Abstract
The human herpesvirus Epstein-Barr virus (EBV) establishes latency and promotes the long-term survival of its host B cell by targeting the molecular machinery controlling cell fate decisions. The cellular antiapoptotic bfl-1 gene confers protection from apoptosis under conditions of growth factor deprivation when expressed ectopically in an EBV-negative Burkitt's lymphoma-derived cell line (B. D'Souza, M. Rowe, and D. Walls, J. Virol. 74:6652-6658, 2000), and the EBV latent membrane protein 1 (LMP1) and its cellular functional homologue CD40 can both drive bfl-1 via an NF-κB-dependent enhancer element in the bfl-1 promoter (B. N. D'Souza, L. C. Edelstein, P. M. Pegman, S. M. Smith, S. T. Loughran, A. Clarke, A. Mehl, M. Rowe, C. Gélinas, and D. Walls, J. Virol. 78:1800-1816, 2004). Here we show that the EBV nuclear antigen 2 (EBNA2) also upregulates bfl-1. EBNA2 trans-activation of bfl-1 requires CBF1 (or RBP-Jκ), a nuclear component of the Notch signaling pathway, and there is an essential role for a core consensus CBF1-binding site on the bfl-1 promoter. trans-activation is dependent on the EBNA2-CBF1 interaction, is modulated by other EBV gene products known to interact with the CBF1 corepressor complex, and does not involve activation of NF-κB. bfl-1 expression is induced and maintained at high levels by the EBV growth program in a lymphoblastoid cell line, and withdrawal of either EBNA2 or LMP1 does not lead to a reduction in bfl-1 mRNA levels in this context, whereas the simultaneous loss of both EBV proteins results in a major decrease in bfl-1 expression. These findings are relevant to our understanding of EBV persistence, its role in malignant disease, and the B-cell developmental process.
The human herpesvirus Epstein-Barr virus (EBV) infects resting B lymphocytes, transforming them into permanently growing lymphoblastoid cell lines (LCLs) (for a review, see reference 27). Efficient virus-driven immortalization of B cells requires the expression of a subset of the thirteen known EBV latent genes, including several nuclear antigens (EBNAs), EBNA1, -2, -3A, -3B, -3C, and -LP, and an integral membrane protein, LMP1 (28). Uncovering the mechanisms by which these viral proteins function is essential to understanding EBV biology and the association of this agent with human malignancies, including African endemic Burkitt's lymphoma (BL), anaplastic nasopharyngeal carcinoma, Hodgkin's disease, and lymphoproliferative disorders in immunosuppressed individuals (for a review, see reference 25), and may also yield insights into the molecular mechanisms that govern the normal process of B-cell activation.
EBNA2 and EBNA-LP are the earliest latent-cycle proteins to be detected following EBV infection of primary B cells (1). EBNA2 is essential for B-cell immortalization, and its principal role therein is likely to be as a regulator of transcription, since the domains of this protein essential for both growth transformation and transcriptional activation are the same (8). EBNA2 functions as a transcriptional trans-activator to regulate the pattern of EBV latent-gene expression in B cells and to modify cellular gene expression with a resultant stimulation of G0 to G1 cell cycle progression. The mechanism by which EBNA2 modulates the expression of its target genes is complex and dependent on the cell context. EBNA2 does not bind DNA directly but is recruited to its sites of action through interactions with cellular proteins, including CBF1 (also known as RBP-Jκ), Spi-1/PU.1 and Spi-B-related proteins of the Ets family of transcription factors, and ATF-CRE (for a review, see reference 71). CBF1 is a DNA-binding nuclear adaptor protein from the cellular Notch signaling pathway that interacts directly with the intracellular domain of Notch proteins, a family of highly conserved transmembrane receptors. The EBNA2-CBF1 interaction is essential for the immortalization of primary B cells by EBV (for a review, see reference 20). Upon ligand binding, the Notch receptor is cleaved, and an intracellular fragment (Notch-IC) is then translocated to the nucleus, where it binds to CBF1 and modulates the expression of target genes. EBNA2 may therefore be considered in part as a functional equivalent of Notch-IC. The promoters of EBV latent genes (EBNA Cp, LMP1, LMP-2A, and LMP-2B) (29, 49, 70) and that of the cellular gene CD23 (48) have been shown to contain functional CBF1-binding motifs and to be targets of EBNA2-CBF1. EBV targets CBF1 by more than one mechanism. EBNA3A, EBNA3B, and EBNA3C have been shown to limit EBNA2-mediated transcriptional activation from the LMP2A, LMP1, and Cp promoters by directly contacting CBF1 and destabilizing its binding to DNA (20, 71). The EBV gene product known as RPMS1, which is translated from an EBV complementary strand transcript (CST) (or Bam A rightward transcripts [BART]), has been shown to act as a negative regulator of Notch/EBNA2 signaling by interacting with CBF1 and associated members of a histone deacetylase repressor complex (60, 67).
A central component of the overall EBV strategy and its role in the development of related malignant disease is its capacity to suppress the cellular apoptotic program. EBV-negative or EBV-positive BL-derived cell lines which express only EBNA1 (referred to as the EBV “latency program”) can easily be induced to undergo apoptosis; however, lymphoblastoid cell lines (LCLs) and EBV-positive BL-derived cell lines expressing a complete set of EBV latent proteins (the “growth program”) display increased resistance to apoptosis induced by a variety of triggers (17). The cellular antiapoptotic bfl-1 gene (also known as A1) is highly expressed in EBV-positive B-cell lines expressing the growth program and conferred protection against apoptosis induced by growth factor deprivation when expressed ectopically in an EBV-positive BL-derived cell line expressing the latency program (6, 11). bfl-1 encodes a protein of the Bcl-2 family whose preferential expression in hematopoietic and endothelial cells is controlled by inflammatory stimuli such as tumor necrosis factor and interleukin-1 (7, 30, 31). Bfl-1 suppresses p53-mediated apoptosis and can inhibit the proapoptotic activities of other Bcl-2 members (9, 10, 64, 66). bfl-1 is an established transcriptional target of latent membrane protein 1 (LMP1), the signaling pathway being initiated through interactions of this EBV protein with components of the cellular tumor necrosis factor receptor/CD40-signaling pathway and in which there is an essential role for the transcription factor NF-κB. The bfl-1 promoter contains a NF-κB-like binding motif that mediates trans-activation by LMP1, CD40, and the NF-κB subunit protein p65 (12).
Here we show that bfl-1 expression is also upregulated by EBNA2 in EBV-negative BL-derived cell lines and that EBNA2 trans-activates the bfl-1 promoter in this cell context by a mechanism that is dependent both on the presence of CBF1 and the EBNA2-CBF1 interaction. We demonstrate an essential role for a novel consensus CBF1-binding site on the bfl-1 promoter and show that upregulation is inhibited by coexpression of EBNA3A, EBNA3B, EBNA3C, or RPMS1 or by a dominant-negative mutant of CBF1 and does not involve activation of NF-κB. bfl-1 expression is induced and maintained at high levels by EBNA2 in a conditional LCL, and withdrawal of either EBNA2 or LMP1 does not lead to a decrease in bfl-1 expression in this context, whereas the simultaneous loss of both EBV proteins leads to a dramatic decrease in the level of bfl-1 mRNA. To our knowledge, this is the first report of a cellular apoptosis-related gene whose expression is transcriptionally modulated by EBNA2 (and not indirectly via LMP1). These findings are evidence that EBNA2 may provide a cell survival advantage during B-cell infection and also contribute to the development of EBV-associated diseases by driving the expression of a cellular antiapoptotic gene.
MATERIALS AND METHODS
Cell lines, transfections and reporter assays.
DG75, BL41, and BJAB are EBV-negative BL-derived cell lines (5, 52). IARC-171 is an LCL established from the same patient from whom the BL41 cell line was derived (2). MUTU I is an EBV-positive BL-derived cell line expressing the EBV latent program (18), and X50-7 is an LCL. The cell lines were cultured as described previously (12). DG75-tTA-EBNA2, BL41-K3, BL41/P3HR1-9A, EREB2.5, SM295 D6, and SM296 D3 and associated EBNA2 induction methods have been described elsewhere (14, 35, 36, 51). cl2/p1480.4 (1480.4) is a conditional LCL established from resting B cells by infection with a mini-EBV plasmid carrying an ER-EBNA2 gene fusion in conjunction with an LMP1 gene expressed from the simian virus 40 promoter (69). The 1852.4 cell line is a conditional LCL generated by using a mini-EBV plasmid that expresses wild-type EBNA2 and from which LMP1 expression is dependent on the presence of tetracycline (1 μg/ml) (39). Transfections and reporter assays have been described elsewhere (12), and each transfection result shown was compiled from three independent experiments.
RNase protection assays, Northern blotting, and reverse transcription real-time PCR.
Total cellular RNA was prepared, using RNA isolator solution (Genosys) according to the manufacturer's specifications. RNase protection assays (RPAs) were performed using the Riboquant multiprobe RNase protection assay system (hAPO-2C template set; Becton Dickinson). RNase protection assays and Northern blotting methods are described elsewhere (11). Following reverse transcription of total RNA, bfl-1- and GAPDH-derived cDNAs were detected by real-time PCR using primers and dual-labeled (FAM/TAMRA) fluorogenic probes (Applied Biosystems assay reagents Hs00187845 and Hs99999905, respectively). The amplification reaction volumes were 25 μl and consisted of 12.5 μl of 2× Taqman Universal Mastermix (Applied Biosystems), 1.25 μl of assay reagent, 9.25 μl of water, and 2 μl of cDNA. Following activation of AmpliTaq Gold (10 min at 95°C), amplification was performed for 40 cycles (15 s at 95°C and 60 s at 60°C), and real-time monitoring of changes in the fluorescence intensity of Taqman probes was done, using an ABI 7500 sequence detection system (Applied Biosystems). Data was analyzed by the comparative cycle threshold method(50).
Western blot analysis.
To detect EBNA2, protein lysates were prepared by boiling for 10 min in 2% sodium dodecyl sulfate (SDS), 100 mM NaCl, 0.01 M Tris-HCl, 5% β-mercaptoethanol, 1 mM EDTA, 100 μg/ml phenylmethylsulfonyl fluoride, and 2 μg/ml leupeptin and were then briefly sonicated on ice. The lysates were clarified by centrifugation at 13,000 rpm for 10 min at room temperature. Protein from 5 × 105 cells was separated by 5% to 10% discontinuous SDS-polyacrylamide gel electrophoresis (PAGE) and blotted onto nitrocellulose filters. The filters were probed with the anti-EBNA2 antibody PE2 (Dako), diluted to a ratio of 1:50 in BLOTTO (5% skimmed milk-0.1% Tween-20 in Tris-buffered saline), and incubated overnight at 4°C. Immunocomplexes were detected with alkaline-phosphatase-conjugated anti-mouse immunoglobulin G (Promega) and visualized with BCIP/NBT liquid substrate (Sigma). LMP1 was detected as described previously (12). To detect Bfl-1, cell extracts were prepared in lysis buffer {150 mM NaCl, 10 mM HEPES [pH 7.4], 1% 3-[(3-cholamidopropyl)-diamethylammonio]-1-propanesulfonate; Calbiochem} with protease inhibitors and processed as above. Two hundred micrograms of protein extracts was fractionated on 17% SDS-PAGE gels and transferred to 0.2-micron nitrocellulose, using a semidry blotting apparatus. The filters were probed, using a chicken anti-human Bfl-1 antibody preparation (M. J. Simmons and C. Gélinas, unpublished data) overnight at 4°C, followed by horseradish peroxidase-conjugated rabbit anti-chicken immunoglobulin Y (item no. 31401; Pierce Biotechnology) at a ratio of 1:5,000, followed by enhanced chemiluminescent detection.
Apoptosis assays.
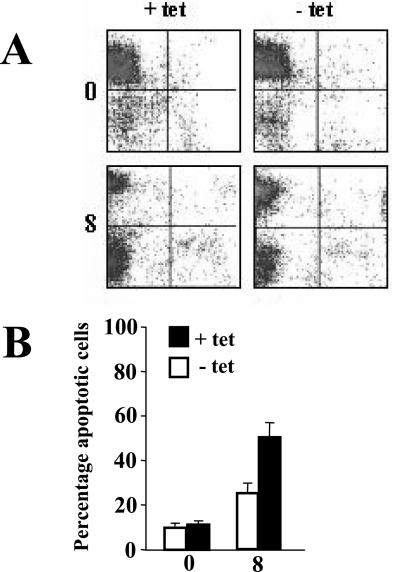
DG75-tTA-EBNA2 cells, both uninduced and induced to express EBNA2, were seeded at a density of 5 × 104 cells per ml in RPMI medium supplemented with either 10% fetal bovine serum (FBS) or 0.5% FBS plus ionomycin (1 μg per ml) to induce apoptosis. The cell culture medium was changed after 4 days, and cell population analyses were performed by flow cytometry (FacsCalibur; Beckton Dickinson) following staining with Syto 16 (Invitrogen) and propidium iodide (Sigma-Aldrich) as described elsewhere (34). Data for 20,000 cells were collected for each analysis made, and two-dimensional plots of Syto 16 versus propidium iodide were generated (Fig. 2A).
FIG. 2.
Comparative analysis of the apoptotic response of DG75-tTA-EBNA2 cells uninduced and induced to express EBNA2. (A) Representative fluorescence-activated cell sorter profiles of Syto 16 fluorescence (y axis) versus propidium iodide (PI) fluorescence (x axis) in DG75-tTA-EBNA2 uninduced (+ tet; left panels) and induced to express EBNA2 (− tet; right panels) for 2 days, followed by 0 and 8 days (as indicated on the left) of partial serum deprivation combined with ionomycin treatment. Viable cells (Syto 16-positive, PI-negative) are represented in each top left quadrant, apoptotic (Syto 16-negative, PI-negative) cells in the bottom left quadrants, and necrotic (Syto 16-negative, PI-positive) cells in the bottom right quadrants. (B) Summary of the average percentage of apoptotic cells in three repeat experiments, including that shown in panel A. The times at which samples were taken are indicated (in days) underneath the graph. Error bars indicate standard deviations.
Plasmids and mutagenesis.
The luciferase reporter constructs −1374/+81-luc, −1240/+81-luc, −367/+81-luc, and −129/+81-luc have been described elsewhere (12). In site-directed mutagenesis experiments with the bfl-1 promoter, the oligonucleotide 5′-CCTTGTAGATTGCTGGTTCTAGAGTGAAATGGTACAACCC-3′was used to introduce sequence changes (base substitutions are underlined) to a consensus CBF1-binding site at −250/−243, using the Altered Sites mutagenesis kit (Promega). The XL Quickchange site-directed mutagenesis kit (Stratagene) was used with the following oligonucleotide pairs to mutate candidate Ets1/PU.1-like binding sites: for −213/−204 (Ets1), 5′-GGATTCTAATTTCTCCACCTGCA GCATTTAAGACTTGCAAAGCTG-3′ and 5′-CAGCTTTGCAAGTCTTAAATGCTGCAGGTGGAGAAATTAGAATCC-3′; for −176/−163 (Ets1), 5′-GCAAAGCTGAATTAATCACAGGCTGCAGAAGTGGCTTCTCTG-3′and 5′-CAGAGAAGCCACTTCTGCAGCCTGTGATTAATTCAGCTTTGC-3′; and for −143/−134 (PU.1), 5′-GGAAGTGGCTTCTCTGAAACATCTGCAGCTTTCACATTTT-3′and 5′AAAATGTGAAAGCTGCAGATGTTTCAGAGAAGCCACTTCC-3′. The 3Enh-luc reporter construct contains three κB elements upstream of a minimal conalbumin promoter linked to the luciferase gene (3). The plasmid pSGEBNA2 (pPDL151) expresses EBNA2 from the B95-8 strain of EBV, and its derivative, pSG5EBNA2ww323sr, expresses an EBNA2 that does not bind to CBF1 (47). pCMV, pCMV-EBNA3A, EBNA3B, EBNA3C, and pSGLMP1 have been described elsewhere (26, 45). Other vectors used were pGa981-6 (53), pEFBOSneo-RBP(R218H) (R218H) (33) (57), pcDNA3-RPMS-1/FLAG (60), pCEP4Ets-(DN) (40), pSG5-HA-mNotch1IC(1751-2294), and pSG5-HA-mNotch1IC-E2TANLS (23).
RESULTS
EBNA2 trans-activates bfl-1 in BL-derived cell lines by a mechanism that is dependent on CBF1.
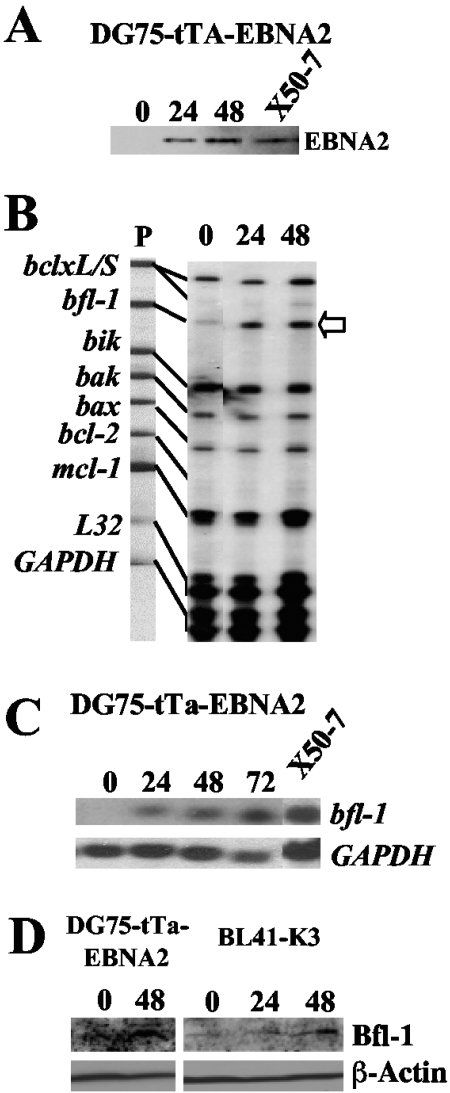
We investigated a role for EBNA2 in regulating the expression of bcl-2 family members by multiprobe RPA, using an established tightly regulatable tetracycline-based (tet-off) system to express EBNA2 in the EBV-negative BL cell line DG75 (DG75-tTA-EBNA2) (14). In this experiment, the level of induced EBNA2 at 48 h approximated that seen in the reference control LCL (X50-7) (Fig. 1A). It can be seen that the steady-state level of bfl-1 mRNA increased significantly in response to EBNA2 induction and was clearly detectable at 24 h (Fig. 1B). Densitometric scanning indicated that the degree of induction of bfl-1 mRNA at 24 and 48 h was about fivefold (relative to the two internal-control mRNAs). Furthermore, bfl-1 was the only gene from the panel (which also included bclxL, bclxS, bik, bak, bax, bcl-2, and mcl-1) to show significant modulation of expression in response to EBNA2.
FIG. 1.
Expression of EBNA2 leads to increased bfl-1 mRNA and protein levels in BL-derived cell lines. (A) Western blot of DG75-tTA-EBNA2 cells induced to express EBNA2 by reculturing cells in the absence of tetracycline. Cells were harvested and analyzed for EBNA2 expression at various time points (indicated in hours above each lane); also included is the reference LCL X50-7. (B) RPA autoradiogram in which mRNA levels from the apoptosis-related genes bcl-x L/S, bfl-1, bik, bak, bax, bcl-2, and mcl-1 from the same experiment as that in panel A were analyzed. Unprotected 32P-labeled antisense riboprobes (5,000 cpm, lane P) were loaded alongside RPA-processed samples and are shown linked to their smaller RNase-protected fragments, which correspond to the steady-state levels of the corresponding mRNA in the samples. An increase in the steady-state level of bfl-1 mRNA (open arrow) is seen upon induction of EBNA2 expression. (C) Northern blot analysis of bfl-1 mRNA levels from a repeat EBNA2 induction experiment using DG75-EBNA2-tTA. Thirty-microgram samples of total RNA were loaded onto the gel, which was then blotted and probed with antisense bfl-1 riboprobe. The lower panel shows the same blot stripped and reprobed with a GAPDH antisense riboprobe. (D) Western blots showing upregulation of Bfl-1 expression in DG75-tTA-EBNA2 cells in response to EBNA2 induction following tetracycline withdrawal and in BL41-K3 cells in which ER-EBNA2 function was activated by addition of β-estradiol. The times postinduction/activation of EBNA2 (expressed in hours) are given above each lane. The lower panel shows the same blots probed with antibody to β-actin.
Northern blotting was carried out in order to confirm the RPA result and to determine the size of the bfl-1 transcript induced by EBNA2 (Fig. 1C). In this experiment, only one band was detectable at 0.8 to 0.85 kilobases, in agreement with the previously reported size of the bfl-1 transcript (38). The level of bfl-1 mRNA did not attain that seen in X50-7 cells, which showed levels typical of LCLs and BL cell lines expressing the EBV growth program (11). In repeat experiments, the levels of induced bfl-1 mRNA seen at 48 h postinduction of EBNA2 were maintained for at least 96 h and did not increase further (not shown). Similar results were obtained by Northern blotting/RPA using RNA prepared from the established BL-derived cell lines BL41-K3 and BL41/P3HR1-9A (35, 37), which express an estrogen receptor-EBNA2 fusion protein (ER-EBNA2) whose function is dependent on β-estradiol. Addition of β-estradiol led to the induction of bfl-1 mRNA in both cell lines, rising to a maximum induction of 5.2- and 5.8-fold after 24 h for BL41-K3 and BL/P3HR1-9A, respectively (data not shown). In the latter case, activation of ER-EBNA2 did not lead to any detectable expression of LMP1 from the endogenous EBV P3HR-1 genome. An increase in the level of Bfl-1 protein was observed in both the DG75-tTA-EBNA2 and BL41-K3 cell lines at 48 h postinduction and activation of EBNA2, respectively (Fig. 1D).
We also investigated whether EBNA2 could activate bfl-1 in DG75 cells in which the CBF1 gene had been inactivated by somatic knockout (51). bfl-1 mRNA levels were therefore examined by reverse transcription real-time PCR in DG75 clones that had been stably transfected with ER-EBNA2 and in which the CBF1 gene had (SM296 D3) or had not (SM295 D6) been inactivated. Addition of β-estradiol to SM295 cells led to a significant transient increase in the level of bfl-1 mRNA (peaking at 8.7-fold after 6 h) (Table 1), an effect that was not seen upon knockout of the CBF1 gene. The induction of bfl-1 mRNA levels in SM295 was transient, in contrast to that for both DG75-tTA-EBNA2 and BL41-K3, where elevated bfl-1 mRNA levels persisted for at least 48 h postinduction of EBNA2 (Fig. 1) (also by real-time PCR; not shown). This discrepancy might be due to a difference in the expression levels or properties of wild-type EBNA2 (DG75-tTA-EBNA2) versus ER-EBNA2 (SM295) in the DG75 background, clonal variations that arose during the generation of these stably transfected derivatives of DG75, or factors particular to the different induction systems in the DG75 cell context. We did not observe any significant change in the stability of bfl-1 mRNA in DG75-tTA-EBNA2 cells in response to EBNA2 induction (not shown). Together, these experiments demonstrate that EBNA2 upregulates bfl-1 and that CBF1 is required for this effect.
TABLE 1.
Upregulation of bfl-1 by EBNA2 requires CBF1
| Samplea | Time after addition of estrogen (h) | Transcript | Average CTb | Fold induction (2ΔΔCT)c |
|---|---|---|---|---|
| SM295 D6 | 0 | bfl-1/GAPDH | 23.91/16.12 | 1.00 ± 0.12 |
| SM295 D6 | 6 | bfl-1/GAPDH | 21.10/16.44 | 8.76 ± 0.35 |
| SM295 D6 | 12 | bfl-1/GAPDH | 22.92/16.90 | 3.42 ± 0.25 |
| SM295 D6 | 48 | bfl-1/GAPDH | 23.40/16.66 | 2.07 ± 0.14 |
| SM296 D3 | 0 | bfl-1/GAPDH | 24.28/16.19 | 0.81 ± 0.02 |
| SM296 D3 | 6 | bfl-1/GAPDH | 23.43/16.30 | 1.58 ± 0.15 |
| SM296 D3 | 12 | bfl-1/GAPDH | 23.72/16.36 | 1.34 ± 0.42 |
| SM296 D3 | 48 | bfl-1/GAPDH | 23.97/16.96 | 1.71 ± 0.24 |
RNA was prepared from cells harvested at various times after the addition of estrogen to the culture medium. The cells were reverse transcribed, and cDNAs were amplified by real-time PCR as described in Materials and Methods.
Average CT, number of PCR cycles at the threshold.
Relative (n-fold) induction was calculated by using the comparative CT method (50) for relative quantitation; relative transcript induction was normalized to that of GAPDH and then expressed as a ratio to that of the SM295 D6 0-h sample.
Assays for apoptosis on DG75 cells induced to express EBNA2.
In order to investigate if EBNA2-associated Bfl-1 upregulation coincided with an increase in resistance to apoptosis, DG75-tTA-EBNA2 cells, both uninduced and induced to express EBNA2, were cultured under reduced serum conditions in combination with ionomycintreatment to trigger apoptosis. At 8 days postinduction, cultures were dually stained with Syto 16 to distinguish viable cells from dead cells and with propidium iodide to distinguish necrotic cell death from apoptosis (34). At 8 days postinduction of EBNA2, there was consistently less apoptotic death (∼24%) in cultures expressing EBNA2 relative to their uninduced counterparts (∼49%) (Fig. 2). Activation of EBNA2 function leads to cytostasis in BL41-K3 cells (37), but growth inhibition does not occur in DG75-tTA-EBNA2 cells in response to EBNA2 induction (14).
EBNA2, but not Notch-IC, trans-activates the bfl-1 promoter in EBV-negative BL-derived cell lines.
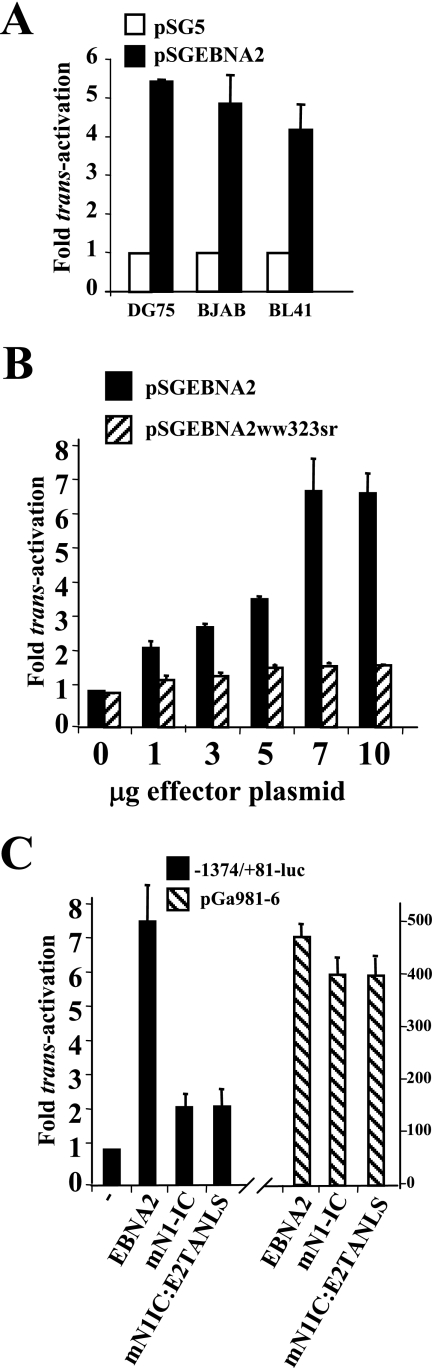
Transient transfections of the EBV-negative BL-derived cell lines DG75, BJAB, and BL41 showed that cotransfection of the bfl-1 promoter reporter construct −1374/+81-luc (12) with 5 μg of EBNA2 expression vector (pSGEBNA2) led to increases of 5.5-, 5.0-, and 4.8-fold, respectively, in luciferase activity at 48 h posttransfection relative to values obtained when pSG5 was used as the cotransfected plasmid (Fig. 3A). The effect of EBNA2 was mediated by the bfl-1 promoter sequence, since EBNA2 expression did not affect basal luciferase levels produced by the corresponding promoterless vector (pGL2basic) (data not shown). In DG75, the use of increasing quantities of EBNA2 plasmid led to a dose-dependent increase in promoter activity up to a maximum of 7.5-fold when 7 μg was used (Fig. 3B). It can also be seen that replacement of pSGEBNA2 with pSGEBNA2ww323sr, which expresses an EBNA2 molecule with amino acid substitutions in residues from conserved region 6 that are critical for binding CBF1 (47), greatly impaired the ability of the protein to trans-activate the bfl-1 promoter (reduced from 7.5- to 1.8-fold) (Fig. 3B). These results indicate that EBNA2 directly trans-activates the bfl-1 promoter in EBV-negative BL-derived cell lines and that the ability of EBNA2 to bind CBF1 is critical for this effect. In contrast, substitution of the EBNA2 expression plasmid with vectors expressing constitutively active Notch-IC fragments did not reveal significant trans-activation of the bfl-1 promoter from the same reporter construct. These included mouse Notch1-IC (mN1-IC), a mouse Notch1-IC chimera in which the trans-activation domain had been substituted with that of EBNA2 (mN1IC-E2TANLS) (23) (Fig. 3C), and human Notch1-IC and Notch2-IC (data not shown). In the same experiments, these effector molecules were seen to drive luciferase expression from pGa981-6, a reporter construct used as a control for CBF-1-driven promoter activity (which contains a hexamerized 50-bp EBNA2 response element from the LMP2A promoter in front of the minimal β-globin promoter [53]).
FIG. 3.
EBNA2, but not Notch-IC, trans-activates the bfl-1 promoter in BL-derived cell lines. (A) DG75, BJAB, and BL41 cells were cotransfected with 7 μg of either pSG5 or pSGEBNA2 together with 1 μg of the bfl-1 promoter-reporter construct −1374/+81-luc. Cells were harvested at 24 h posttransfection and analyzed for luciferase activities, which were then normalized for transfection efficiency (based on β-galactosidase activity measured from acotransfected pCMVlacZ reporter which was included in all transfections). Luciferase values obtained from cotransfections with pSG5 were arbitrarily assigned a value of 1.0, and activation represents the relative normalized luciferase activities obtained upon cotransfection with pSGEBNA2. (B) Dose-dependent trans-activation of the bfl-1 promoter by EBNA2 but not EBNA2ww323sr, a mutant of EBNA2 that cannot bind CBF1. DG75 cells were cotransfected with increasing amounts of either pSGEBNA2 or pSGEBNA2ww323sr (effector plasmids; quantities are indicated underneath) and 1 μg of −1374/+81-luc. Cells were harvested at 24 h posttransfection and analyzed for luciferase activities, which were normalized and presented as described for panel A. (C) DG75 cells were transfected with 1 μg of either −1374/+81-luc or pGa981-6 and 5 μg of plasmid expressing the effector molecules indicated underneath each bar. The chimeric protein mNotch1IC:E2TANLS binds CBF1 via mNotch1IC and contains the additional trans-activation domain and nuclear localization sequence of EBNA2. Higher quantities of any of these effector plasmids (up to 12 μg) did not lead to increased bfl-1 promoter trans-activation (not shown). Cells were harvested at 24 h posttransfection and analyzed for luciferase activities, which were normalized and presented as described for panel A.
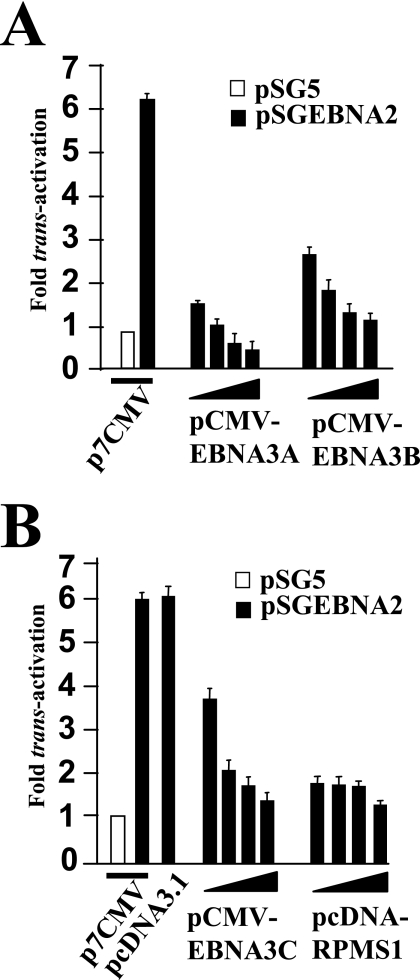
EBNA2-associated trans-activation of the bfl-1 promoter is inhibited by EBNA3A, EBNA3B, EBNA3C, and the EBV gene product RPMS1.
In order to investigate if other CBF1-contacting proteins could repress EBNA2-mediated transcription from the bfl-1 promoter, vectors expressing EBNA3A, EBNA3B, EBNA3C, and the product of the RPMS1 gene were included in EBNA2-bfl-1 promoter trans-activation assays in DG75 cells. Cotransfection with increasing quantities of each individual expression vector led to dose-dependent decreases in bfl-1-driven luciferase values (Fig. 4). In the case of EBNA3A, cotransfection with more than 3 μg of expression vector led to a decrease to below basal levels (absence of EBNA2 expression vector), whereas near-basal levels of promoter activity were restored with 7 μg of either the EBNA3B or EBNA3C expression vector (Fig. 4A and B). Cotransfection with as little as 1 μg of RPMS1 expression vector (pcDNA3-RPMS1/FLAG) led to the almost complete loss of EBNA2-associated bfl-1 promoter activation (Fig. 4B). The level of EBNA2 expressed from pSGEBNA2 is not affected by cotransfection with these effector plasmids (45, 68; data not shown).
FIG. 4.
EBNA2-mediated trans-activation of the bfl-1 promoter is inhibited by EBNA3A, EBNA3B, EBNA3C, and the EBV latent gene product RPMS1. DG75 cells were cotransfected with 1 μg of −1374/+81-luc together with increasing amounts (in all cases 1, 3, 5, and 7 μg consecutively) of the expression vectors pCMV-EBNA3A, pCMV-EBNA3B, pCMV-EBNA3C, or pcDNA3-RPMS1/FLAG. Cells were harvested at 24 h posttransfection, and normalized luciferase values were expressed as activation levels (n-fold) relative to those of the corresponding control transfected cells in which the effector plasmids were pSGEBNA2 together with either p7CMV (5 μg) or pcDNA3.1 (5 μg).
bfl-1 promoter activation by EBNA2 is inhibited by coexpression of a dominant-negative mutant form of CBF1 and does not involve activation of NF-κB.
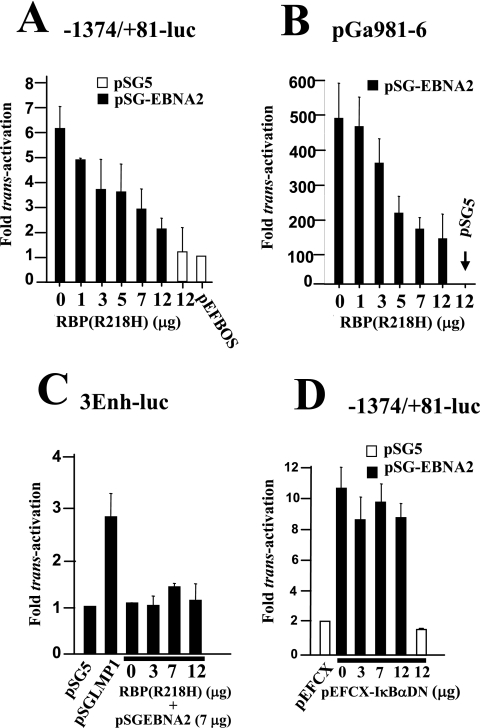
A direct role for CBF1 in mediating EBNA2 responsiveness on the bfl-1 promoter was investigated, using a non-DNA-binding mutant of CBF1 [RBP(R218H)]. This protein has been shown to act as a dominant-negative suppressor of activation of the EBV LMP2A and cellular HES-1 promoters by Notch-IC and is likely to exert such an effect by competing with Notch-IC (or EBNA2) or unknown cofactors for binding to CBF1 (32, 33). Cotransfections were performed using DG75 with −1374/+81-luc and various amounts of RBP(R218H) expression vector [pEFBOSneo-RPB(R218H)]. It can be seen from this experiment that expression of RBP(R218H) efficiently inhibited EBNA2-associated bfl-1 promoter trans-activation in DG75 cells in a dose-dependent manner to a maximum of 70% when 12 μg of pEFBOSneo-RPB(R218H) was used (Fig. 5A). Coexpression of RBP(R218H) also inhibited EBNA2-driven luciferase expression from pGa981-6 to a degree similar to that seen with the bfl-1 promoter (Fig. 5B).
FIG. 5.
trans-activation of the bfl-1 promoter by EBNA2 is inhibited by coexpression of a dominant-negative mutant form of CBF1 and does not involve activation of NF-κB. The names of the reporter constructs are given above each figure. (A and B) Inhibition of EBNA2-mediated trans-activation of the bfl-1 promoter by a non-DNA-binding mutant of CBF1 [RBP(R218H)]. DG75 cells were cotransfected with 1 μg of −1374/+81-luc (A) or the reporter construct pGa981-6, in which transcription of the luciferase gene is regulated by CBF1 (B), and 7 μg of pSGEBNA2 together with increasing amounts of pEF-BOSneo-RBP(R218H) [indicated as RBP(R218H) underneath each bar]. (C) Overexpression of either EBNA2 or RBP(R218H) does not lead to increased NF-κB-dependent transcriptional activation in DG75 cells. Here, NF-κB activation was monitored by using the established NF-κB-dependent reporter construct, 3Enh-luc. (D) Coexpression of an IκBα mutant that inhibits activation of NF-κB did not significantly affect trans-activation of the bfl-1 promoter by EBNA2. In all cases (A to D), cells were harvested at 24 h posttransfection, and normalized luciferase values were expressed as activation levels (n-fold) relative to those of the corresponding control transfected cells with the relevant empty vectors as appropriate in each case.
bfl-1 is an established NF-κB-responsive gene, and there is an increasing body of evidence to show cell context-specific, antagonistic, or synergistic connections between the NF-κB and CBF1-mediated Notch signal transduction pathways (4, 19, 63). Furthermore, it has recently been shown that basal expression of IκBα, and as a consequence NF-κB activity, is under CBF1 control (55). We therefore used a known NF-κB-dependent reporter construct (3Enh-luc) in assays to monitor the relative levels of activated NF-κB in DG75 when either EBNA2 or RBP(R218H) was expressed (Fig. 5C). It can be seen that a 3.5-fold activation of NF-κB occurred upon cotransfection with the LMP1 expression vector but that the basal level of activated NF-κB did not change significantly in response to cotransfection with expression vectors encoding EBNA2 and/or RBP(R218H). Further cotransfections were carried out using a vector that expressed a “super-repressor” mutant form of IκBα (pEFCX-IκBαDN), in which serine residues at positions 32 and 36 of that protein have been replaced with alanines (46). The resulting IκBα mutant can no longer be phosphorylated and subsequently proteolyzed, thus effectively retaining NF-κB in the cytoplasm and blocking its function as a regulator of transcription. Coexpression of IκBαDN only resulted in a marginal decrease (<10%) in EBNA2-associated trans-activation of the bfl-1 promoter, even when up to 12 mg of expression vector was used (Fig. 4D), in contrast to its ability to inhibit LMP1-associated trans-activation of the same promoter (12). We also observed that trans-activation of the bfl-1 promoter by EBNA2 occurred just as efficiently when the −52/−43 enhancer element that mediates trans-activation by LMP1/CD40/p65 was mutated (not shown). These results are further evidence of a role for CBF1 in the activation of the bfl-1 promoter by EBNA2 and show that this process does not involve activation of the transcription factor NF-κB.
Identification of a CBF1-like binding site on the bfl-1 promoter that mediates trans-activation by EBNA2.
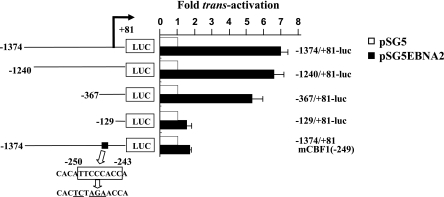
In order to identify DNA sequence elements on the bfl-1 promoter that mediate trans-activation by EBNA2, a series of reporter constructs in which the luciferase gene was driven by bfl-1 promoter fragments of different lengths was used (Fig. 6). It can be seen that in DG75 cells, EBNA2 trans-activated the −1374/+81, −1240/+81, and −367/+81 promoter fragments by 7-, 6.5-, and 5.5-fold, respectively, with a significant decrease to 1.5-fold being observed when only the −129/+81 promoter sequence was retained. This implied that the sequence between −367 and −129 contained DNA sequence element(s) that mediated a high proportion of the observed EBNA2/CBF1-dependent trans-activation. A search of the available bfl-1 upstream transcriptional regulatory region led to the identification of one copy of the sequence motif 5′-GGTGGGAA-3′ at position −243 to −250 on the inverse strand. This element is part of the consensus binding sequence for CBF1 (CGTGGGAA), and site-directed mutagenesis was used to introduce base substitutions into the core of this motif in the reporter plasmid −1374/+81-luc so as to eliminate potential binding by CBF1 (Fig. 6) (48, 62). It can be seen that this mutation led to the nearly complete loss of trans-activation by EBNA2. This result correlated with the observation of a similar reduction in EBNA2-associated trans-activation upon deletion of the −367 to −129 region (Fig. 6). Similar results were obtained with this series of reporter constructs when the BL41 cell line was used (data not shown). These experiments show that a consensus CBF1-binding element at −243/−250 in the bfl-1 transcription regulatory region is essential for mediating trans-activation by EBNA2.
FIG. 6.
Identification of a CBF1-like binding site on the bfl-1 promoter that mediates trans-activation by EBNA2. The figure shows a series of bfl-1-promoter-reporter constructs in which the promoter sequence has been progressively deleted from the 5′end (the coordinate of the 5′ end is given to the left of each construct). They all share a common 3′ terminus at 81 bp downstream from the transcription initiation site (indicated by a bent arrow), at which point they are joined to the luciferase gene (LUC). The relative location (black box) and sequence (open box) of a consensus CBF1-binding site is indicated, and the base changes made to this motif by site-directed mutagenesis to generate −1374/+81-luc(mCBF1) are underlined. DG75 cells were transfected with 7 μg of either pSG5 or pSGEBNA2 together with 2 μg of the individual reporter constructs (the names are given on the right). Cells were harvested at 24 h posttransfection and assayed for luciferase activity as previously described. Each normalized luciferase value was expressed as the activation level (n-fold) relative to the corresponding value obtained for each reporter construct when cotransfected with control pSG5.
Ets family transcription factors are involved in trans-activation of the bfl-1 promoter by EBNA2.
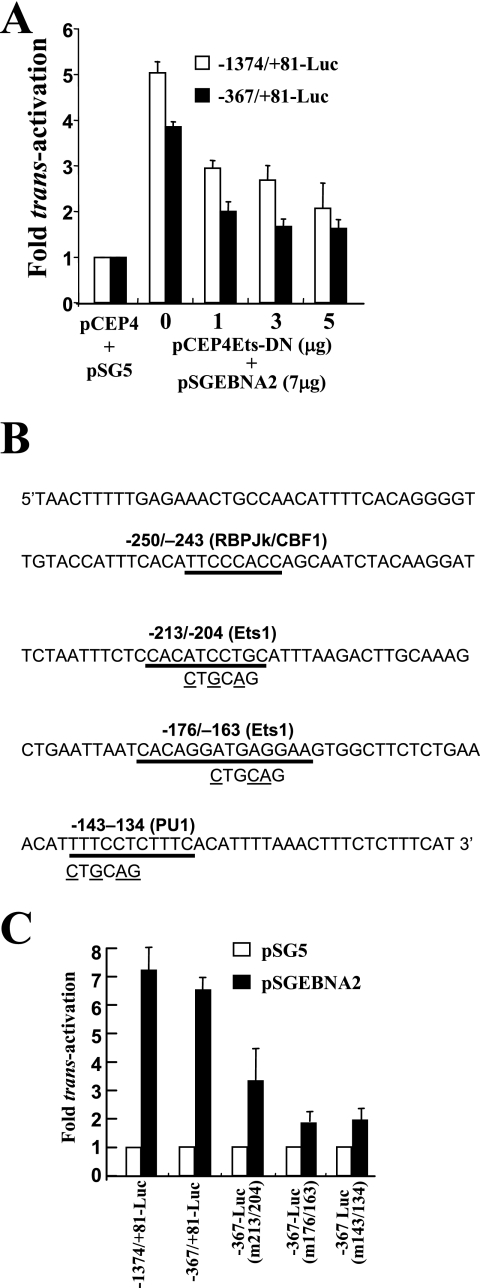
In order to investigate if Ets proteins played a role in EBNA2-mediated trans-activation of the bfl-1 promoter, we performed cotransfections in DG75 cells, which are known to express Spi-1/PU.1 (42), using a vector (pCEP4Ets-DN) which expresses a dominant-negative human Ets-1 mutant (Ets-DN) in which the trans-activation domain has been deleted (41, 54). Cotransfection with increasing amounts of Ets-DN expression vector led to a reduction in trans-activation by EBNA2 of up to 40% when −1374/+81-luc and −367/+81-luc were used as reporter plasmids (Fig. 7A). Potential Ets-binding motifs (58) were identified in the bfl-1 promoter region, using Transcription Element Search software (TESS), and site-directed mutagenesis was then used to introduce base substitutions (Fig. 7B) into their core sequences in the reporter plasmid −367/+81-luc. It can be seen that mutation of the Ets-like binding sites at −213/−202, −176/−163, and −143/−134 led to significantly less trans-activation by EBNA2 (approximately 3.4-, 1.9-, and 1.95-fold, respectively) compared to that for −367-Luc (6.7-fold) (Fig. 7C). These experiments indicate a potentially important role for Ets family members in the activation of the bfl-1 promoter by EBNA2.
FIG. 7.
A role for Ets-family transcription factors in bfl-1 promoter trans-activation by EBNA2. (A) Sequence of the −301 to −111 region of the bfl-1 promoter showing the locations and sequences of candidate binding sites for relevant transcription factors. Three further reporter constructs were made by introducing base changes (underlined) to these sequences in −367/+81-Luc, thus eliminating each motif in turn. (B) DG75 cells were transfected with 7 μg of either pSGEBNA2 or pSG5, together with 2 μg of a reporter construct (indicated underneath). Cells were harvested 24 h posttransfection and assayed for luciferase activity as previously described. Each normalized luciferase value was expressed as an activation level (n-fold) relative to the corresponding value obtained for each reporter construct when cotransfected with control pSG5. (C) DG75 cells were cotransfected with 7 μg of pSGEBNA2 and 2 μg of either −1374/+81-luc or −367/+81-luc plus increasing quantities of pCEP4Ets-DN. Each normalized luciferase value was expressed as an activation level (n-fold) relative to the corresponding value obtained for each reporter construct when cotransfected with control pSG5 and pCEP4 (5 and 7 μg, respectively).
EBNA2 induces and maintains the high levels of bfl-1 transcript seen in the conditional LCL EREB2.5.
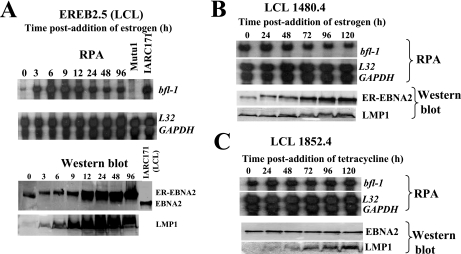
In EBV-immortalized, permanently growing LCLs, EBNA2 is the main activator of EBV latent genes, including LMP1. EREB2.5 is a conditional LCL in which EBNA2 function is dependent on the presence of β-estradiol (36). In this cell line, the proliferative and growth transformation effects of EBV are reversed upon withdrawal of β-estradiol from the medium. We used this system to investigate if there was an association between the EBV growth program and the high levels of bfl-1 mRNA seen in LCLs. It can be seen that the removal of β-estradiol coincided with a dramatic reduction in the steady-state level of bfl-1 mRNA (Fig. 8A, upper panels). Upon reactivation of EBNA2, the bfl-1 transcript level rapidly increased, peaking at 6 h and declining to an elevated steady-state level after 24 h. These results show that bfl-1 expression is induced and maintained at high levels by EBV latent gene products in an LCL context. The decreases in electrophoretic mobility and level of expression of ER-EBNA2 upon β-estradiol withdrawal are known features of this system and are most likely due to changes in its posttranslational modification and the loss of its ability to positively regulate its own promoter (Fig. 8A, lower panels). Induction of bfl-1 occurred prior to the increase in the level of ER-EBNA2, and as expected, inactivation of ER-EBNA2 led to the loss of LMP1 (Fig. 8A, lower panel), a rise in which was detectable as early as 0.5 h after the addition of the hormone (data not shown).
FIG. 8.
bfl-1 is driven by EBV latent proteins in an LCL context, and the loss of both EBNA2 and LMP1 is required before bfl-1 mRNA levels decline significantly. Estrogen-starved EREB2.5 cells (A) and LCL 1480.4 cells (B) were treated with β-estradiol and sampled for RNA and protein analysis at the time points indicated above each panel. bfl-1, L32, and GAPDH mRNA levels were assayed by RPA (upper panels); levels of ER-EBNA2 and LMP1 were determined by Western blotting (lower panels). (C) LCL 1852.4 cells were treated with tetracycline and processed for RNA and protein analysis at the time points indicated.
EBNA2 maintains elevated bfl-1 mRNA levels in an LCL context in the absence of LMP1.
In order to assess the relative contributions of EBNA2 and LMP1 to bfl-1 expression in an LCL context, we used an established LCL in which the expression of both EBV proteins can be uncoupled. In 1480.4 cells (69), LMP1 is constitutively expressed and EBNA2 function is lost in the absence of β-estradiol (Fig. 8B, upper panels). The removal of β-estradiol from this cell line did not lead to a decrease in the level of the bfl-1 transcript, which in fact increased by a factor of two relative to the steady-state level in this cell line (Fig. 8B, lower panels). In 1852.4 cells (39), wild-type EBNA2 is constitutively expressed, whereas LMP1 expression may be regulated by tetracycline (Fig. 8C, upper panels). Here the loss of LMP1 expression led to only a marginal decrease in bfl-1 mRNA levels. These results show that the loss of both LMP1 and EBNA2 is necessary before the level of the bfl-1 transcript declines significantly in an LCL context.
DISCUSSION
In addition to its central role as a regulator of EBV latent genes, the modulation of cellular gene expression by EBNA2 is likely to make a key contribution to B-cell immortalization. The upregulation of bfl-1 is an EBV-host cell interaction that promotes resistance to apoptosis in type 1 BL cells, a fact that lends support to a role for Bfl-1 in promoting the survival of germinal-center B cells. Here we show that expression of EBNA2 alone leads to an increase in the level of Bfl-1 protein and correlates with increased resistance to apoptosis in DG75 cells. The targeting of bfl-1 by EBNA2 may therefore be a mechanism to raise the apoptotic barrier during primary EBV infection of B cells prior to the expression of LMP1.
Several observations pointed to a key role for CBF1 in this process. CBF1 is a repressor that recruits histone deacetylases to the chromatin of target genes (24). Here we observed that the somatic knockout of CBF1 in DG75 cells did not lead to increased endogenous bfl-1 mRNA levels, indicating that this gene is not repressed by CBF1 in the absence of EBNA2. Coexpression of EBNA3A, EBNA3B, EBNA3C, or RPMS1 inhibited EBNA2-associated bfl-1 promoter trans-activation, and in the absence of EBNA2 they all showed some small degree of inhibition of its basal level of activity (data not shown). trans-activation of the bfl-1 promoter by EBNA2 is mechanistically different from that of LMP1, with neither NF-κB activation nor a crucial enhancer motif at −52/−43 that mediates trans-activation by p65 being required (12). Furthermore, EBNA2 failed to mediate trans-activation of a heterologous minimal promoter sequence linked to the LMP-1-dependent enhancer (data not shown). These experiments do not rule out, however, the possibility that NF-κB subunit proteins may play a role in mediating trans-activation of bfl-1 by EBNA2.
Analyses of the bfl-1 5′ transcriptional regulatory region showed that CBF1- and Ets-like binding motifs play key roles in trans-activation by EBNA2. Base substitutions in the GGG triplet of the former almost completely eliminated promoter trans-activation by EBNA2, although we have not yet been able to demonstrate a direct interaction between CBF1 and this motif by band shift assays. An Spi-1 binding site in the LMP1 promoter is essential for trans-activation by EBNA2, and EBNA3C has also been shown to activate LMP1 by a cooperative mechanism involving EBNA2 and Spi-1/Spi-B transcription factors (29, 42, 59, 68). Spi-1 and Spi-B play important roles in the development and differentiation of B lymphocytes by regulating the expression of a variety of genes critical for the functions of hematopoietic-lineage cells (reviewed in reference 56). Unlike EBNA2, the failure of Notch-IC proteins to drive the bfl-1 promoter may be due to a requirement for interactions with B-cell-specific factors, as is the case with the LMP1 promoter (22), or to a weaker interaction with the CBF1-binding motif in its context. We did not observe EBNA2 trans-activation of the bfl-1 promoter in several non-B-cell lines, including Jurkat (T cell), C33A (epithelial), or vascular smooth muscle cells (data not shown). Elsewhere, trans-complementation experiments in which estrogen-starved EREB2.5 cells were “rescued” by transduction with a recombinant lentivirus expressing human Notch1-IC showed that when directly compared to EBNA2, Notch1-IC was a far less efficient inducer of LMP1 and CD23 in that LCL (16). In the present study, the lack of significant bfl-1 promoter trans-activation by mN1IC-E2TANLS indicated that the general failure of Notch-IC proteins in this regard was not due solely to the relative weakness of the Notch-IC trans-activation domain compared to that of EBNA2.
An EBNA2 with a double amino acid substitution that abolishes binding to CBF1 almost completely failed to trans-activate the bfl-1 promoter. It has been shown elsewhere that, when incorporated into the viral genome, a mutation within the EBNA2 CBF1-binding motif resulted in a nonimmortalizing EBV (65). A crucial role for the EBNA2/CBF1 interaction is further underscored by the observation that a cell-permeable EBNA2 peptide that blocks EBNA2-CBF1-binding can prevent EBV immortalization of primary B cells in vitro (13). EBNA2 is a key regulator of LMP1 expression, and EBNA2 activity is largely under viral control and at the level of the CBF1 complex. In EREB2.5, activation of EBNA2 mimics the early events that occur in the process of EBV-driven B-cell growth transformation. Here we show that activation of EBNA2 leads to a rapid increase in bfl-1 mRNA, which attains the level typically seen in established LCLs. Although LMP1 appears to be a more-robust transactivator of bfl-1 in the BL cell line induction systems that we have used (a 20-fold induction level with LMP1 [11] versus a 5-fold induction level with EBNA2) (Fig. 1), the elevated levels of bfl-1 mRNA that remain in 1480.4 and 1852.4 cells upon withdrawal of EBNA2 and LMP1, respectively (Fig. 8B and C) indicate that the expression of this gene is maintained to a similar degree in the absence of either EBV protein. Upon withdrawal of β-estradiol, EREB2.5 cells undergo apoptosis, and this is coincident with a significant decrease in bfl-1 expression. However, the conditional LCLs p1480.4 and 1852.4 both fail to initiate apoptosis in the absence of β-estradiol and tetracycline (due to the continued presence of EBNA2 and LMP1, respectively). Under these circumstances, however, both cell lines undergo growth arrest, indicating that the maintenance of an elevated bfl-1 mRNA level is not sufficient to maintain cell proliferation in the absence of either one of these EBV proteins.
The majority of EBV-associated tumors do not express EBNA2, presumably due to its immunogenicity and that of the EBNA3s whose expression is under EBNA2 control. EBNA2 is expressed in EBV-associated tumors arising in immunocompromised patients such as posttransplant patients, patients with lymphoproliferative disease, and AIDS patients with primary central nervous system lymphomas (25), and its antiapoptotic activity may be a factor in the development of these malignancies. Overexpression of EBNA2 in transgenic mice has been shown to lead to the formation of kidney adenocarcinomas, which clearly demonstrates the oncogenic potential of EBNA2 (61). Bfl-1 has been shown to possess oncogenic cooperative transforming potential (9), and it will be interesting to determine if bfl-1 is a transcriptional target of EBNA2 in that context.
EBV has been thought to counter apoptosis during latency through LMP1-mediated activation of NF-κB-regulated genes (11, 15, 21). EBNA2 has, however, been shown to prevent Nur77-mediated cell death by binding via its conserved region 4 to Nurr77 and preventing the mitochondrial targeting of that protein (43, 44). The identification of bfl-1 as a CBF1-dependent transcriptional target of EBNA2 is additional evidence that this viral protein contributes to cell survival, as well as to the proliferative response that occurs upon infection.
Acknowledgments
We are grateful to M. Rowe for DG75-tTA-EBNA2, W. Hammerschmidt for LCL 1852.4, U. Zimber-Strobl for LCL 1480.4, D. Hayward for pSG5EBNA2ww323sr, T. Honjo for pEFBOSneo-RBP(R218H), P. Farrell for pcDNA3-RPMS-1/FLAG, and H. Sato for pCEP4Ets-(DN).
This work was funded by grants from the Health Research Board (HRB, Ireland) (D.W.) and Cancer Research Ireland (CRI02WAL; D.W. and B.N.D.) and a travel award from Dublin City University Educational Trust (P.M.P.). We also acknowledge support from Deutsche Forschungsgemeinschaft (SFB455 to B.K.), Wilhelm-Sander Stiftung (B.K.), Deutsche Krebshilfe (B.K.), and National Institutes of Health grant CA83937 (C.G.) and partial support from an NIH predoctoral training grant in Biochemistry and Molecular Biology (GM08360 to M.J.S.).
REFERENCES
- 1.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, M. L., N. J. Stam, G. Klein, H. L. Ploegh, and M. G. Masucci. 1991. Aberrant expression of HLA class-I antigens in Burkitt lymphoma cells. Int. J. Cancer 47:544-550. [DOI] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos, F., P. Turpin, M. Rodriguez, D. Thomas, R. T. Hay, J. L. Virelizier, and C. Dargemont. 1997. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J. Cell Sci. 110:369-378. [DOI] [PubMed] [Google Scholar]
- 4.Bash, J., W. X. Zong, S. Banga, A. Rivera, D. W. Ballard, Y. Ron, and C. Gelinas. 1999. Rel/NF-kappaB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors. EMBO J. 18:2803-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75). Int. J. Cancer 19:27-33. [DOI] [PubMed] [Google Scholar]
- 6.Cahir-McFarland, E. D., D. M. Davidson, S. L. Schauer, J. Duong, and E. Kieff. 2000. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 97:6055-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, S. S., I. C. Park, J. W. Yun, Y. C. Sung, S. I. Hong, and H. S. Shin. 1995. A novel Bcl-2 related gene, Bfl-1, is overexpressed in stomach cancer and preferentially expressed in bone marrow. Oncogene 11:1693-1698. [PubMed] [Google Scholar]
- 8.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Sa-Eipper, C., and G. Chinnadurai. 1998. Functional dissection of Bfl-1, a Bcl-2 homolog: anti-apoptosis, oncogene-cooperation and cell proliferation activities. Oncogene 16:3105-3114. [DOI] [PubMed] [Google Scholar]
- 10.D'Sa-Eipper, C., T. Subramanian, and G. Chinnadurai. 1996. bfl-1, a bcl-2 homologue, suppresses p53-induced apoptosis and exhibits potent cooperative transforming activity. Cancer Res. 56:3879-3882. [PubMed] [Google Scholar]
- 11.D'Souza, B., M. Rowe, and D. Walls. 2000. The bfl-1 gene is transcriptionally upregulated by the Epstein-Barr virus LMP1, and its expression promotes the survival of a Burkitt's lymphoma cell line. J. Virol. 74:6652-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Souza, B. N., L. C. Edelstein, P. M. Pegman, S. M. Smith, S. T. Loughran, A. Clarke, A. Mehl, M. Rowe, C. Gelinas, and D. Walls. 2004. Nuclear factor κB-dependent activation of the antiapoptotic bfl-1 gene by the Epstein-Barr virus latent membrane protein 1 and activated CD40 receptor. J. Virol. 78:1800-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrell, C. J., J. M. Lee, E. C. Shin, M. Cebrat, P. A. Cole, and S. D. Hayward. 2004. Inhibition of Epstein-Barr virus-induced growth proliferation by a nuclear antigen EBNA2-TAT peptide. Proc. Natl. Acad. Sci. USA 101:4625-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floettmann, J. E., K. Ward, A. B. Rickinson, and M. Rowe. 1996. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 223:29-40. [DOI] [PubMed] [Google Scholar]
- 15.Fries, K. L., W. E. Miller, and N. Raab-Traub. 1996. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 70:8653-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordadze, A. V., R. Peng, J. Tan, G. Liu, R. Sutton, B. Kempkes, G. W. Bornkamm, and P. D. Ling. 2001. Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus. J. Virol. 75:5899-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory, C. D., C. Dive, S. Henderson, C. A. Smith, G. T. Williams, J. Gordon, and A. B. Rickinson. 1991. Activation of Epstein-Barr virus latent genes protects human B cells from death by apoptosis. Nature 349:612-614. [DOI] [PubMed] [Google Scholar]
- 18.Gregory, C. D., M. Rowe, and A. B. Rickinson. 1990. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt's lymphoma cell line. J. Gen. Virol. 71:1481-1495. [DOI] [PubMed] [Google Scholar]
- 19.Guan, E., J. Wang, J. Laborda, M. Norcross, P. A. Baeuerle, and T. Hoffman. 1996. T cell leukemia-associated human Notch/translocation-associated Notch homologue has I kappa B-like activity and physically interacts with nuclear factor-kappa B proteins in T cells. J. Exp. Med. 183:2025-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayward, S. D. 2004. Viral interactions with the Notch pathway. Semin. Cancer Biol. 14:387-396. [DOI] [PubMed] [Google Scholar]
- 21.Henderson, S., M. Rowe, C. Gregory, D. Croom-Carter, F. Wang, R.Longnecker, E. Kieff, and A. Rickinson. 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65:1107-1115. [DOI] [PubMed] [Google Scholar]
- 22.Hofelmayr, H., L. J. Strobl, C. Stein, G. Laux, G. Marschall, G. W. Bornkamm, and U. Zimber-Strobl. 1999. Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J. Virol. 73:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu, J. L., and S. L. Glaser. 2000. Epstein-Barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit. Rev. Oncol. Hematol. 34:27-53. [DOI] [PubMed] [Google Scholar]
- 26.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-kappa B and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 27.Izumi, K. M. 2004. Epstein-Barr virus signal transduction and B-lymphocyte growth transformation. Prog. Mol. Subcell. Biol. 36:269-288. [DOI] [PubMed] [Google Scholar]
- 28.Izumi, K. M. 2001. Identification of EBV transforming genes by recombinant EBV technology. Semin. Cancer Biol. 11:407-414. [DOI] [PubMed] [Google Scholar]
- 29.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karsan, A., E. Yee, and J. M. Harlan. 1996. Endothelial cell death induced by tumor necrosis factor-alpha is inhibited by the Bcl-2 family member, A1. J. Biol. Chem. 271:27201-27204. [DOI] [PubMed] [Google Scholar]
- 31.Karsan, A., E. Yee, K. Kaushansky, and J. M. Harlan. 1996. Cloning of human Bcl-2 homologue: inflammatory cytokines induce human A1 in cultured endothelial cells. Blood 87:3089-3096. [PubMed] [Google Scholar]
- 32.Kato, H., T. Sakai, K. Tamura, S. Minoguchi, Y. Shirayoshi, Y. Hamada, Y. Tsujimoto, and T. Honjo. 1996. Functional conservation of mouse Notch receptor family members. FEBS Lett. 395:221-224. [DOI] [PubMed] [Google Scholar]
- 33.Kato, H., Y. Taniguchi, H. Kurooka, S. Minoguchi, T. Sakai, S. Nomura-Okazaki, K. Tamura, and T. Honjo. 1997. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124:4133-4141. [DOI] [PubMed] [Google Scholar]
- 34.Kelly, G. L., A. E. Milner, R. J. Tierney, D. S. Croom-Carter, M. Altmann, W. Hammerschmidt, A. I. Bell, and A. B. Rickinson. 2005. Epstein-Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt's lymphoma cells and with increased resistance to apoptosis. J. Virol. 79:10709-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kempkes, B., M. Pawlita, U. Zimber-Strobl, G. Eissner, G. Laux, and G. W. Bornkamm. 1995. Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-J kappa in a conditional fashion. Virology 214:675-679. [DOI] [PubMed] [Google Scholar]
- 36.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kempkes, B., U. Zimber-Strobl, G. Eissner, M. Pawlita, M. Falk, W.Hammerschmidt, and G. W. Bornkamm. 1996. Epstein-Barr virus nuclear antigen 2 (EBNA2)-oestrogen receptor fusion proteins complement the EBNA2-deficient Epstein-Barr virus strain P3HR1 in transformation of primary B cells but suppress growth of human B cell lymphoma lines. J. Gen. Virol. 77:227-237. [DOI] [PubMed] [Google Scholar]
- 38.Kenny, J. J., T. J. Knobloch, M. Augustus, K. C. Carter, C. A. Rosen, and J. C. Lang. 1997. GRS, a novel member of the Bcl-2 gene family, is highly expressed in multiple cancer cell lines and in normal leukocytes. Oncogene 14:997-1001. [DOI] [PubMed] [Google Scholar]
- 39.Kilger, E., A. Kieser, M. Baumann, and W. Hammerschmidt. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 17:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, K. R., T. Yoshizaki, H. Miyamori, K. Hasegawa, T. Horikawa, M. Furukawa, S. Harada, M. Seiki, and H. Sato. 2000. Transformation of Madin-Darby canine kidney (MDCK) epithelial cells by Epstein-Barr virus latent membrane protein 1 (LMP1) induces expression of Ets1 and invasive growth. Oncogene 19:1764-1771. [DOI] [PubMed] [Google Scholar]
- 41.Kita, D., T. Takino, M. Nakada, T. Takahashi, J. Yamashita, and H. Sato. 2001. Expression of dominant-negative form of Ets-1 suppresses fibronectin-stimulated cell adhesion and migration through down-regulation of integrin alpha5 expression in U251 glioma cell line. Cancer Res. 61:7985-7991. [PubMed] [Google Scholar]
- 42.Laux, G., B. Adam, L. J. Strobl, and F. Moreau-Gachelin. 1994. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 13:5624-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, J. M., K. H. Lee, C. J. Farrell, P. D. Ling, B. Kempkes, J. H. Park, and S. D. Hayward. 2004. EBNA2 is required for protection of latently Epstein-Barr virus-infected B cells against specific apoptotic stimuli. J. Virol. 78:12694-12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, J. M., K. H. Lee, M. Weidner, B. A. Osborne, and S. D. Hayward. 2002. Epstein-Barr virus EBNA2 blocks Nur77-mediated apoptosis. Proc. Natl. Acad. Sci. USA 99:11878-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Roux, A., B. Kerdiles, D. Walls, J. F. Dedieu, and M. Perricaudet. 1994. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology 205:596-602. [DOI] [PubMed] [Google Scholar]
- 46.Liljeholm, S., K. Hughes, T. Grundstrom, and P. Brodin. 1998. NF-kappaB only partially mediates Epstein-Barr virus latent membrane protein 1 activation of B cells. J. Gen. Virol. 79:2117-2125. [DOI] [PubMed] [Google Scholar]
- 47.Ling, P. D., and S. D. Hayward. 1995. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJk. J. Virol. 69:1944-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling, P. D., J. J. Hsieh, I. K. Ruf, D. R. Rawlins, and S. D. Hayward. 1994. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J. Virol. 68:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ling, P. D., D. R. Rawlins, and S. D. Hayward. 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90:9237-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−7ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 51.Maier, S., M. Santak, A. Mantik, K. Grabusic, E. Kremmer, W. Hammerschmidt, and B. Kempkes. 2005. A somatic knockout of CBF1 in a human B-cell line reveals that induction of CD21 and CCR7 by EBNA-2 is strictly CBF1 dependent and that downregulation of immunoglobulin M is partially CBF1 independent. J. Virol. 79:8784-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menezes, J., W. Leibold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276-284. [PubMed] [Google Scholar]
- 53.Minoguchi, S., Y. Taniguchi, H. Kato, T. Okazaki, L. J. Strobl, U. Zimber-Strobl, G. W. Bornkamm, and T. Honjo. 1997. RBP-L, a transcription factor related to RBP-Jκ. Mol. Cell. Biol. 17:2679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakada, M., J. Yamashita, Y. Okada, and H. Sato. 1999. Ets-1 positively regulates expression of urokinase-type plasminogen activator (uPA) and invasiveness of astrocytic tumors. J. Neuropathol. Exp. Neurol. 58:329-334. [DOI] [PubMed] [Google Scholar]
- 55.Oakley, F., J. Mann, R. G. Ruddell, J. Pickford, G. Weinmaster, and D. A. Mann. 2003. Basal expression of IkappaBalpha is controlled by the mammalian transcriptional repressor RBP-J (CBF1) and its activator Notch1. J. Biol. Chem. 278:24359-24370. [DOI] [PubMed] [Google Scholar]
- 56.Oikawa, T., and T. Yamada. 2003. Molecular biology of the Ets family of transcription factors. Gene 303:11-34. [DOI] [PubMed] [Google Scholar]
- 57.Sakai, T., and T. Honjo. 1997. [Transcriptional activity of EBNA2 through RBP-J.]. Nippon Rinsho. 55:293-298. [PubMed] [Google Scholar]
- 58.Sementchenko, V. I., and D. K. Watson. 2000. Ets target genes: past, present and future. Oncogene 19:6533-6548. [DOI] [PubMed] [Google Scholar]
- 59.Sjoblom, A., A. Nerstedt, A. Jansson, and L. Rymo. 1995. Domains of the Epstein-Barr virus nuclear antigen 2 (EBNA2) involved in the transactivation of the latent membrane protein 1 and the EBNA Cp promoters. J. Gen. Virol. 76:2669-2678. [DOI] [PubMed] [Google Scholar]
- 60.Smith, P. R., O. de Jesus, D. Turner, M. Hollyoake, C. E. Karstegl, B. E. Griffin, L. Karran, Y. Wang, S. D. Hayward, and P. J. Farrell. 2000. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J. Virol. 74:3082-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tornell, J., S. Farzad, A. Espander-Jansson, G. Matejka, O. Isaksson, and L. Rymo. 1996. Expression of Epstein-Barr nuclear antigen 2 in kidney tubule cells induce tumors in transgenic mice. Oncogene 12:1521-1528. [PubMed] [Google Scholar]
- 62.Tun, T., Y. Hamaguchi, N. Matsunami, T. Furukawa, T. Honjo, and M. Kawaichi. 1994. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 22:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, J., L. Shelly, L. Miele, R. Boykins, M. A. Norcross, and E. Guan. 2001. Human Notch-1 inhibits NF-kappa B activity in the nucleus through a direct interaction involving a novel domain. J. Immunol. 167:289-295. [DOI] [PubMed] [Google Scholar]
- 64.Werner, A. B., E. de Vries, S. W. Tait, I. Bontjer, and J. Borst. 2002. Bcl-2 family member Bfl-1/A1 sequesters truncated bid to inhibit its collaboration with pro-apoptotic Bak or Bax. J. Biol. Chem. 277:22781-22788. [DOI] [PubMed] [Google Scholar]
- 65.Yalamanchili, R., X. Tong, S. Grossman, E. Johannsen, G. Mosialos, and E. Kieff. 1994. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology 204:634-641. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, H., S. W. Cowan-Jacob, M. Simonen, W. Greenhalf, J. Heim, and B. Meyhack. 2000. Structural basis of BFL-1 for its interaction with BAX and its anti-apoptotic action in mammalian and yeast cells. J. Biol. Chem. 275:11092-11099. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, J., H. Chen, G. Weinmaster, and S. D. Hayward. 2001. Epstein-Barr virus BamHi-A rightward transcript-encoded RPMS protein interacts with the CBF1-associated corepressor CIR to negatively regulate the activity ofEBNA2 and NotchIC. J. Virol. 75:2946-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimber-Strobl, U., B. Kempkes, G. Marschall, R. Zeidler, C. Van Kooten, J. Banchereau, G. W. Bornkamm, and W. Hammerschmidt. 1996. Epstein-Barr virus latent membrane protein (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 15:7070-7078. [PMC free article] [PubMed] [Google Scholar]
- 70.Zimber-Strobl, U., E. Kremmer, F. Grasser, G. Marschall, G. Laux, and G. W. Bornkamm. 1993. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 12:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zimber-Strobl, U., and L. J. Strobl. 2001. EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes. Semin. Cancer Biol. 11:423-434. [DOI] [PubMed] [Google Scholar]