3′.A UL32 glycosylation site substitution mutant, the Ser921Ala mutant (pON2785), was constructed with the primers 5′-CCGCCCTCGGTCCCCGTGGCCGGCAGCGCGCCGGGTCGCCTG-3′and 5′-CAGGGCGACCCGGCGCGCTGCCGGCCAGGGGACCGAGGGCGG-3′. A second UL32 glycosylation site substitution mutant, the Ser952Ala mutant (pON2786), was constructed with primers 5′-CCACCGTTTACCCACCGTCGGCGACGGCCAAAAGCAGCGTATCAAATGCGCCGCCTGTGGCCTC-3′and 5′-GAGGCCACAGGCGGCGCATTTGATACGCTGCTTTTGGCCGTCGCCGACGGTGGGTAAACGGTGG-3′.The SCMV UL32 homolog was previously described (5) and was kindly provided by Wade Gibson. The murine cytomegalovirus (MCMV) UL32 homolog (pON4888) was constructed with the primers 5′-CGCGGATCCCCACCATGTCCGCTCGAGGGCGCGCG-3′and 5′-CCTTTAGCGGCCGCTCACAAGTCTTCTTCAGAAATCAGCTTTTGTTCGTGAGACGACGATTTTTTTTAC-3′to amplify the M32 ORF from MCMV DNA (strain K181 derivative RM427). The fragment was cleaved with BamHI and NotI and cloned into pcDNA3.1zeo+. All of the substitution mutants were constructed with a QuikChange mutagenesis kit (Stratagene, La Jolla, CA), using pON2780 as a template.
Generation of UL32 stable cell line.
A pp150-expressing cell line (UL32-HFF) was produced by cotransfection of pON2780 (UL32 ORF cloned into pLNCX retroviral vector under the HCMV immediate-early promoter) along with plasmids LTRVSVG, JK3, and CMVtat into Phoenix cells as described previously (4, 29). pp150 expressing retrovirus isolated from the supernatant of transfected Phoenix cells was used to infect low-passage HFF. Stable UL32-HFF lines were selected with 400 μg/ml Geneticin (Gibco/Invitrogen, Carlsbad, CA).
Antibodies.
Monoclonal antibody (MAb) 36-14 was used to detect pp150 and has been previously described (47). MAb 28-4 was used to detect HCMV major capsid protein (MCP), and MAb 28-19 was used to detect pp65 (kindly supplied by William J. Britt). Fluorescein isothiocyanate (FITC)-conjugated MAb 810 antibody (Chemicon, Temecula, CA) was used to detect HCMV immediate-early protein 1 (IE1)/IE2. HCMV UL44 was detected with MAb 1202-01 (GICR, Plantation, FL). HCMV pp28 was detected with MAb CA004-100 (Virusys, Sykesville, MD). HCMV gB was detected with a MAb from ViroGen Corporation (Watertown, MA). To detect 5-bromodeoxyuridine (BrdU), incorporation into replicated DNA of an Alexa Fluor 594-conjugated anti-BrdU MAb (Molecular Probes, Eugene, OR) was used.
To detect pp150 c-Myc-tagged proteins by immunoblotting and immunofluorescence assay (IFA), an anti-c-Myc MAb (Santa Cruz Biotechnology, Santa Cruz, CA) was used. A peroxidase-labeled anti-mouse immunoglobulin G (Vector Laboratories Inc., Burlingame, CA) secondary antibody was used for immunoblot analysis. For IFA, Alexa Fluor 594 (Molecular Probes, Eugene, OR)-conjugated anti-mouse secondary antibody was used. To assess protein gel sample loading, an antiactin rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used, followed by a horseradish peroxidase-conjugated anti-rabbit secondary antibody (Dako, Denmark).
Secondary spread assay.
HFFs were transfected by calcium phosphate as described previously (41), with some modifications. HFFs were split to a ratio of 1:2 into a 24-well plate (5.5 × 104 cells/well) and incubated overnight. Roughly 2 h prior to transfection, the medium was changed (0.5 ml). Five-tenths micrograms of BAC DNA, 0.5 μg of the pp71 expression plasmid pON2788 (29), which has previously been shown to augment the infectivity of HCMV DNA transfected into HFF (3), and 0.5 μg of a UL32 expression plasmid DNA (or empty plasmid as a control) were added to sterile H2O along with 6.2 μl of 2 M CaCl2 to a final volume of 50 μl. Next, 50 μl of 2× Hanks balanced salt solution (280 mM NaCl, 1.4 mM Na2HPO4, 10 mM KCl, 5.6 mM glucose, 20 mM HEPES, pH 7.05) was added in drops, along with gentle agitation of the solution in between drops. The mixture was then added to HFF, followed by incubation for 4 to 6 h at 37°C in a 5% CO2 incubator. The medium was then replaced with a glycerol shock solution (15% glycerol, 1× Hanks balanced salt solution) for 90 s at 37°C. The cells were then washed four times in complete medium. Transfection experiments carried out in either 12- or 6-well dishes were scaled up based on well surface area. Transfected cells were grown in medium that contained pooled human gamma globulin (Aventis Behring), which contains neutralizing anti-HCMV antibodies, at a 1:1,000 dilution to limit virus spread through the medium but allow cell-to-cell spread. The medium was changed every 2 days. Transfections were done in triplicate, with each individual transfection producing at least 30 GFP-positive cells. ΔUL32 virus was isolated from the supernatant of cells that had been cotransfected with ΔUL32-BAC and wild-type (WT) pp150 expression plasmid (in the absence of pooled gamma globulin) and used to infect HFF. The supernatant was harvested on days 5, 7, and 9 and used to infect HFF after centrifugation at 3,000 × g to remove cells and debris. GFP fluorescence was used to identify ΔUL32 virus-infected cells.
Immunoblotting.
To determine the levels of expression of the pp150 plasmids, 293T cells were transfected with 2 μg of each construct with TransIT transfection reagent (Mirus Co., Madison, WI). Forty-eight hours later, cells were lysed in disruption buffer containing 2% sodium dodecyl sulfate (SDS) and 5% β-mercaptoethanol and then heated for 5 min at 95°C. Samples were separated on a 10% SDS-polyacrylamide gel, followed by transfer to a nitrocellulose membrane. Detection of pp150 was done with an anti-c-Myc MAb followed by a secondary anti-mouse horseradish peroxidase-conjugated antibody. Protein bands were detected with ECL Western blot detection reagent (Amersham, Piscataway, NJ) on Kodak MR film (Kodak, Rochester, NY).
Immunofluorescence microscopy.
HFFs were split to 50% confluence 24 h prior to CaPO4 transfection in a 24-well dish containing a coverslip. The IFA protocol was adapted from a previous study (30). At days 5 to 10 posttransfection with BAC DNA, cells were washed in phosphate-buffered saline (PBS) and then fixed in 3.7% formaldehyde-PBS for 10 min, followed by three washes in PBS. Cells were then permeabilized in 0.1% Triton X-100 in PBS for 15 min, followed by a 1-h block in PBS containing 1% bovine serum albumin and 20% FBS at 37°C. Following a wash, the primary antibody was added in 1% bovine serum albumin and 1% FBS in PBS for 1 h at 37°C. Secondary antibodies were added after three washes in PBS for 30 min at 37°C. The cells were washed and incubated for 5 min in PBS containing a 1:3,000 dilution of Hoechst 33342 (Molecular Probes). The coverslips were then inverted and fixed to glass microscope slides with Prolong Gold antifade reagent (Molecular Probes). The slides were viewed under an Olympus BX60 fluorescent microscope (Melville, NY), and images were captured on a charge-coupled-device camera by use of Image-Pro Plus software (Media Cybernetics, Inc., Silver Spring, MD). HFF were also fixed in a 3:1 solution of methanol:acetic acid at −20°C for 20 min and then washed three times in PBS. This was followed by the same blocking and antibody treatment described above.
The BrdU protocol was adapted from a procedure listed on the Chemicon website (www.chemicon.com/techsupp/Protocol/BrdUProtocol.asp). Briefly, viral DNA replication analyses were done by exposing cells to a 10 μM concentration of BrdU for 2 h on day 5 posttransfection. The cells were then washed in media, followed by a 48-h chase period. The cells were fixed on glass coverslips in a 3:1 solution of methanol:acetic acid at −20°C for 20 min and then washed three times in PBS. DNA was denatured in 2 M HCl for 1 h at 37°C, followed by three washes in neutralizing borate buffer (pH 8.5). The blocking procedure and the subsequent IFA steps were identical to those mentioned above. This experiment was done twice, with at least two separate coverslips used each time.
RESULTS
Transient complementation of ΔUL32-BAC.
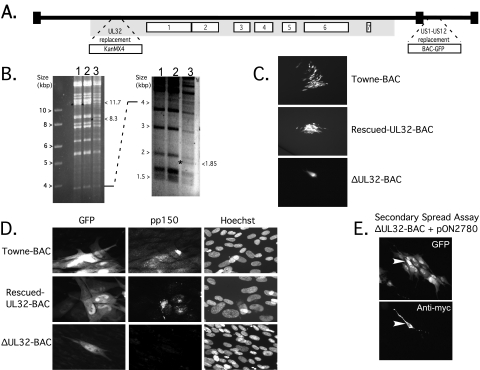
ΔUL32-BAC employed here was previously reported (13) to be a Towne-BAC derivative carrying a KanMX4 cassette replacing the UL32 ORF located within the betaherpesvirus-conserved gene cluster of the HCMV genome (35), as depicted in Fig. 1A. When digested with HindIII restriction endonuclease, ΔUL32-BAC DNA yielded the expected pattern, with two additional fragments at 8.3 kbp and 1.85 kbp in place of the 11.7-kbp fragment of Towne-BAC or rescued-UL32-BAC DNA (Fig. 1B). Towne-BAC, rescued-UL32-BAC, and ΔUL32-BAC also displayed the expected characteristics (13) when transfected into HFF using pp71 enhancement (3). GFP-positive cells were first detected roughly 4 days posttransfection and remained as single cells in ΔUL32-BAC-transfected cultures. These cells were followed from day 4 up to day 15 posttransfection. As expected, GFP-positive foci formed in both Towne-BAC- and rescued-UL32-BAC-transfected cultures (Fig. 1C). Importantly, foci were readily detected by GFP expression from the BAC-GFP cassette in each of these Towne-BAC-derived viruses (Fig. 1A). ΔUL32-BAC-transfected cells failed to express pp150 (Fig. 1D), whereas rescued-UL32-BAC- or Towne-BAC-derived foci expressed pp150 in the expected juxtanuclear pattern (Fig. 1D) (17, 47, 50). When a myc-tagged pp150 expression plasmid (pON2780) was cotransfected with ΔUL32-BAC, foci formed starting about day 5 posttransfection and reached a maximum of 10 to 20 GFP-positive cells by days 8 to 10 posttransfection (Fig. 1E). Thus, replication took place in the initial, myc-tagged pp150-positive cotransfected cell and transiently complemented ΔUL32 virus spread into surrounding cells, whereas the absence of pp150 resulted in a replication block, demonstrating complementation and permitting the analysis of the ΔUL32 phenotype in secondarily infected cells. Although pp71 enhancement was used to increase the frequency of transfection, the phenotype of ΔUL32 was the same in experiments where pp71 was not included (data not shown).
FIG. 1.
UL32-BAC structure and complementation. (A) Schematic of the HCMV TownevarATCCshort strain, Towne-BAC-derived ΔUL32-BAC genome, with the locations of the BAC-GFP insertion replacing US1 to US12 and the KanMX4 insertion replacing the entire betaherpesvirus-conserved UL32 ORF (13) indicated by expanded regions. The numbered open boxes indicate the locations of herpesvirus core genes, and the gray shaded area indicates the location of the betaherpesvirus-conserved genes (35, 36). (B) Ethidium bromide-stained images of electrophoretically separated (larger fragments, left; smaller fragments, right) HindIII restriction digests of 5 μg of Towne-BAC (lane 1), rescued-UL32-BAC (lane 2), or ΔUL32-BAC (lane 3). The 11.7-kbp fragment present in Towne-BAC or rescued-UL32-BAC is replaced by 8.3- and 1.85-kbp fragments in ΔUL32-BAC, as indicated by an asterisk on the left of the DNA fragment and by the sizes on the right side of each panel. (C) GFP expression following transfection of HFFs with rescued-UL32-BAC (a focus is shown at ×50 magnification), Towne-BAC (a focus is shown at ×50 magnification), and ΔUL32-BAC (a single cell is shown at ×100 magnification) at day 10 posttransfection. (D) GFP expression following transfection (as described for panel C) with Towne-BAC, rescued-UL32-BAC, and ΔUL32-BAC. Detection of GFP, pp150 antigen, and DNA (Hoechst) was performed on day 8 posttransfection (×300 magnification). A merge is shown to localize the pp150 accumulation. (E) Detection of GFP (top panel) and myc-tagged pp150 (bottom panel) in a secondary spread assay at 8 days following cotransfection of HFFs with ΔUL32-BAC and WT pp150 expression plasmid (pON2780) (×100 magnification). The arrowhead identifies the myc epitope-tagged pp150-positive cell.
Requirement for amino- and carboxyl-terminal regions of pp150 in replication.
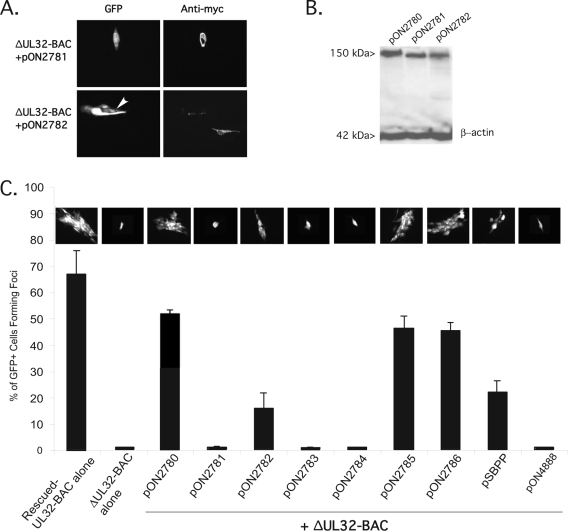
A series of pp150 mutant expression plasmids were tested by a secondary spread assay (Fig. 1E) to identify regions of the protein playing critical roles during infection. As shown in Fig. 2, a 71-amino-acid (aa) deletion of the amino terminus (pON2781) expressing a truncated protein (Fig. 2B) failed to support secondary spread of ΔUL32-BAC (Fig. 2A, top row). The amino terminus of pp150 contained conserved regions found in other betaherpesvirus homologs that have been shown to be important for binding to capsids in vitro (5). A 69-aa deletion of the carboxyl terminus (pON2782) partially complemented ΔUL32-BAC spread (Fig. 2A, bottom row), resulting in foci 3 to 10 cells in size (Fig. 2A, bottom left) as opposed to the 10- to 20-cell foci formed following cotransfection with the WT pp150 expression plasmid (Fig. 1). Furthermore, this carboxyl-terminal truncation mutant promoted cell-to-cell spread in only about 15% of GFP-positive cells, threefold fewer than arose with the WT pp150 expression plasmid (Fig. 2C). The amino- and carboxyl-terminal pp150 truncation mutants (Fig. 2B, lanes 2 and 3) were expressed at similar levels and exhibited the expected faster migration compared to that of WT pp150 by an immunoblot analysis. In all of the secondary spread assays, transient WT or mutant pp150 expression was largely coincident with GFP-positive cells receiving ΔUL32-BAC. Few pp150-positive cells were GFP negative and few GFP-positive cells were pp150 negative following cotransfection. On rare occasions, two cells in one ΔUL32-BAC complement-generated focus contained pp150, suggesting that the cells may have been in the process of dividing at the time of transfection. These results showed that transient complementation of ΔUL32-BAC with WT or mutant forms of pp150 provided information on gene function and implicated the amino terminus containing CR1 in replication. In addition to large amino- and carboxyl-terminal deletions, several site-directed mutants (Table 1) were tested for activity in the secondary spread assay by use of a day-10-posttransfection endpoint when complemented ΔUL32-BAC-derived foci had reached maximal size. Figure 2C graphically represents the percentage of GFP-positive cells that developed into foci and includes a representative GFP fluorescent image depicting the extent of spread following either transfection or cotransfection of ΔUL32-BAC with pp150 expression constructs. Roughly 66% of transfected, GFP-positive cells developed into foci following transfection with either Towne-BAC (data not shown) or rescued-ΔUL32-BAC (Fig. 2C). A slightly lower percentage (roughly 50%) of cells with the ΔUL32-BAC mutant and complementing WT pp150 expression plasmid formed secondary foci. Disruption of either CR1 (pON2783) or CR2 (pON2784), sequence blocks of 11 and 9 aa in length, respectively, that have been implicated in the binding of the SCMV UL32 homolog to B and C capsids in vitro (5), resulted in pp150 expression plasmids that failed to support secondary spread of mutant virus. Two additional substitution mutations were made in predicted O-linked glycosylation sites (16). Ser921Ala (pON2785) and Ser952Ala (pON2786) both complemented ΔUL32-BAC spread at levels comparable to that of the WT pp150 expression plasmid, and both of these expression plasmids expressed pp150 at WT levels (data not shown). These results show that CR1 and CR2 were critical domains within pp150 and that without them secondary spread of mutant virus is blocked.
FIG. 2.
Complementation of ΔUL32-BAC by mutant forms of pp150. (A) IFA of pON2781-encoded amino-terminal truncation of pp150 (top panels) and pON2782-encoded carboxyl-terminal truncation (bottom panels) following cotransfection with ΔUL32-BAC in the secondary spread assay at 10 days posttransfection. A small focus of cells that was generated following cotransfection of the carboxyl-terminal deletion mutant is identified by an arrowhead. Magnification,×80.(B) Immunoblot of 293T cell lysates at 48 h posttransfection with 2 μg of pON2780 (lane 1), pON2781 (lane 2), and pON2782 (lane 3). Proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to a nylon membrane, and immunoblotted for myc-tagged pp150. β-Actin served as a loading control. (C) IFA of secondary spread, noting the percentage of GFP-positive single cells that developed into foci as described for panel A. Error bars represent the standard deviations of the means of triplicate samples. Photomicrographs above each bar in the graph show representative images of foci or single cells (where foci do not form) for cotransfection combinations indicated. Magnification, ×20.
TABLE 1.
pp150 expression plasmids
| Plasmid | Description |
|---|---|
| pON2780 | Wild-type pp150 |
| pON2781 | Amino-terminal 71-aa deletion of pp150 |
| pON2782 | Carboxyl-terminal 67-aa deletion of pp150 |
| pON2783 | Leu58Ile Trp59Asp pp150 |
| pON2784 | Asn201Ala Lys202Ile Leu203Asp Val204Ala Tyr205Ala pp150 |
| pON2785 | Ser921Ala pp150 |
| pON2786 | Ser952Ala pp150 |
| pON4888 | MCMV UL32 homolog |
| pSBPP | SCMV UL32 homolog |
UL32 homologs from SCMV and MCMV were tested for activity in the secondary spread assay. The SCMV homolog (5), exhibiting 49% aa identity and 71% aa similarity to HCMV pp150 (http://www.ncbi.nih.gov), complemented ΔUL32-BAC secondary spread but produced significantly smaller foci developing from only 22% of cotransfected cells (Fig. 2C), less than half the number that arose when complementation was carried out with the WT pp150 expression plasmid. In contrast, MCMV M32, exhibiting only 27% aa identity and 44% similarity to HCMV UL32 (40), failed to complement ΔUL32-BAC at all (Fig. 2C). The ability of primate CMV UL32 homologs to complement replication is consistent with previously established binding qualities (5) dependent on CR1 and CR2 as well as additional conservation necessary to support replication of HCMV.
Expression of IE, delayed-early, and late viral proteins in the absence of functional pp150.
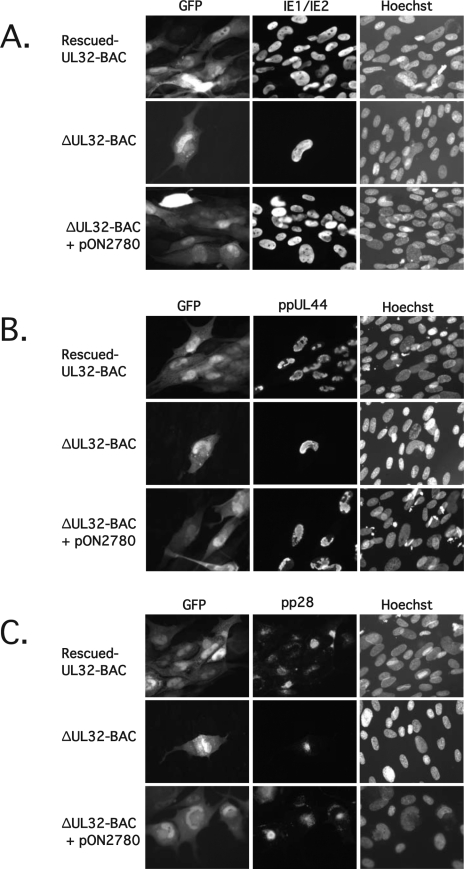
In order to investigate the replication block in the absence of pp150, we examined the expression of viral gene products in secondary spread foci, comparing ΔUL32-BAC complemented with WT pp150 (Fig. 3A, B, and C, bottom rows) to rescued-UL32-BAC (Fig. 3A, B, and C, top rows). Viral gene expression was also evaluated directly in single cells following transfection of ΔUL32-BAC in the absence of a complementing expression plasmid (Fig. 3A, B, and C, middle rows). Initially, we compared the levels of viral proteins representing the three temporal classes (IE, or α; delayed early, or β; and late, or γ) by using an indirect IFA. Figure 3A shows that IE1/IE2 (α) antigen accumulated with similar patterns in the rescued-UL32-BAC-derived and complemented ΔUL32-BAC-derived foci, as well as in noncomplemented ΔUL32-BAC-derived single cells. Expression of HCMV ppUL44, a delayed-early (β) antigen associated with viral DNA replication (36), also showed similar patterns in all settings (Fig. 3B). The localization pattern of the HCMV tegument protein pp28 (ppUL99), a true late (γ2) protein involved in cytoplasmic maturation and egress (9, 22, 52), was detected in its expected juxtanuclear position in rescued-UL32-BAC-derived or in complemented ΔUL32-BAC-derived foci as well as in noncomplemented ΔUL32-BAC-transfected cells (Fig. 3C). pp71 has been shown to be a strong viral transactivator (3, 8, 18, 27). When analyzing gene expression patterns in this assay, it should be remembered that pp71 transactivation occurs only in the transfected cells and not in secondarily infected cells. Therefore, these data show that all three kinetic classes of HCMV gene expression occur in ΔUL32 virus-infected cells. A general defect in gene expression does not account for the mutant replication phenotype.
FIG. 3.
Expression of IE, early, and late viral antigens by ΔUL32-BAC. HFFs were transfected with rescued-UL32-BAC, ΔUL32-BAC, or ΔUL32-BAC plus WT pp150 expression plasmid pON2780. (A) Immunofluorescent analysis of cells fixed 8 days posttransfection and stained with FITC-conjugated MAb 810 to detect IE1/IE2. (B) Immunofluorescent analysis of cells fixed 8 days posttransfection and stained with an HCMV ppUL44 MAb followed by detection with Alexa Fluor 594-conjugated secondary antibody. (C) Immunofluorescent analysis of cells fixed 10 days posttransfection and stained with HCMV pp28 (UL99) MAb, followed by secondary detection with Alexa Fluor 594-conjugated secondary antibody. Hoechst staining was used to localize nuclear DNA. Magnification, ×300.
Viral DNA synthesis and translocation of DNA to the cytoplasm proceed in the absence of functional pp150.
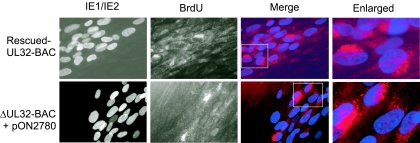
Detection and normal localization of the cytoplasmic egress tegument protein pp28 in ΔUL32-BAC-infected cells suggested that all stages of viral replication, including viral DNA synthesis, had proceeded normally. Antisense inhibition of UL32 expression by 80 to 90% was previously reported not to disrupt viral DNA replication (33). We undertook a BrdU pulse-chase analysis (39), using the secondary spread assay to determine whether viral DNA replication as well as packaging and translocation to the cytoplasm proceeded in the absence of pp150. Five days after transfection, when foci were first forming, cells were pulsed with BrdU for 2 h, washed, and then chased for 48 h in the absence of this nucleoside. Figure 4 shows dual labeling of HFF with an FITC-conjugated MAb to IE1/IE2 along with an Alexa Fluor 594-conjugated MAb to BrdU. The foci arising from secondary spread of complemented mutant virus showed a BrdU localization pattern similar to that for rescued-UL32-BAC. In each ΔUL32-BAC-derived focus analyzed, more than five cells incorporated BrdU into viral DNA that moved to a cytoplasmic, juxtanuclear position over the 2-day chase period, consistent with ongoing viral DNA replication and normal translocation of DNA-containing virus particles from the nucleus to the cytoplasm. Thus, pp150 was not required for DNA synthesis or the nuclear egress steps in virion maturation.
FIG. 4.
Localization of newly synthesized viral DNA. (A) Immunofluorescent images of HFFs transfected with rescued-UL32-BAC (top panels) or ΔUL32-BAC plus WT pp150 (bottom panels), followed by a BrdU (10 μM) pulse for 12 h and a 48-h chase. BrdU was localized with an Alexa Fluor 594-conjugated MAb. A false color merge was used to localize BrdU-stained viral DNA (red) in a juxtanuclear position relative to IE1/IE2-positive nuclei (blue). The merged images contained in boxes are enlarged in column 4. Cells were fixed in methanol:acetic acid (3:1). Magnification, ×400 for columns 1 to 3 and ×1,000 for column 4.
MCP and gB accumulate in the cytoplasm in the absence of functional pp150.
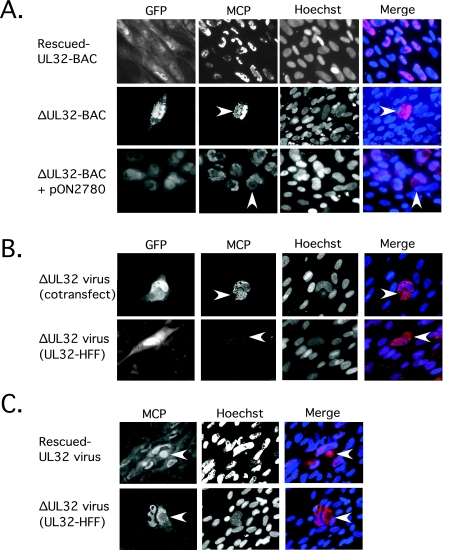
The pattern of capsid maturation was further evaluated by IFA, following MCP localization and accumulation in transfected and infected cells. MCP localized almost exclusively to the nucleus in rescued-UL32-BAC-transfected cultures (Fig. 5A, top row) observed on day 10, as expected for a WT virus (46). Surprisingly, cytoplasmic as well as nuclear localization was observed in ΔUL32-BAC-transfected single cells (Fig. 5A, middle row) as well as in foci that formed in the secondary spread assay (Fig. 5A, bottom row). This striking pattern was also observed in HFFs that were infected with ΔUL32 virus recovered from supernatants of cotransfected cultures (Fig. 5B, top row) as well as when ΔUL32 virus recovered from the supernatants of UL32-HFFs was used to infect HFFs (Fig. 5B, bottom row). When UL32-HFFs were used to complement ΔUL32-BAC growth, cell-to-cell spread proceeded slowly over many weeks, with only small amounts of virus being produced. This was apparently due to low expression of pp150 within these cells (data not shown). Use of a small amount of infectious virus to initiate infection produced the same pattern of cytoplasmic MCP accumulation in HFFs as in other settings where mutant virus was evaluated. Interestingly, when methanol:acetic acid (3:1) fixative was used instead of 3.7% formaldehyde prior to the IFA, MCP was readily detected in the cytoplasmic inclusion of rescued-UL32 virus-infected cells (Fig. 5C, top row) as well as that of mutant-virus-infected cells (Fig. 5C, bottom row). Thus, MCP staining in mutant-virus-infected cells was independent of the fixation method, but staining was dependent on methanol:acetic acid fixation in rescued-UL32-infected cells. When the latter fixative was used, MCP detected in the cytoplasm of rescued-UL32-infected cells appeared more distinctly punctate. Also, MCP was distributed around the perimeter of the cytoplasmic inclusion in mutant-virus-infected cells, whereas this antigen was coincident with the inclusion in rescued-UL32-infected cells. MCP accumulation in the cytoplasm in ΔUL32 virus-infected cells surrounded the cytoplasmic inclusion, a pattern observed with other virion proteins that have been studied (Fig. 3C). Ultrastructural evaluation of capsid accumulation in mutant-infected cells has not yet yielded information, due to low-level-infection conditions that have been possible with the mutant virus, although this issue deserves further analysis.
FIG. 5.
Localization of MCP within the cytoplasm of ΔUL32-BAC-transfected or -infected cells. (A) Immunofluorescent images of HFFs fixed with 3.7% formaldehyde at day 10 following transfection with rescued-UL32-BAC (top panels), ΔUL32-BAC (middle panels), and ΔUL32-BAC plus WT pp150 (bottom panels). (B) Immunofluorescent images of HFFs infected with supernatant virus from ΔUL32-BAC plus WT pp150-cotransfected cells [ΔUL32 virus (cotransfect)] fixed on day 10 postinfection (top panels) and HFFs infected with supernatant virus recovered from the ΔUL32-BAC-transfected pp150-expressing HFF line [ΔUL32 virus (UL32-HFF)] fixed at day 7 postinfection (bottom panels). (C) Immunofluorescent images of MCP accumulation at day 8 postinfection with rescued-UL32 (top panels) and ΔUL32 (bottom panels) virus fixed with methanol:acetic acid (3:1). GFP-positive cells are shown in the far left panel for all samples fixed with 3.7% formaldehyde (methanol:acetic acid fixation destroys GFP detection). All staining for MCP was done with MAb 28-4 along with Alexa Fluor 594 secondary antibody. Hoechst was used to stain nuclei, and a false color merge with MCP antigen localization is shown. The arrowhead indicates cytoplasmic localization of MCP. All images were collected with equivalent exposure times. Magnification, ×200.
HCMV gB, which is involved in viral entry (11), is an abundant, herpesvirus-conserved virion envelope glycoprotein (36). gB localization to the cytoplasmic inclusion was evident in HFFs following cotransfection or during infection with ΔUL32-BAC (Fig. 6, top row, and data not shown). A similar pattern is observed with rescued-UL32-BAC-derived foci (Fig. 6, bottom row) as well as with other WT virus strains (47). Like MCP, gB appeared to accumulate with more intense IFA staining in ΔUL32 virus-infected cells than in rescued-UL32 virus-infected cells. Apparently, in the absence of pp150, virion capsid envelope proteins accumulated in the cytoplasmic inclusion. This was most consistent with a block to either maturation or egress and manifested as a failure in cell-to-cell spread based on GFP or IE1/IE2 expression in cells infected with mutant virus. Despite localization studies suggesting that pp150 initially interacts with capsids in the nucleus and continues to associate with nucleocapsids in the cytoplasm (17, 45), our data are most consistent with a role for UL32 in cytoplasmic events in virion maturation.
FIG. 6.
Localization and accumulation of gB in the cytoplasm of ΔUL32-BAC-transfected and -infected cells. Immunofluorescent images of HFFs from a secondary spread assay at day 10 posttransfection with rescued-UL32-BAC or ΔUL32-BAC plus WT pp150 (pON2780). Detection of gB was accomplished with an anti-HCMV gB MAb along with an Alexa Fluor 594 secondary antibody. Three individual plaques are shown for each construct. All images were collected with equivalent exposure times. Magnification, ×105.
Cell-to-cell transfer of pp65 is independent of pp150 function.
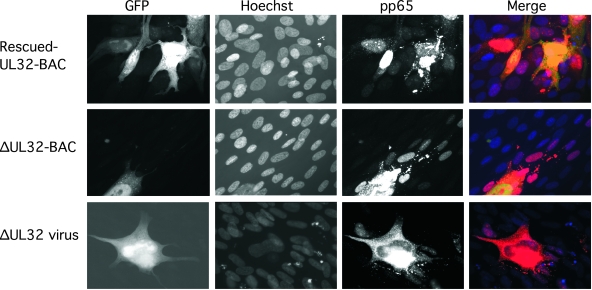
Like pp28, MCP, and gB, expression of the most abundant tegument protein, pp65, accumulated in the cytoplasmic inclusion and was not reduced in ΔUL32-BAC-transfected or -infected cells (Fig. 7). Somewhat surprisingly, though, pp65 was also detected in nuclei of cells surrounding ΔUL32-BAC-transfected or -infected cells (Fig. 7), even though pp150 was completely absent from the experiment. These adjacent pp65-positive cells remained negative for IE1/IE2 antigen as well as for other viral antigens (Fig. 3 and data not shown). The delivery of this abundant tegument protein to adjacent cells and presence in the nuclei of adjacent cells are indications of cell-to-cell spread of pp65-positive particles. Given the absence of any other viral antigens in adjacent cells, this result suggests that particles lacking viral capsids and viral DNA, most likely dense bodies, are responsible for the delivery of pp65 antigen to adjacent cells. Dense bodies are composed largely of the tegument protein pp65, with small amounts of other tegument proteins, including some pp150 (57), surrounded by a lipid bilayer that is similar to the virion envelope (37), and retain the capacity to fuse with uninfected cells(49). Our data suggest that dense bodies may follow a maturation pathway that is independent of pp150 function, consistent with their formation in the cytoplasm and absence of capsid antigen or viral DNA (37). If confirmed, our data implicate pp150 in the maturation of capsid-bearing viral particles but not dense bodies. An alternative possibility is that pp150 plays some role in uncoating as well as in maturation. Thus far, electron micrographs have not yielded evidence sufficient to assess particle formation or movement, due to the inherently low infection conditions that have been available. Taken together, our data on localization patterns and pp150 mutants suggest that this viral protein is crucial for cytoplasmic events that impact virion maturation but may be dispensable for the release of pp65-containing dense bodies.
FIG. 7.
Localization and distribution of pp65 in ΔUL32-BAC-transfected and ΔUL32 virus-infected cells. Immunofluorescent images of rescued-UL32 (top panels) or ΔUL32 (middle panels) transfection at day 10 posttransfection compared to ΔUL32 virus infection (bottom panels) at day 7 postinfection. Detection of pp65 was accomplished with MAb 28-19 followed by Alexa Fluor 594 secondary antibody. Magnification, ×400.
DISCUSSION
This study implicates a betaherpesvirus-conserved virion tegument phosphoprotein in maturation. By demonstrating a requirement for the UL32 gene product pp150 in virion maturation, our data provide insights into previous reports of novel capsid binding and structural characteristics associated with pp150 (5, 10, 56). The pp150-deficient ΔUL32-BAC virus proceeds through most of the replication cycle to late gene expression, encoding normal levels of viral antigens pp28, MCP, gB, and pp65, synthesizing normal levels of viral DNA, and accumulating viral DNA as well as MCP and gB at a prominent cytoplasmic inclusion without release or cell-to-cell spread. This inclusion has long been believed to be the site of the final stages of virion maturation, and our data suggest that pp150 is critical during this final phase of replication. As a tegument protein that interacts with capsids but has a phenotype that blocks maturation in the cytoplasm as well as release, it appears that pp150 may direct the proper assembly of tegument layers in the cytoplasm. This phenotype, which is supported by complementation studies with WT and mutant derivatives of pp150, strongly supports the role of CR1 and CR2 mediating interactions between nucleocapsids and machinery controlling maturation events in the cytoplasmic inclusion. We believe that pp150 is most likely to be necessary for tegumentation in advance of final envelopment as proposed in the envelopment/deenvelopment/reenvelopment model of herpesvirus maturation (32). This suggestion is consistent with the binding interactions of pp150 (5) as well as speculation from ultrastructural data on the location of pp150 in the virion particle (10, 56). While our studies cannot formally preclude an impact of CR1 or CR2 mutations on overall protein conformation, the choice of site-directed mutagenesis and the independent, consistent growth phenotype of either CR1 or CR2 argue against this possibility. The continued maturation and cell-to-cell spread of pp65-containing particles reinforce the precision of the block in pp150 mutant-infected cells.
The late replication defect observed with ΔUL32-transfected or -infected cells is reminiscent of HCMV UL99 mutant virus (9, 22, 52) or alphaherpesvirus mutants disrupting the homolog of this myristolated tegument protein (1, 2, 24, 48) that controls cytoplasmic egress. Although both pp28 and pp150 localize to the cytoplasmic inclusion and mutations in these tegument protein genes cause maturation defects with increased accumulation of structural antigens in the inclusion (9, 22, 52), there are apparent differences in the impacts of these two tegument proteins on maturation pathways within the cytoplasmic inclusion. Whereas UL99 mutants are not as tightly blocked and fail to release cell-free virus but allow cell-to-cell spread, as demonstrated by expression of viral gene products in adjacent cells (51), UL32 is required for release as well as cell-to-cell spread of virus but does not control cell-to-cell spread of pp65-containing dense bodies. Localization of pp65 to nuclei of cells surrounding a ΔUL32-transfected or -infected cell results from particle delivery and not from expression of this viral gene, because abundant, readily detected early viral antigens such as IE1/IE2 remained undetectable in pp65-positive cells. Thus, ΔUL32 virus exhibited a pattern distinct from that of UL99 mutants, although both are required for maturation events that occur at the juxtanuclear cytoplasmic inclusion. Our data thus reinforce the importance of this inclusion in late maturation and release of all types of viral particles.
pp150 localization and function are associated with the prominent cytoplasmic inclusion that is a hallmark of HCMV infection. Incisive colocalization studies have suggested that pp150 first associates with capsids in the nucleus and accompanies capsids during egress and maturation (45). Though our results differ from a report that employed antisense inhibition of UL32 expression but did not completely eliminate pp150 (33), they are consistent with the suggestion that pp150 associates with capsids in the nucleus even though this protein does not function as a chaperone necessary for escorting nucleocapsids from the nucleus to the cytoplasm. It is likely that the low level of accumulation of pp150 in the nuclei of WT-virus-infected cells reflects the efficient translocation with capsids to the cytoplasm for final envelopment.
Although we recovered only low levels of complemented ΔUL32 virus following cotransfection into HFFs or on UL32-HFFs, complemented virus showed the same defects as direct transfection of ΔUL32-BAC. Failure to express pp150 had no impact on nucleocapsid translocation from the nucleus to the cytoplasm but dramatically reduced virion release and cell-to-cell spread. Future studies will require complementing systems to allow greater production of ΔUL32 virus in order to infect greater numbers of HFFs so that ultrastructural analysis may be employed in order to investigate the behaviors of mutant and WT virus particles more directly.
The secondary spread assay we employed allows rapid screening of mutations for function. This assay allows study of viral mutants where complementation is difficult as well as screening of specific mutations for function. Transient complementation, for example, has facilitated unambiguous investigation of IE1 mutants to identify functional domains (41). The use of IFA to investigate the block in replication revealed an accumulation of gB and MCP as well as viral DNA within the cytoplasm independent of pp150 function. The greater accumulation of both MCP and gB in mutant-virus-infected cells is consistent with a defect in virion envelopment. The altered localization of MCP in ΔUL32 virus-infected cells, with a broader, punctate pattern surrounding the region of the cytoplasmic inclusion, suggested a block in capsid targeting to the appropriate cytoplasmic destination in the absence of pp150 even though the inclusion that formed in mutant-infected cells was indistinguishable from that of WT-infected cells and contained both pp65 and pp28. The impact of fixation on detection of MCP in WT-virus-infected cells raised the possibility that the MAb epitope was masked in a pp150-dependent manner. The result using methanol:acetic acid fixation, which revealed the presence of MCP in the cytoplasmic inclusion as well as in the nuclei of WT-virus-infected cells, does not favor masking. Furthermore, the different fixation protocols did not affect the localization of MCP in ΔUL32-transfected or -infected cells. Although MCP can be visualized in the cytoplasmic inclusion of rescued-UL32 virus-infected cells, the intensity of IFA was much less than after mutant virus infection, where nearly every infected cell accumulated intensive MCP antigen signal irrespective of fixative. This pattern was seen consistently in ΔUL32-BAC-transfected and virus-infected cells as well as in the cells tested by a secondary spread assay.
Cryoelectron microscopy of CMV capsids has suggested the presence of a regularly organized tegument protein very close to the capsid surface, apparently in association with the capsid triplex structure (10, 56). Tegument proteins are added after packaging of viral DNA in the nucleus but before final envelopment. Here, analysis of pp150 facilitated the functional evaluation of both CR1 and CR2, regions that have been implicated in capsid binding activity (5), as necessary for virion maturation. CR1, which is contained within an amino-terminal pp150 truncation that failed to complement mutant virus, and CR2, which lies outside of this deletion, are independently involved in pp150 function. The importance of both CR1 and CR2 in HCMV replication was suggested by earlier studies where HCMV and SCMV homologs were demonstrated to bind to HCMV or SCMV capsids (5). Here capsid binding via CR1 and CR2 was related to maturation and the SCMV homolog was shown to supplant the need for pp150. The M32 homolog is more highly diverged and may have failed to complement due to a failure in CR1/CR2 binding or other activities that are required to support cytoplasmic maturation. The two major O-linked N-acetylglucosamine glycosylation sites, thought to possibly regulate protein complex formation or interaction with capsid or membrane proteins during maturation (6, 16), are dispensable for replication. No matter whether pp150 is added to capsids in the nucleus during primary tegumentation or in the cytoplasm during final envelopment steps that occur in the inclusion body, the phenotype of this betaherpesvirus-conserved tegument protein mutant virus is most consistent with a critical role at reenvelopment during virus maturation.
Acknowledgments
We appreciate the generous assistance of Ying Dong for cell culture work. We acknowledge Walter Dunn and Fenyong Liu for providing virus clones.
This work was supported by PHS grant RO1 AI20211, by PHS training grants MSTP GM07365 (to G. B. Smith), F30NS051109 (to G. B. Smith), 1F32AI56959 (to C. D. Meiering), and 5T32AI07328 (to D. P. AuCoin), and by a Stanford School of Medicine Dean's Postdoctoral Fellowship (to D. P. AuCoin).
REFERENCES
- 1.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. [DOI] [PubMed] [Google Scholar]
- 5.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benko, D. M., R. S. Haltiwanger, G. W. Hart, and W. Gibson. 1988. Virion basic phosphoprotein from human cytomegalovirus contains O-linked N-acetylglucosamine. Proc. Natl. Acad. Sci. USA 85:2573-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt, W. J., M. Jarvis, J. Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 11.Compton, T., D. M. Nowlin, and N. R. Cooper. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193:834-841. [DOI] [PubMed] [Google Scholar]
- 12.Dohner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 16.Greis, K. D., W. Gibson, and G. W. Hart. 1994. Site-specific glycosylation of the human cytomegalovirus tegument basic phosphoprotein (UL32) at serine 921 and serine 952. J. Virol. 68:8339-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hensel, G., H. Meyer, S. Gartner, G. Brand, and H. F. Kern. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591-1601. [DOI] [PubMed] [Google Scholar]
- 18.Homer, E. G., A. Rinaldi, M. J. Nicholl, and C. M. Preston. 1999. Activation of herpesvirus gene expression by the human cytomegalovirus protein pp71. J. Virol. 73:8512-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homman-Loudiyi, M., K. Hultenby, W. Britt, and C. Soderberg-Naucler. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis, M. A., T. R. Jones, D. D. Drummond, P. P. Smith, W. J. Britt, J. A. Nelson, and C. J. Baldick. 2004. Phosphorylation of human cytomegalovirus glycoprotein B (gB) at the acidic cluster casein kinase 2 site (Ser900) is required for localization of gB to the trans-Golgi network and efficient virus replication. J. Virol. 78:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, T. R., and S. W. Lee. 2004. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 78:1488-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landini, M. P., B. Severi, L. Badiali, E. Gonczol, and G. Mirolo. 1987. Structural components of human cytomegalovirus: in situ localization of the major glycoprotein. Intervirology 27:154-160. [DOI] [PubMed] [Google Scholar]
- 27.Lin, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting sites. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormick, A. L., C. D. Meiering, G. B. Smith, and E. S. Mocarski. 2005. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J. Virol. 79:12205-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormick, A. L., A. Skaletskaya, P. A. Barry, E. S. Mocarski, and V. S. Goldmacher. 2003. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology 316:221-233. [DOI] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 32.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer, H. H., A. Ripalti, M. P. Landini, K. Radsak, H. F. Kern, and G. M. Hensel. 1997. Human cytomegalovirus late-phase maturation is blocked by stably expressed UL32 antisense mRNA in astrocytoma cells. J. Gen. Virol. 78:2621-2631. [DOI] [PubMed] [Google Scholar]
- 34.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-982, 984-986, 989-990. [PMC free article] [PubMed] [Google Scholar]
- 35.Mocarski, E. S. Comparative analysis of betaherpesvirus-common proteins. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis, in press. Cambridge Press, Cambridge, United Kingdom. [PubMed]
- 36.Mocarski, E. S. Comparative analysis of herpesvirus-common proteins. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis, in press. Cambridge Press, Cambridge, United Kingdom. [PubMed]
- 37.Mocarski, E. S., Jr., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 38.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 39.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 40.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhardt, J., G. B. Smith, C. T. Himmelheber, J. Azizkhan-Clifford, and E. S. Mocarski. 2005. The carboxyl-terminal region of human cytomegalovirus IE1491aa contains an acidic domain that plays a regulatory role and a chromatin-tethering domain that is dispensable during viral replication. J. Virol. 79:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roby, C., and W. Gibson. 1986. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J. Virol. 59:714-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses, p. 2399-2509. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 45.Sampaio, K. L., Y. Cavignac, Y. D. Stierhof, and C. Sinzger. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 79:2754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez, V., P. C. Angeletti, J. A. Engler, and W. J. Britt. 1998. Localization of human cytomegalovirus structural proteins to the nuclear matrix of infected human fibroblasts. J. Virol. 72:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schimmer, C., and A. Neubauer. 2003. The equine herpesvirus 1 UL11 gene product localizes to the trans-Golgi network and is involved in cell-to-cell spread. Virology 308:23-36. [DOI] [PubMed] [Google Scholar]
- 49.Schmolke, S., P. Drescher, G. Jahn, and B. Plachter. 1995. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J. Virol. 69:1071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholl, B. C., J. Von Hintzenstern, B. Borisch, B. Traupe, M. Broker, and G. Jahn. 1988. Prokaryotic expression of immunogenic polypeptides of the large phosphoprotein (pp150) of human cytomegalovirus. J. Gen. Virol. 69:1195-1204. [DOI] [PubMed] [Google Scholar]
- 51.Silva, M. C., J. Schroer, and T. Shenk. 2005. Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc. Natl. Acad. Sci. USA 102:2081-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 56.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zipeto, D., F. Baldanti, E. Percivalle, G. Gerna, and G. Milanesi. 1993. Identification of a human cytomegalovirus mutant in the pp150 matrix phosphoprotein gene with a growth-defective phenotype. J. Gen. Virol. 74:1645-1648. [DOI] [PubMed] [Google Scholar]