Abstract
The RNA-dependent RNA polymerase of influenza virus is a heterotrimer formed by the PB1, PB2, and PA subunits. Although PA is known to be required for polymerase activity, its precise role is still unclear. Here, we investigated the function of the N-terminal region of PA. Protease digestion of purified recombinant influenza virus A/PR/8/34 PA initially suggested that its N-terminal region is folded into a 25-kDa domain. We then systematically introduced point mutations into evolutionarily conserved amino acids in the N-terminal region of influenza virus A/WSN/33. Most alanine-scanning mutations between residues L109 and F117 caused PA degradation, mediated by a proteasome-ubiquitin pathway, and as a consequence interfered with polymerase activity. Three further PA mutations, K102A, D108A, and K134A, were investigated in detail. Mutation K102A caused a general decrease both in transcription and replication in vivo, whereas mutations D108A and K134A selectively inhibited transcription. Both the D108A and K134A mutations completely inhibited endonuclease activity in vitro, explaining their selective defect in transcription. K102A, on the other hand, resulted in a significant decrease in both cap binding and viral RNA promoter-binding activity and consequently inhibited both transcription and replication. These results suggest that the N-terminal region of PA is involved in multiple functions of the polymerase, including protein stability, endonuclease activity, cap binding, and promoter binding.
Influenza A virus is a negative-sense virus belonging to the family Orthomyxoviridae (25). The genome of its segmented, single-stranded RNA is transcribed and replicated by a virus-encoded RNA-dependent RNA polymerase in infected cells (12). Transcription requires a capped RNA primer in the initiation step, which is derived by cleavage of host cell RNA polymerase II transcripts. This “cap snatching” is performed by a cap-dependent endonuclease activity of the RNA polymerase, generating short, 9- to 17-nucleotide (nt) capped RNA primers. On the other hand, replication is independent of a primer. The negative-sense viral RNA (vRNA) is copied to a full-length positive-sense RNA (cRNA) (first step of replication), which serves as a template for the synthesis of full-length negative-sense vRNA (second step of replication).
Influenza virus RNA polymerase is a trimeric complex comprising three different subunits: PB1, PB2, and PA (12, 25). Electron microscopy shows that it is very compact, with no apparent boundaries (1), although no high-resolution structure is available. All three subunits are generally found to be required for both transcription and replication (13, 33, 35), although some reports disagree (19, 30). The PB1 subunit contains the S-D-D motif characteristic of polymerases and is directly involved in RNA synthesis (12, 28). The PB2 subunit is responsible for cap binding (10, 12, 25). Although early studies suggested that PB2 harbors endonuclease activity (2, 38), more recently it was reported that the endonuclease active site is located in PB1 (28).
The precise function of the PA subunit is, however, not well established. PA has been suggested to be implicated in replication, not in transcription (19, 24, 29). However, recent studies have indicated that PA is involved in transcription as well as replication (13, 33, 35). The PA mutation H510A impaired the endonuclease activity and selectively inhibited transcription (13). Several other PA mutations decreased RNA synthesis of mRNA, cRNA, and vRNA, suggesting that PA is involved in both transcription and replication (13)
The promoter vRNA binding sites on PB1 have been mapped, although their precise locations are controversial (17, 22, 27). However, both PA and PB2 have been reported to interact with the promoter (5, 14, 15, 22, 34). Efficient binding to the 5′ end of the promoter has been shown to be dependent on the formation of a PB1-PA dimeric complex with or without PB2 (5, 26), suggesting that PA is necessary for a stable interaction between the polymerase and the promoter.
Because the precise mechanism by which PA participates in transcription and replication is not well defined, we decided to analyze the function of PA by a systematic mutational study of its N-terminal region. This analysis extends our previous study of the C-terminal region of PA (13). Here, we found that a region within an N-terminal region of PA is critical for polymerase structure, as mutations in this region caused protein degradation mediated by the proteasome-ubiquitin pathway. Importantly, other residues within the N-terminal region of PA are also involved in endonuclease activity, cap binding, and vRNA promoter binding.
MATERIALS AND METHODS
Plasmids.
The plasmids expressing PB1, PB2, PA, and NP of influenza virus A/WSN/33 (pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, pcDNA-PB2-TAP, and pcDNA-PA-TAP) have been described previously (5, 13). The plasmid pPOLI-vCAT-RT was used to express the vRNA-like chloramphenicol acetyltransferase (CAT) RNA reporter gene. Mutants of the PA plasmids were prepared by site-directed mutagenesis and were fully sequenced. pcDNA-PA-TAP mutants were constructed by replacing the PA sequence between the ClaI and BsrGI sites of pcDNA-PA-TAP with the corresponding sequence of the pcDNA-PA mutant. Sequences of the mutagenic primers are available upon request.
Proteolytic digestion of PA.
Proteolytic digestion of PA was performed by using purified PA that was expressed from Sf21 (Spodoptera frugiperda) insect cells infected with recombinant baculovirus-encoding PA of influenza virus A/PR/8/34, as described previously (18). Initially, 8 μg of purified PA was incubated with increasing amounts of trypsin (10, 40, and 100 ng) at 37°C for 30 min. The reaction products were heated to 95°C for 5 min in an equal volume of sodium dodecyl sulfate (SDS) sample buffer (200 mM Tris-HCl [pH 8.8], 4% SDS, 10% glycerol, 0.02% bromophenol blue), analyzed by 12% SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by staining with Coomassie blue. Intact, undigested PA and tryptic fragments were sequenced by Edman degradation after separation on a 4 to 12% Nu-PAGE (Invitrogen) gel in MES (morpholineethanesulfonic acid) buffer.
Preparation of purified TAP-tagged recombinant polymerase.
293T cells were transfected with the plasmids pcDNA-PB1, pcDNA-PB2-TAP, and pcDNA-PA (wild type or mutant) and harvested 2 days posttransfection. Recombinant polymerase was partially purified by the tandem affinity purification (TAP) method, as described previously (5). The partially purified TAP-tagged polymerase was mixed with glycerol, giving a final concentration of 40% glycerol, and stored at −80°C. Partially purified TAP-tagged polymerase or cell lysates were analyzed by 7.5% SDS-PAGE, followed by Western blotting using polyclonal rabbit anti-PB1, anti-PB2, or anti-PA antibody, as described previously (9).
Protease inhibitor treatment.
293T cells (approximately 90 to 100% confluence in 9-cm dishes) were transfected with pcDNA-PA-TAP (wild type or mutant). After incubation at 37°C for 24 h, the proteasome inhibitor MG132 (Sigma), dissolved in dimethyl sulfoxide (DMSO), was added at a final concentration of 5 μM and further incubated for 16 h. An equal volume of DMSO was used as a negative control. Cells were harvested, and PA-TAP was partially purified by the TAP method (5) and analyzed by silver staining after 7.5% SDS-PAGE. Ubiquitinated proteins were detected by Western blotting using monoclonal antiubiquitin antibody (Santa Cruz).
RNA isolation and primer extension assay.
293T cells were transfected with pcDNA-PB1, pcDNA-PB2, pcDNA-PA (wild type or mutant), pcDNA-NP, and pPOLI-vCAT-RT plasmids by using Lipofectamine 2000 (Invitrogen). Cells were harvested 24 h posttransfection, and total RNA was isolated with TRIzol reagent (Invitrogen). A primer extension assay was performed essentially as described previously (13) except that the 32P-labeled primer 5′ ACCCTGCTTAGCTTCCGAGA 3′ was used to detect 5S rRNA as an internal control, for which the expected size of the product was 62 nt. Transcription products were separated on 7% polyacrylamide gels containing 7 M urea in Tris-borate-EDTA (TBE) buffer and were detected by autoradiography. Primer extension assays for each mutant were carried out at least three times with independently transfected cells.
Transcription assay in vitro.
Globin mRNA-primed transcription and capped RNA-primed transcription assays were performed as described previously (13), except that reactions were carried out in a total reaction volume of 6 μl. Three microliters of TAP-purified polymerase was mixed with 4 pmol of the 5′ end of vRNA (5′ AGUAGAAACAAGGCC 3′) (Dharmacon), 4 pmol of the 3′ end of the vRNA promoter (5′ GGCCUGCUUUUGCU 3′) (Dharmacon), and 10 ng of globin mRNA (Sigma) in 3 μl containing 1 μCi of [α-32P]GTP, 1 mM ATP, 0.5 mM CTP, 2.5 mM MgCl2, 1.7 mM dithiothreitol (DTT), and 5 U of RNasin (Promega). After incubation at 30°C for 90 min, the transcription products were analyzed by 16% PAGE containing 7 M urea in TBE buffer, detected by autoradiography, and quantitated by phosphorimaging analysis. For the capped RNA-primed transcription assay, globin mRNA and [α-32P]GTP were replaced with an 11-nt 32P-labeled 2′-O-methylated capped RNA (5′ m7G32pppAmAmAmUACUCAAG 3′) (3) and 0.5 mM unlabeled GTP, respectively.
Endonuclease assay.
The endonuclease assay was performed essentially as described previously (13), except that the reaction was carried out in a total reaction volume of 6 μl containing 3 μl of TAP-purified polymerase, 32P-labeled poly(A)-tailed capped RNA (5′ m7G32pppAmAmAmUACUCAAGAn 3′), 4 pmol of the 5′ end of the vRNA promoter, 4 pmol of the 3′ end of the vRNA promoter, 2.5 mM MgCl2, 1 mM DTT, and 10 U of RNasin at 30°C for 90 min. The products were separated on a 16% polyacrylamide gel containing 7 M urea in TBE buffer, autoradiographed, and quantitated by phosphorimaging analysis.
UV cross-linking assay.
For cross-linking to the vRNA promoter, a UV cross-linking assay was performed as described previously (13), except that 5 μl of TAP-purified polymerase was mixed with approximately 0.5 pmol (100,000 dpm) of 32P-labeled 3′ end of the vRNA promoter in a total reaction volume of 10 μl containing 4 pmol of the 5′ end of vRNA, 10 mM HEPES (pH 7.5), 100 mM KCl, 2 mM MgCl2, 0.5 mM EGTA, 1 mM DTT, 8 U of RNasin, and 10% glycerol and incubated at 30°C for 30 min. After UV irradiation (254 nm), the cross-linked products were analyzed by 7.5% SDS-PAGE and detected by autoradiography. For cross-linking to the capped RNA, 32P-labeled 3′ end of the vRNA promoter was replaced with unlabeled 3′ end of the vRNA promoter and 32P-labeled capped RNA (100,000 dpm) (see above). Each polymerase subunit cross-linked with the vRNA promoter or capped RNA was identified by immunoprecipitation with polyclonal rabbit anti-PB1, anti-PB2, and anti-PA antibody, as described previously (4).
Replication assay in vitro.
The replication activity of the polymerase was analyzed by the dinucleotide replication initiation assay, as described previously (6), except that ATP was replaced with adenosine (T. Deng, J. Sharps, and G. G. Brownlee, personal communication). TAP-purified polymerase (1.5 μl) was mixed with 3 μl of reaction buffer containing 0.4 μCi of [α-32P]GTP, 0.6 mM adenosine, 2.5 mM MgCl2, 1.7 mM DTT, 3 U of RNasin, 4 pmol of the 5′ end of vRNA, and 4 pmol of the 3′ end of vRNA (see above). After incubation at 30°C for 15 to 16 h, the reaction products were analyzed on a 25% polyacrylamide gel containing 6 M urea in TBE buffer, detected by autoradiography, and quantitated by phosphorimaging analysis.
RESULTS
Limited trypsin proteolysis of PA.
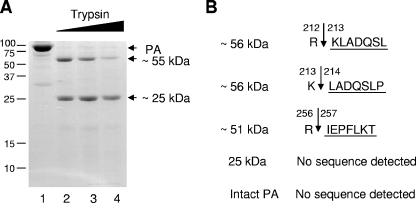
In order to gain some structural information on PA, we initially carried out a limited proteolysis with trypsin. A purified recombinant PA from influenza virus A/PR/8/34 that was expressed in insect cells was digested with increasing amounts of trypsin. PA yielded two major fragments of ∼55 kDa and ∼25 kDa (Fig. 1A). The larger, ∼55 kDa fragment resolved into a doublet of ∼56 kDa and ∼51 kDa fragments on a 4 to 12% polyacrylamide gradient gel (results not shown). These fragments were separately N-terminally sequenced by Edman degradation (see Materials and Methods). The ∼56-kDa fragment was composed of two fragments that matched the PA sequence, extending from amino acid 213 or 214 (Fig. 1B). The ∼51-kDa fragment matched the sequence of PA from amino acid 257 (Fig. 1B). The molecular masses of the ∼56-kDa and ∼51-kDa fragments suggested that all fragments extended to the C terminus. The ∼25-kDa fragment of PA failed to show any N-terminal degradation product, suggesting that the N terminus is modified. Intact PA was also resistant to Edman degradation, a result consistent with this hypothesis (Fig. 1B). These results suggest that PA has two distinct domains: one extending from amino acid 1 to 212, the other from amino acid 212 to the C terminus. However, the domain extending from position 212 to the C terminus is more susceptible to further protease degradation than is the N-terminal domain (Fig. 1).
FIG. 1.
Digestion of the PA subunit by trypsin. (A) Purified PA (8 μg) was incubated with increasing amounts of trypsin (10, 40, and 100 ng; lanes 2 to 5) or without trypsin (lane 1) at 37°C for 30 min. The reaction products were resolved by 12% SDS-PAGE and stained with Coomassie blue. The positions of marker proteins are indicated, in kilodaltons, on the left. (B) N-terminal amino acid sequences of the two ∼56-, the ∼51-, and the ∼25-kDa fragments determined by Edman degradation (underlined). The identity of the neighboring amino acids is also shown. The arrows represent the predicted trypsin cleavage sites.
Selection of N-terminal amino acids for mutagenesis.
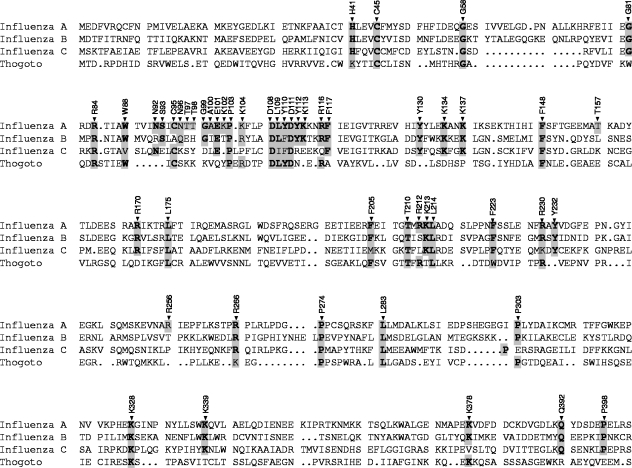
Next, we aligned amino acids 1 to ∼400 of the PA subunit of typical strains of influenza A, B, and C viruses and Thogoto virus to identify evolutionarily conserved amino acids (Fig. 2). Firstly, amino acids identical in all viruses were selected. Secondly, we included basic (R, K, and H) amino acids conserved in two or three viruses, as well as some aromatic (F, Y, and W), hydrophobic (P), and hydrophilic (E and Q) amino acids. Thirdly, we focused on residues 92 to 117 because preliminary experiments had suggested that amino acids important for the polymerase activity were clustered in this region despite a lower, or indeed no, conservation of some amino acids between influenza A, B, and C viruses and Thogoto virus. We included PA 100 (A→V) because V is typical of avian influenza A virus strains at this position, whereas A is typical of human influenza A virus strains, including the 1918 Spanish influenza virus (39). Fourthly, R212 and R256 were included because the domain study of PA (Fig. 1) suggested that these amino acids were readily accessible by trypsin. In addition, T157 was included because it has been implicated in replication activity (20). Thus, we introduced alanine mutations at 51 amino acid positions, except for residue 100, where valine was introduced (Fig. 1).
FIG. 2.
Alignment of residues 1 to 402 of the PA subunits of the RNA polymerases of influenza A, B, and C viruses and Thogoto virus. Sequence accession numbers are X17336 for influenza virus A/WSN/33, AF102022 for influenza virus B/Victoria/2/87, M28062 for influenza virus C/JJ/50, and AF006073 for Thogoto virus. Sequences were aligned with Omiga software, version 2.0 (Oxford Molecular Ltd., Oxford, United Kingdom). Numbers refer to amino acid positions in the A/WSN/33 sequences, and amino acids selected for mutagenesis are highlighted. Identical amino acids are boldfaced.
Effects of PA mutations on the polymerase activity in 293T cells.
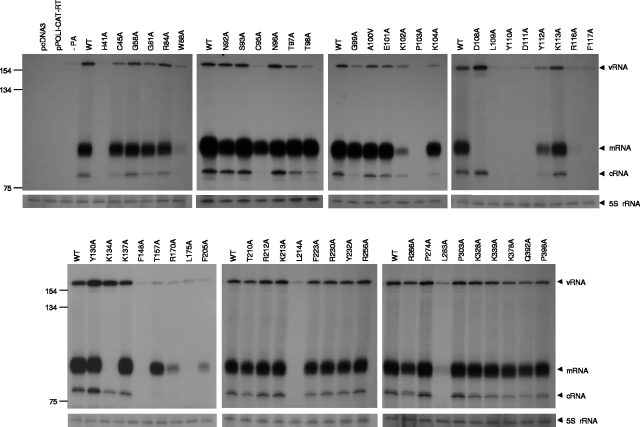
To test whether mutations in the N-terminal region of PA affect the polymerase activity in 293T cells, we performed primer extension assays using reconstituted viral RNPs. The plasmids expressing mutant PAs were transfected in 293T cells with plasmids expressing PB1, PB2, NP, and a vRNA-like CAT RNA reporter. Cell extracts were tested for polymerase activity 24 h posttransfection by measuring the levels of vRNA, mRNA, and cRNA. Three primers specific for negative-sense RNA (vRNA), positive-sense RNA (cRNA and mRNA), and 5S rRNA as an internal control were used in the same primer extension reaction (see Materials and Methods).
Thirty-one of the 51 mutants showed either no effect or only a slight decrease in the synthesis of CAT reporter vRNA, cRNA, and mRNA compared to the wild type, indicating that these amino acids are not critical for the function of the polymerase in this assay (Fig. 3). For mutations W88A, K102A, Y112A, R170A, and F205A, all three RNA species were significantly decreased (<50%) compared to the wild type. These results indicate that mutations at these positions are critical for both transcription and replication activities of the polymerase. Mutants D108A and K134A differed and showed nearly wild-type levels of cRNA and vRNA, but mRNA synthesis was not detected, suggesting that these mutations selectively inhibit transcription. On the other hand, mutations C95A and T157A preferentially decreased the synthesis of cRNA and vRNA but did not decrease mRNA synthesis by as much. For mutants H41A, P103A, L109A, Y110A, D111A, R116A, F117A, F148A, L175A, L214A, and L283A, there was no detectable mRNA or cRNA; only background levels of input vRNA were visible.
FIG. 3.
Effects of mutations in the PA subunit on RNA synthesis in vivo. 293T cells were transfected with plasmids expressing vRNA-like CAT RNA reporter, NP, PB1, PB2, and either a wild-type (WT) PA or mutant PAs. Cell extracts were tested for polymerase activity 24 h posttransfection by measuring the levels of vRNA, mRNA, and cRNA by primer extension. Sizes are indicated, in nucleotides, on the left. Positions of vRNA, mRNA, and cRNA signals are indicated on the right. The extension product of the 5S rRNA, with an expected size of 62 nt, is indicated on the right as an internal control.
Amino acids L109, Y110, D111, R116, and F117 are required for PA stability.
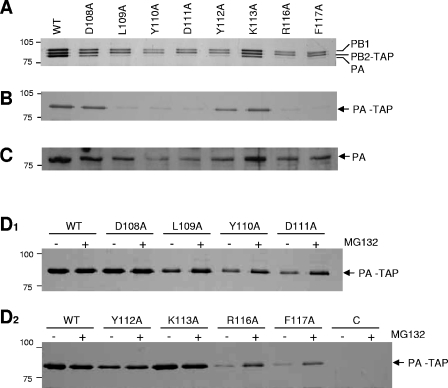
As all mutations from L109 to F117, inclusive, except for K113A, inhibited polymerase activity severely (Fig. 3), we decided to characterize these PA mutants further. In order to test whether these mutations affect the functional assembly of the trimeric complex of PB1, PB2, and PA, a TAP-tagged PB2 polymerase complex was partially purified (see Materials and Methods) from 293T cells transiently expressing the three subunits. Significantly, the expression of PA was lower in the L109A, Y110A, D111A, Y112A, R116A, and F117 mutants, although in the D108A and K113A mutants it was comparable to the wild type (Fig. 4A). A significant decrease in the PA level was also observed when PA was expressed alone, in the absence of PB1 and PB2 (Fig. 4B [TAP purified] and C [crude lysate]), except in the case of Y112, where ∼50% of the wild-type level was observed. These observations suggest that the lack of polymerase activity in vivo (Fig. 3) is primarily due to the low levels of PA.
FIG. 4.
Effects of mutations in the PA subunit on the assembly of the influenza virus RNA polymerase complex. (A) Expression and assembly of the polymerase subunit PB1, TAP-tagged PB2, and PA mutants in 293T cells. (B) Expression of TAP-tagged PA mutants alone in 293T cells. (C) Expression of PA mutants alone in total cell lysates of 293T cells. TAP-tagged proteins were partially purified from lysates of transfected 293T cells 24 h posttransfection with immunoglobulin G (IgG)-Sepharose, and bound proteins were cleaved by tobacco etch virus protease. The samples were analyzed by silver-stained 7.5% SDS-PAGE. (D1 and D2) Effects of the proteasome inhibitor on PA mutants. TAP-tagged PA was purified from cell lysates with IgG Sepharose and analyzed by 7.5% SDS-PAGE and silver staining. The positions of PB1, PB2-TAP, PA, and PA-TAP are shown on the right. The positions of marker proteins are indicated, in kilodaltons. WT, wild type.
We postulated that mutations L109A, Y110A, D111A, Y112A, R116A, and F117A might have altered the structure of the expressed protein and induced its degradation. To evaluate the possibility that mutant PAs were degraded via the proteasome pathway, we examined the effect of the proteasome-specific inhibitor MG132. Transfected cells, expressing PA mutants, in the absence of the PB1 and PB2 subunits, were treated with MG132 and analyzed by SDS-PAGE. MG132 inhibited the degradation of PA (Fig. 4D1 and D2) and led to increased expression levels in mutants L109A, Y110A, D111A, R116A, and F117A, but not Y112A, although wild-type levels were not always reached. In addition, ubiquitinated PA could be detected in TAP-purified PA in the presence of MG132 by immunoblotting with antiubiquitin monoclonal antibody (results not shown). These results suggest that the degradation of those PA mutants is mediated by the proteasome-ubiquitin pathway and that amino acids L109, Y110, D111, R116, and F117 might be critical for the correct folding of PA. It seems unlikely that PA mutations L109A, Y110A, D111A, R116A, and F117A would result in increased transcription and replication activity in the primer extension assay (Fig. 3) if polymerase containing these mutations were prepared from cells grown in MG132, since increased stability did not always approach 100% (Fig. 4D). Nevertheless, this remains to be formally tested.
In vitro transcription activity of three selected mutants: K102A, D108A, and K134A.
The polymerase activity of mutants K102A, D108A and K134A was severely inhibited (Fig. 3), although the assembly of the trimeric complex appeared to be similar to that of the wild type (Fig. 5A). Mutation K102A caused a general decrease in both transcription and replication, whereas mutations D108A and K134A selectively inhibited transcription (Fig. 3). To address the question why these three mutations inhibited the polymerase activity severely, we analyzed them in more detail in vitro.
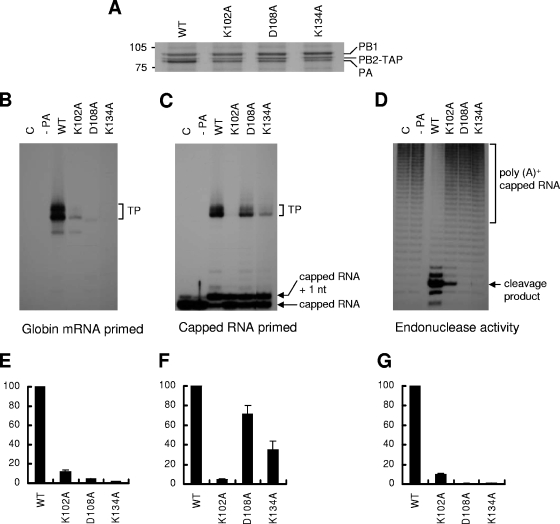
FIG. 5.
Transcription initiation activity of PA mutants in vitro. 293T cells were transfected with plasmids expressing PB1 and PB2-TAP, either without PA (−PA), with a wild-type PA (WT), or with mutant PAs (K102A, D108A, and K134A). TAP-tagged polymerases were partially purified from cell extracts by immunoglobulin G-Sepharose and used in vitro. As a negative control (C), cell extracts from untransfected cells were prepared. (A) Silver staining of purified TAP-tagged polymerases separated by 7.5% SDS-PAGE. The positions of PB1, PB2-TAP, and PA are shown on the right. (B) Transcription activity on a model vRNA promoter of mutant polymerases primed with a globin mRNA primer. The positions of the 27- or 28-nt-long transcription products (TP) are indicated. (C) Transcription activity of mutant polymerases primed with an 11-nt 32P-labeled capped RNA primer (see Materials and Methods). The positions of the 27- or 28-nt-long transcription products (TP) of capped RNA primer are indicated. (D) Endonuclease activity (see Materials and Methods) of mutant polymerases. TAP-purified polymerases were incubated with [32P]poly(A)+-capped RNA. The positions of the poly(A)+-capped RNA and specific 11-nt cleavage product are indicated on the right. (D, E, and F) Quantification of results obtained in panels B, C, and D, respectively, by phosphorimaging analysis. Data are expressed as percentages relative to the wild type (mean ± standard deviation; n = 3).
The initiation of viral mRNA synthesis requires endonuclease activity to generate short, capped RNA to use as a primer. To distinguish which component of transcription was affected, i.e., cap binding, endonuclease cleavage, or capped-primer extension, we tested the transcription activity of TAP-purified polymerase by three different assays: (i) a globin mRNA-primed transcription assay that tested both endonuclease activity and transcription by the resultant globin mRNA primers; (ii) a capped RNA-primed transcription assay in which a capped 11-nt primer primed transcription, thus bypassing the need for endonuclease cleavage; and (iii) a direct endonuclease assay (see Materials and Methods).
For K102A, the transcription activity with globin mRNA was significantly decreased, to about 11% of wild-type activity (Fig. 5B and E). This reduced activity correlated well with the reduction (10%) in endonuclease activity (Fig. 5D and G). The capped RNA-primed transcription assay, however, showed that this mutation could not extend the capped RNA primer (Fig. 5C and F). Because the capped RNA-primed transcription activity of the polymerase depends on its ability to bind capped RNA, these results suggest that the K102A mutation affects cap-binding activity.
On the other hand, for D108A and K134A, the activity with globin mRNA was decreased even further (<4% of wild-type activity [Fig. 5B and E]). No significant endonuclease activity was detected (Fig. 5D and G), indicating that these mutations completely inhibited endonuclease activity. The D108A mutant polymerase apparently could, however, extend the capped RNA, although less efficiently than the wild type (Fig. 5C and F). Transcription products were also detected in the K134A mutant polymerase in capped RNA-primed transcription, though the activity was only 35% of that of the wild type (Fig. 5C and F). These results suggested that the transcription defect in the D108A and K134A mutants was mainly due to a loss of endonuclease activity.
Capped RNA binding, vRNA promoter binding, and replication initiation activity with a model vRNA or cRNA promoter.
The results of in vitro transcription assays (see above) suggested that mutation K102A might inhibit cap binding activity, but D108A and K134A differed and were capable of binding caps but were defective in endonuclease activity. To test this directly, we analyzed capped RNA binding activity by a UV cross-linking assay. The K102A mutant had a dramatically reduced (10% of the wild type [Fig. 6A, upper panel, and Fig. 6C]) signal of cross-linked PB1 and PB2 compared to the wild type. A reduction in cap-binding activity was also observed in the D108A and K134A mutants (44% and 29%, respectively). These results suggest that K102 is mainly involved in cap binding and that consequently the K102A mutation reduces transcription. However, D108 and K134 are primarily involved in the endonuclease activity, since the D108A and K134A mutants could still bind capped RNA.
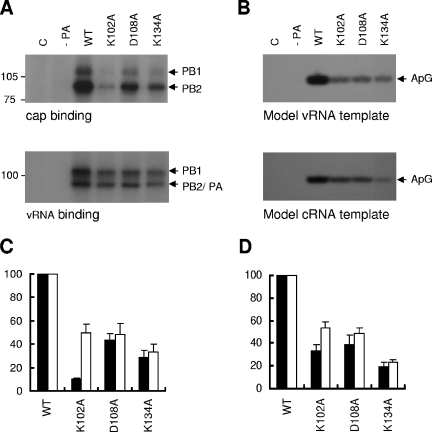
FIG. 6.
RNA binding activity and replication initiation activity of PA mutants. (A) UV cross-linking analysis of the binding of RNA with polymerases containing wild-type PA (WT) or mutant PAs (K102A, D108A, and K134A). Purified TAP-tagged polymerases were incubated with 32P-labeled capped RNA in the presence of both 5′ vRNA and 3′ vRNA (upper panel) or with 32P-labeled 3′ vRNA in the presence of 5′ vRNA (lower panel). The reaction mixtures were subsequently exposed with UV light and analyzed by 7.5% SDS-PAGE. The positions of the cross-linked products are indicated on the right. (B) Replication initiation activity of the polymerases containing WT PA or mutant PAs (K102A, D108A, and K134A). The position of the specific ApG product is indicated on the right. (C and D) Quantification of results obtained in panels A and B, respectively, by phosphorimaging. Data are expressed as percentages relative to the wild type (mean ± standard deviation; n = 3). (C) Black bars, cap binding; white bars, model vRNA promoter binding. (D) Black bars, model vRNA template; white bars, model cRNA template.
We next tested the binding of mutants K102A, D108A, and K134A to a model vRNA promoter by UV cross-linking (see Materials and Methods). As shown in Fig. 6A (lower panel) and C, all three mutant polymerases showed significantly reduced vRNA promoter binding (49%, 48%, and 33%, respectively). We expected that this reduction in the vRNA binding activity would lead to decreased activity in a primer-independent initiation-of-replication assay. To confirm this, the TAP-purified mutant polymerases were incubated with adenosine, 32P-labeled GTP, the 5′ end of vRNA, and the 3′ end of vRNA in the absence of primer. As expected, all three mutant polymerases synthesized reduced yields of ApG (Fig. 6B, upper panel, and D). Interestingly, the D108A and K134A mutants showed decreased levels of replication initiation in vitro (39% and 20%, respectively), whereas they did not show a significant decrease in vivo (Fig. 3) (see Discussion). The reduced replication initiation activity of the D108A and K134A mutant polymerases appears to be correlated with their reduced vRNA binding activity (compare Fig. 6C and D), suggesting that reduced replication directly reflects reduced promoter binding. A similar reduction in the replication activity of mutants K102A, D108A, and K134A was observed when a model cRNA promoter was used (Fig. 6B, lower panel, and D). This result suggests that there is no differential effect in the efficiencies of polymerase binding to the vRNA or cRNA promoter. Thus, we conclude that amino acids K102, D108, and K134 are also involved in vRNA and cRNA promoter binding.
DISCUSSION
The aim of this study was to investigate the precise functions of the N-terminal region of PA both in transcription and replication. Initially, we performed limited proteolysis of purified recombinant PA, isolated from influenza virus A/PR/8/34, in order to identify any structural domains present. We found that it had two distinct domains: one from amino acid 1 to 212 (domain 1), the other from amino acid 212 to the C terminus (domain 2) (Fig. 1). This new finding suggests a domain structure of PA, but further experiments, outside the scope of this study, on purified heterotrimeric polymerase are required to prove whether these PA domains are retained in the heterotrimer. However, an unexpected finding was that the N terminus of PA was blocked to Edman degradation, suggesting that PA is posttranslationally modified by N-terminal acetylation—a common modification in eukaryotic proteins (32).
We next systematically introduced alanine mutations at conserved amino acids (Fig. 2) between influenza A, B, and C viruses and Thogoto virus, assuming that such residues were likely to be essential for structure and/or function. Screening of mutants by primer extension demonstrated that 20 of the 51 selected amino acids were essential for transcription and/or replication (Fig. 3). These results confirm that PA is required for both transcription and replication. A number of individual mutations, all at highly evolutionarily conserved residues, were severely affected in transcription and replication, i.e., at H41, P103, F148, L175, L214, and L283 (Fig. 3). They presumably affected PA structure, stability, nuclear transport, and/or assembly with PB1 or PB2 and were not pursued further. However, a region from D108 to F117 contained a cluster of amino acids critical for polymerase activity, suggesting the importance of this discrete region for polymerase function. In agreement with this, a double mutation of PA (D111A, Y112A) has recently been reported to interfere with transcription and replication and to prevent isolation of viable virus containing these mutations (35). Therefore, we initially focused on this region.
Many of the mutations in the region from L109 to F117, inclusive, caused proteasome-mediated degradation (Fig. 4), suggesting that L109, Y110, D111, R116, and F117 must be important for protein stability. However, the mechanism by which these mutations result in PA degradation is less clear. Theoretically, mutation may lead to incorrect PA folding, weaker interaction with the PB1 subunit, or inhibition of interaction of PA with cellular factors, e.g., hCLE (21), kinases (37), or nuclear transport receptors. However, the most likely hypothesis is that these mutations interfered with correct PA folding, since they were still unstable when expressed alone, in the absence of PB1 and PB2 (Fig. 4). Our results cannot exclude the possibility that these mutations may additionally interfere with the assembly of the PB1-PA dimer (5) because of weaker interaction with PB1. PB1-PA dimerization is known to be required for efficient nuclear accumulation of PA (16).
We subsequently focused on three other mutations, K102A, D108A, K134A, because polymerase activity was severely decreased (Fig. 3), despite the fact that assembly of the trimeric complex appeared to be similar to that of the wild type (Fig. 5A). Other mutations, e.g., W88A, R170A and F205A, were interesting in that they resulted in a marked reduction in both transcription and replication (Fig. 3); they were not investigated further, however, so that we could focus on our attention on K102A, D108A, and K134A. Primer extension assays showed that the K102A mutation decreased mRNA, vRNA, and cRNA levels, whereas the D108A and K134A mutations selectively impaired mRNA synthesis (Fig. 3). Further analysis of the K102A mutant in vitro confirmed that its most striking feature was a defect in cap binding (Table 1). However, D108A and K134A differed from K102A in showing a complete loss of endonuclease activity (Table 1). K102A also decreased endonuclease activity, but this likely reflected reduced cap binding (Table 1). D108 is completely conserved among influenza A, B, and C viruses and Thogoto virus, and K134 is conserved among influenza A, B, and C viruses, emphasizing their important roles. K102, however, is not conserved and is unique to influenza virus A, suggesting that the role of K102 is not shared by influenza B and C viruses and Thogoto virus.
TABLE 1.
Mutant PA activity
| Parameter | Mean mutant PA activity ± SD (%)a
|
||
|---|---|---|---|
| K102A | D108A | K134A | |
| Globin mRNA-primed transcription | 12 ± 2.1 | 4.1 ± 0.6 | 1.4 ± 0.7 |
| Capped RNA-primed transcription | 4.2 ± 0.8 | 71 ± 9.4 | 35 ± 9.4 |
| Endonuclease activity | 10 ± 0.5 | 0.1 ± 0.8 | 0.7 ± 0.4 |
| Cap-binding activity | 10 ± 1.0 | 44 ± 5.4 | 29 ± 6.1 |
| vRNA-binding activity | 49 ± 7.9 | 48 ± 9.7 | 33 ± 6.9 |
| Replication activity (vRNA) | 34 ± 5.4 | 38 ± 9.0 | 20 ± 3.5 |
| Replication activity (cRNA) | 54 ± 5.3 | 49 ± 4.6 | 23 ± 2.5 |
Data are expressed as percentages of wild-type activity from three independent experiments.
The conclusion that the defect in K102A is primarily a defect of cap binding (Table 1) is the first time, to our knowledge, that a specific residue in PA has been implicated in cap binding. However, whereas cap structures are known to cross-link to the PB2 and PB1 subunits, no cross-linking of caps to PA has been previously observed (10). This suggests that the effect of the K102A mutation is likely to be indirect, possibly by inducing a conformational change that influences the cap binding site formed by the PB1 and PB2 subunits. The D108A and K134A mutations also reduced cap binding as well as endonuclease activity (Table 1). However, because cap binding is obviously a prerequisite for the endonuclease activity, these results are to be expected (12). An unexpected finding for the D108A mutation was the discrepancy between the value of 44% in cap-binding activity compared with 71% activity in cap-primed transcription (Table 1). An unknown component or condition of the capped primed transcription assay must have favored cap binding and extension compared to the UV cross-linking assay.
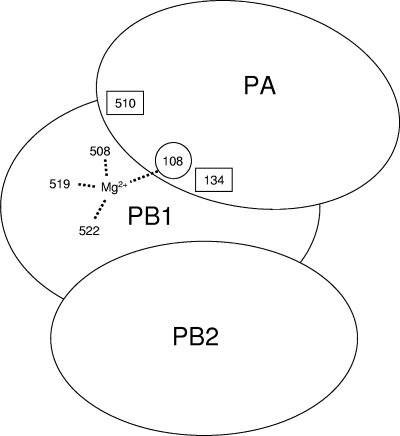
The observations here that residues D108 and K134 are directly or indirectly involved in endonuclease activity extends and confirms previous conclusions, based on a study of H510, that PA is involved in this activity (13). Thus, it is necessary to modify the conclusion that the PB1 subunit is solely responsible for endonuclease activity, catalyzed by three acidic amino acids (E508, E519, and D522) at its active site (28). In general, highly conserved acidic amino acids of endonucleases play an important role in coordinating one or two divalent metal ions to stabilize the transition site and promote the phosphoryl-transfer reaction (8, 11, 31). Thus, we speculate that four acidic amino acids, three in PB1 (E508, E519, and D522) and one in PA (D108), might interact with one of the Mg2+ ions (Fig. 7). Alternatively, two independent active sites involving these acidic residues may exist (7).
FIG. 7.
Model of the endonuclease active site of the influenza virus RNA polymerase. Amino acids 508, 519, and 522 derive from the PB1 subunit, while amino acids 108, 134, and 510 are from PA. Acidic amino acids in PB1 (E508, E519, and D522) and PA (D108) interact with a divalent metal ion (Mg2+) in the endonuclease active site. Two other amino acids in PA (K134 and H510) are also involved (see the text for details).
Lysine is also conserved in the active site of the type II restriction endonucleases (31), and a nearby histidine was suggested to be important for endonuclease activity (40). Thus, it is possible that K134 and H510 of the PA of influenza virus polymerase might also play a critical role in the endonuclease active site (Fig. 7), although we cannot exclude the possibility that K134 and H510 might indirectly affect activity.
The PA mutations K102A, D108A, and K134A significantly decreased binding to the model vRNA promoter (Fig. 6). Several reports have shown that PA binds both vRNA and cRNA promoters (14, 15, 22), although the precise binding site has not yet been identified. Our UV cross-linking data suggest that K102, D108, and K134 are involved in vRNA promoter binding, although, interestingly, significant promoter binding is still retained (Table 1). Clearly, reduced promoter binding is consistent with the observed reduced transcription and replication activities in vitro (Table 1).
A new replication initiation assay in vitro showed that mutations D108A and K134A resulted in reduced activity compared to the wild type (39% and 20%, respectively, Fig. 6B), whereas a significant decrease in replication was not observed in 293T cells in vivo (Fig. 3). This discrepancy between the in vivo and in vitro results may be explained as follows. Vreede et al. (41) suggested that the initiation of transcription and replication is regulated stochastically. Thus, the polymerase may preferentially synthesize cRNA and vRNA if mRNA transcription has been selectively impaired. Therefore, successive rounds of replication could overcome a decreased level of replication in 293T cells, resulting in replication levels comparable to that of the wild type.
Although most mutants were selected because of sequence conservation among influenza viruses, a few were studied here because of a special interest. The T157A mutation, characteristic of influenza A viruses, decreased replication significantly more than it decreased transcription (Fig. 3), in agreement with an extensive study in influenza virus A/Victoria/3/75 (20). However, the molecular mechanism by which this mutation causes this effect is uncertain (20). On the other hand, the A100V mutation showed no significant differences in mRNA, vRNA, or cRNA levels from wild-type PA (Fig. 3), suggesting that A100 in the PA of the 1918 Spanish influenza virus is not critical for transcription or replication (39). Similarly, the R212A, K213A, and R256A mutations in influenza virus A/WSN/33, at residues susceptible to tryptic cleavage in intact PA (Fig. 1), showed no differences from the wild type in the primer extension assay (Fig. 3), suggesting that they are not crucial for transcription or replication.
In summary, our study indicates that the N-terminal region of PA is multifunctional. It plays an important role in the initiation of both transcription and replication by participating in PA stability, endonuclease activity, cap binding, and vRNA promoter binding. These data add to the previous knowledge that PA has proteolysis-inducing activity (36) and is involved in nuclear accumulation of PB1 (16), in the assembly of the trimeric complex (23), and in the assembly or release of virions from cells (35).
Acknowledgments
We thank J. Robinson for sequencing; Ervin Fodor, Frank T. Vreede, Ruth Harvey, Jane Sharps, and Helena Maier for helpful discussions; Masanobu Ohuchi and Reiko Ohuchi (Kawasaki Medical School, Japan) for construction of some of PA mutants; and Tony Willis (Biochemistry, Oxford) for Edman degradation.
This work was supported by MRC grants (G9523972 and G9901312) to G.G.B.
REFERENCES
- 1.Area, E., J. Martin-Benito, P. Gastaminza, E. Torreira, J. M. Valpuesta, J. L. Carrascosa, and J. Ortin. 2004. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc. Natl. Acad. Sci. USA 101:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blok, V., C. Cianci, K. W. Tibbles, S. C. Inglis, M. Krystal, and P. Digard. 1996. Inhibition of the influenza virus RNA-dependent RNA polymerase by antisera directed against the carboxy-terminal region of the PB2 subunit. J. Gen. Virol. 77:1025-1033. [DOI] [PubMed] [Google Scholar]
- 3.Brownlee, G. G., E. Fodor, D. C. Pritlove, K. G. Gould, and J. J. Dalluge. 1995. Solid phase synthesis of 5′-diphosphorylated oligoribonucleotides and their conversion to capped m7Gppp-oligoribonucleotides for use as primers for influenza A virus RNA polymerase in vitro. Nucleic Acids Res. 23:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow, M., T. Deng, M. Addley, and G. G. Brownlee. 2004. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 78:6263-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng, T., J. Sharps, E. Fodor, and G. G. Brownlee. 2005. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J. Virol. 79:8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng, T., F. T. Vreede, and G. G. Brownlee. 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 80:2337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doan, L., B. Handa, N. A. Roberts, and K. Klumpp. 1999. Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry 38:5612-5619. [DOI] [PubMed] [Google Scholar]
- 8.Dryden, D. T., N. E. Murray, and D. N. Rao. 2001. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 29:3728-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelhardt, O. G., M. Smith, and E. Fodor. 2005. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 79:5812-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fechter, P., L. Mingay, J. Sharps, A. Chambers, E. Fodor, and G. G. Brownlee. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 278:20381-20388. [DOI] [PubMed] [Google Scholar]
- 11.Fedor, M. J. 2002. The role of metal ions in RNA catalysis. Curr. Opin. Struct. Biol. 12:289-295. [DOI] [PubMed] [Google Scholar]
- 12.Fodor, E., and G. G. Brownlee. 2002. Influenza virus replication, p. 1-29. In C. W. Potter (ed.), Influenza. Elsevier Science, New York, N.Y.
- 13.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodor, E., B. L. Seong, and G. G. Brownlee. 1993. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J. Gen. Virol. 74:1327-1333. [DOI] [PubMed] [Google Scholar]
- 16.Fodor, E., and M. Smith. 2004. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78:9144-9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez, S., and J. Ortin. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara, K., M. Shiota, H. Kido, Y. Ohtsu, T. Kashiwagi, J. Iwahashi, N. Hamada, K. Mizoue, N. Tsumura, H. Kato, and T. Toyoda. 2001. Influenza virus RNA polymerase PA subunit is a novel serine protease with Ser624 at the active site. Genes Cells 6:87-97. [DOI] [PubMed] [Google Scholar]
- 19.Honda, A., K. Mizumoto, and A. Ishihama. 2002. Minimum molecular architectures for transcription and replication of the influenza virus. Proc. Natl. Acad. Sci. USA 99:13166-13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huarte, M., A. Falcon, Y. Nakaya, J. Ortin, A. Garcia-Sastre, and A. Nieto. 2003. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77:6007-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huarte, M., J. J. Sanz-Ezquerro, F. Roncal, J. Ortin, and A. Nieto. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 75:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, T. E., and G. G. Brownlee. 2006. A new promoter-binding site in the PB1 subunit of the influenza A virus polymerase. J. Gen. Virol. 87:679-688. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi, A., T. Naito, and K. Nagata. 2005. Involvement of influenza virus PA subunit in assembly of functional RNA polymerase complexes. J. Virol. 79:732-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krug, R. M., M. Ueda, and P. Palese. 1975. Temperature-sensitive mutants of influenza WSN virus defective in virus-specific RNA synthesis. J. Virol. 16:790-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roitman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Lee, M. T., K. Bishop, L. Medcalf, D. Elton, P. Digard, and L. Tiley. 2002. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, M. L., B. C. Ramirez, and R. M. Krug. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa, Y., K. Oda, and S. Nakada. 1996. The PB1 subunit alone can catalyze cRNA synthesis, and the PA subunit in addition to the PB1 subunit is required for viral RNA synthesis in replication of the influenza virus genome. J. Virol. 70:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perales, B., J. J. Sanz-Ezquerro, P. Gastaminza, J. Ortega, J. F. Santaren, J. Ortin, and A. Nieto. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pingoud, A., and A. Jeltsch. 2001. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29:3705-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polevoda, B., and F. Sherman. 2000. N α-terminal acetylation of eukaryotic proteins. J. Biol. Chem. 275:36479-36482. [DOI] [PubMed] [Google Scholar]
- 33.Portela, A., T. Zurcher, A. Nieto, and J. Ortin. 1999. Replication of orthomyxoviruses. Adv. Virus Res. 54:319-348. [DOI] [PubMed] [Google Scholar]
- 34.Pritlove, D. C., E. Fodor, B. L. Seong, and G. G. Brownlee. 1995. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J. Gen. Virol. 76:2205-2213. [DOI] [PubMed] [Google Scholar]
- 35.Regan, J. F., Y. Liang, and T. G. Parslow. 2006. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol. 80:252-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz-Ezquerro, J. J., S. de la Luna, J. Ortin, and A. Nieto. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69:2420-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz-Ezquerro, J. J., J. Fernandez Santaren, T. Sierra, T. Aragon, J. Ortega, J. Ortin, G. L. Smith, and A. Nieto. 1998. The PA influenza virus polymerase subunit is a phosphorylated protein. J. Gen. Virol. 79:471-478. [DOI] [PubMed] [Google Scholar]
- 38.Shi, L., D. F. Summers, Q. Peng, and J. M. Galarza. 1995. Influenza A virus RNA polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology 208:38-47. [DOI] [PubMed] [Google Scholar]
- 39.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889-893. [DOI] [PubMed] [Google Scholar]
- 40.Tsutakawa, S. E., T. Muto, T. Kawate, H. Jingami, N. Kunishima, M. Ariyoshi, D. Kohda, M. Nakagawa, and K. Morikawa. 1999. Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell 3:621-628. [DOI] [PubMed] [Google Scholar]
- 41.Vreede, F. T., T. E. Jung, and G. G. Brownlee. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78:9568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]