Abstract
Vesicular stomatitis virus (VSV) is being developed for cancer therapy. We created a recombinant replicating VSV (rrVSV) that preferentially infected Her2/neu-expressing breast cancer cells. This rrVSV did not express the native VSV-G glycoprotein (gp). Instead, it expressed a chimeric Sindbis gp which included a single-chain antibody (SCA) directed to the human Her2/neu receptor. The virus infected mouse mammary carcinoma cells (D2F2/E2) expressing Her2/neu 23-fold better than the parent cells (D2F2). However, viral growth in cultured D2F2/E2 cells was curtailed after several cycles, and viral yield was very poor at 2 × 104 infectious doses (ID)/ml. We performed in vitro serial passage in D2F2/E2 cells to evolve a virus with improved growth that could be used for preclinical therapy trials in mice. Fifteen passes generated an adapted virus that progressed through multiple cycles in cultured D2F2/E2 cells until all cells were infected and had a viral yield of 1 × 108 ID/ml. Sequencing of the entire viral genomes found only 2 mutations in the adapted virus. Both mutations occurred in the gp gene segment coding for the SCA. An additional N-glycosylation site was created by one of the mutations. The adapted virus showed higher density of gp on the viral envelope, improved infectivity, much greater stability, higher burst size, and decreased induction of cellular interferon. The specificity for cells expressing the Her2/neu receptor was unchanged. These studies demonstrate that serial passage can be used to rapidly evolve a VSV genome encoding an improved chimeric glycoprotein.
Viruses and viral vectors that kill specific cells are being designed for cancer therapy (31, 34, 41, 46). Important issues are safety, efficacy, and targeting. Vesicular stomatitis virus (VSV) is an excellent candidate for development as an oncolytic virus because it is an efficient cell killer that grows and spreads rapidly and yet is safe for human use (7). We have previously created a recombinant replicating VSV (rrVSV) with an altered surface glycoprotein (gp) that targeted preferentially to breast cancer cells. The key change was replacing the native G gp gene in VSV with a modified gp gene from Sindbis virus (SV). The receptor for G is ubiquitous, and VSV promiscuously infects most cell types. Our attempts to modify the binding characteristics of G to render G more cell type specific were unsuccessful because the site of the binding domain and its relationship to the fusion domain within the G protein are unknown. On the other hand, a binding-defective mutant of another RNA virus, Sindbis virus, had already been identified (10). The surface gp of Sindbis consists of an E1 fusion protein and an E2 binding protein. Deletion of amino acids 72 and 73 within E2 reduces binding and infectivity of the virus >90% (10). Others had shown that targeted Sindbis virus and retroviral vectors could be created by placing an Fc-binding domain of protein A within this site and adding exogenous antibody (24, 29). We placed a gene segment coding for a single-chain antibody (SCA) in this site within the E2 protein (3). The SCA recognizes the Her2/neu receptor, erbb2, that is overexpressed on many breast cancer cells and expressed little or not at all on normal cells. To facilitate our future studies, the viral genome was also modified by the inclusion of genes expressing mouse granulocyte-macrophage colony-stimulating factor (GM-CSF) and green fluorescent protein. The genome of this virus is illustrated in Fig. 1. We showed that rrVSV created from this vector selectively infected, replicated, and killed cells expressing erbb2, including D2F2/E2 cells, a mouse mammary cancer cell line stably transfected to express erbb2 (3). We then wanted to test the therapeutic efficacy in an animal tumor model of implanted D2F2/E2 cells but found that the viral titer on this cell line was only 6 × 105/ml and that virus growth was curtailed in D2F2/E2 cells. In low multiplicities of infection (MOI), the yield of progeny virus was very low. It was essential to develop an rrVSV with improved infection and growth in D2F2/E2 cells that could be used in preclinical efficacy studies in mice.
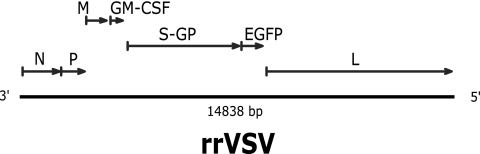
FIG. 1.
rrVSV genome drawn to scale. The VSV-G gene has been deleted and replaced with the hybrid SV gp gene containing the SCA, labeled S-GP. The genes for mouse GM-CSF and EGFP have also been added to the genome as annotated.
Our goal in the present study was to use natural selection to create an improved rrVSV against D2F2/E2 mouse mammary cancer cells. VSV has a high mutation rate, averaging about 1 × 10−4 to 4 × 10−4 per base site and, like most RNA virus populations, exists as a mixture of genetic and phenotypic variants called quasi-species (8, 38). Previous work has shown that genetic diversity within a virus quasi-species population facilitates adaptation to specific cell environments (26). Competition between two VSV populations passed together serially on BHK-21 cells showed slow continuous improvement in fitness of both populations until a vastly superior mutant from one of the populations overgrew and displaced the other virus (5). We demonstrate in this study that serial passage of rrVSV for 15 cycles in D2F2/E2 cells evolved a virus better adapted to productively infect this cell line. We identify the genotypic and phenotypic changes in the chimeric Sindbis glycoprotein that were associated with this adaptation. In addition, we show that endogenous VSV genes and inserted foreign genes did not accumulate any mutations during this process.
MATERIALS AND METHODS
Cells, antibodies, and chemicals.
The following cell lines were obtained from American Type Culture Collection (ATCC) (Rockville, MD) and grown using standard tissue culture techniques in a humidified incubator at 37°C with 5% CO2: SKBR3 human breast adenocarcinoma, COS-7 simian kidney, and BHK-21 hamster kidney. SKBR3 cells are known HER2/neu amplified/overexpressing breast cancer cells (2). D2F2/E2 cells, a mouse mammary tumor line that has been stably transfected with a vector expressing the human Her2/neu gene, and its parent cell line, D2F2, were a generous gift from Wei-Zen Wei, Karmanos Cancer Institute, Wayne State University, Detroit, MI. ldlD cells were a generous gift from Rebecca P. Hughey, University of Pittsburgh School of Medicine. These cells are CHO mutants which are UDP-galactose/UDP-N-acetylgalactosamine 4-epimerase-deficient and express a reversible defect in protein O-glycosylation. Absence of mycoplasma contamination in all cell lines was confirmed by the Gen-Probe rapid detection system (Gen-Probe, Inc., San Diego, CA). VR1248, a mouse polyclonal antibody directed to the Sindbis virus, was obtained from the ATCC. Sheep antiserum to mouse L-cell interferon, cat. no. G-024-501-568, was generously supplied by the NIAID Reference Reagent Repository. The material was prepared by Kurt Paucker, Barbara Dalton, and Clifton A. Ogburn of the Medical College of Pennsylvania, Philadelphia, PA. After reconstitution in 1 ml of sterile 0.9% NaCl, the interferon (IFN) neutralizing titer was 300,000 per ml. Bafilomycin A1 was obtained from Kamiya Biomedical Co. (Seattle, WA).
Vectors, VSV pseudotypes, and recombinant VSV genomes.
Vectors expressing the VSV genome (XN2) and the individual VSV genes P, L, N, and G (pBS-P, L, N, and G, respectively) on a T7 promoter were a very generous gift of John K. Rose, Yale University School of Medicine. A vector expressing VSV-G on a cytomegalovirus (CMV) promoter (CMV-VSV-G) was a generous gift of Paul D. Robbins, University of Pittsburgh School of Medicine. Vectors expressing Sindbis gp (Sindbis) and Sindbis gp modified between amino acids 71 and 74 to express two immunoglobulin G (IgG) binding domains (Sindbis-ZZ) were generously supplied by Irvin S. Y. Chen, University of California, Los Angeles Medical School. VSV pseudotypes were created as previously described (3). In brief, we first constructed a recombinant VSV (rVSV) genome in which the G gene was replaced by a gene encoding enhanced green fluorescent protein (EGFP) (EGFP-ΔG/XN2). This construct was used to create a nonreplicating virus, whose genome consisted of rVSV-EGFP-ΔG and whose surface protein was G, as follows (21). EGFP-ΔG/XN2, pBS-P, pBS-L, pBS-N, and pBS-G were transfected into COS-7 cells and infected with vTF-7, a vaccinia virus expressing T7 polymerase (NIH AIDS Research and Reference Reagent Program, Rockville, MD). Virus were harvested from the supernatant 2 days later and amplified on BHK-21 cells that were transfected with CMV-VSV-G using a standard Lipofectamine protocol. Nonreplicating pseudotype VSV were created by transfecting BHK-21 with DNA encoding one of the chimeric Sindbis gp and, 2 days later, infecting the cells with rVSV-EGFP-ΔG at an MOI of 6. The supernatant was harvested 1 day following infection and stored at −70°C. Titers were determined by incubating the virus and the indicator cell line in one well of a six-well tray (catalog no. 3516; Corning, Inc., Corning, NY) for 2 h, washing, and replacing the media. Green cells were counted in an inverted fluorescent microscope (Axiovert 135; Carl Zeiss, Inc., Thornwood, NY) 1 day later.
Similar techniques were used to create a recombinant replicating VSV whose surface gp was Sindbis-SCA-erbb2 with the mutations found in the adapted rrVSV. First, PCR-based overlap extension mutagenesis was used to place the mutations individually and collectively into the gene expressing Sindbis-SCA-erbb2. This gene was then substituted for the G gene in the XN2 vector expressing the entire VSV genome plus the EGFP gene as previously described (3). This recombinant genome as well as pBS-P, pBS-L, pBS-N, and pBS-G were transfected into COS-7 cells and infected with vTF-7. Virus were harvested from the supernatant 2 days later, amplified once on BHK-21 cells transfected with CMV-VSV-G and then grown once on D2F2/E2 cells. The only surface gp on this virus was Sindbis-SCA-erbb2. Titers were determined as detailed above except that, after a 2-h viral incubation, the medium was replaced with medium containing 30 nM bafilomycin A1 to avoid counting newly produced virus.
RT-PCR, qRT-PCR, and virus sequence determination.
Viral RNA was isolated from rrVSV using the QiaAmp viral RNA extraction kit (catalog no. 52904; QIAGEN, Inc., Valencia, CA). cDNA for sequencing was produced using the Superscript first-strand synthesis system (Invitrogen, Carlsbad, CA). Sequencing was performed from ∼1- to 2-kb PCR products generated with Pfu polymerase (Stratagene). The viral termini were sequenced using rapid amplification of cDNA ends as previously described for the 3′ end (27) and using the RLM-RACE kit (Ambion, Austin, TX) per the manufacturer's instructions for the 5′ end. The sequences of all of the primers used can be found at www.chp.edu/researchsupplements/bergman. Nucleotide sequences were aligned and analyzed using SEQUENCHER software (Gene Codes Corp., Ann Arbor, MI). The sequences derived from the viruses were compared with the sequences of the vectors used to create the original virus (3). Quantitative reverse transcription (qRT)-PCR was performed as previously described (4). Briefly, viral RNA was amplified in a one-step RT-PCR using primers and a probe to a 77-nucleotide internal region of the VSV M gene (13). Results were analyzed on an ABI Prism 7700 sequence detector (PE Applied Biosystems).
Flow cytometry.
BHK-21 cells were transfected with vectors expressing one of the chimeric Sindbis gp using a standard Lipofectamine protocol, incubated for 48 h, and then stained. Flow cytometry was performed by incubating 5 × 105 to 1 × 106 cells with 2 μg of antibody and then staining with a fluorescein isothiocyanate-conjugated goat anti-mouse IgG antibody (catalog no. F-2012; Sigma, St. Louis, MO). Immunofluorescence was quantified using a FACStarPlus cytometer (Becton Dickinson, Mountain View, CA).
Analysis of the protein composition of pseudotype VSV.
VSV pseudotypes were created by transfecting BHK-21 with DNA encoding either the initial gp or the adapted gp and then infecting them with rrVSV whose genome did not contain any gp gene. Preparation of 35S-labeled pseudotype virus was performed as previously described (3). Briefly, BHK-21 cells were transfected with vectors expressing one of the chimeric Sindbis gp using a standard Lipofectamine protocol and incubated at 32°C. Twenty-four hours later, the medium was removed from each well and replaced with 1 ml of cysteine/methionine-free media (Cellgro catalog no. 17-104-L1; Mediatech, Inc., Herndon, VA) containing 20 μCi of [35S]methionine (10 mCi/ml, Trans35S-LABEL, catalog no. 510067; MP Biomedicals, LLC, Irvine, CA). Forty-eight hours after transfection, the cells were infected by the addition of VSV-EGFP-ΔG coated with G gp at an MOI of 5. This virus was adsorbed with the cells for 2 h in Dulbecco's modified Eagle's medium with 2.5% fetal calf serum, and then the medium was replaced with [35S]methionine containing medium as constituted above. Seventy-two hours after transfection, the medium and the cell layer were harvested separately. The medium containing the radiolabeled virus was clarified by centrifugation at 1,000 × g for 5 min. The virus was then pelleted by ultracentrifugation in a swinging bucket rotor at 80,000 × g for 2 h. The drained virus pellet was resuspended in 300 μl of serum-free medium. The cell layer was harvested by incubation in immunoprecipitation buffer consisting of 10 mM Tris, pH 7.4, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100, 100 mM NaCl, and 1 mM EDTA. The cell lysates underwent immunoprecipitation using 2.5 μl of antibody VR1248 with protein A/G plus (Santa Cruz Biotechnical, Inc., Santa Cruz, CA). The purified radiolabeled virus or immunoprecipitated cell lysate was diluted 1:1 with 2× Laemmli sample buffer and subjected to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 10% acrylamide containing 0.09% bis acrylamide (20). The gel was dried under a vacuum and examined by autoradiography. Viral bands were identified by their molecular weights. Similar techniques were used to prepare 35S-labeled rrVSV. Four hours after infection of D2F2/E2 cells at an MOI of 2 in 24-well trays with 200 μl of medium per well, the medium was removed and replaced with 0.5 ml of cysteine/methionine-free media containing 10 μCi of [35S]methionine (Trans35S-LABEL, 10mCi/ml). The medium containing the radiolabeled virus was harvested 24 h after infection, and the virus was harvested as described above. Density analysis was performed using Quantity One software (Bio-Rad Laboratories, Hercules, CA).
Analysis of glycosylation.
BHK-21 and ldlD cells were transfected with vectors expressing one of the chimeric Sindbis gp using a standard Lipofectamine protocol. Twenty-four hours later, cells were labeled for 2 h with [35S]methionine as described above and then incubated for 1 h in complete medium. One set of ldlD cells were supplemented with N-acetylgalactosamine (GalNAc) and galactose (Gal) during the 2-h labeling period. Cells were harvested and underwent immunoprecipitation with VR1248 and gel electrophoresis as outlined above. One set of BHK-21 cell lysates was treated with 500 U PNGase (New England Biolabs, Beverly, MA) for 1 h at 37°C prior to electrophoresis.
Serial passage.
D2F2/E2 cells were plated in a six-well tray (Corning, Inc.) at 4 × 105 cells/well. One well was infected with rrVSV at an MOI of 0.01. Two days later, when all of the cells were infected as indicated by EGFP expression, the supernatant was harvested and diluted 100-fold, and 1 to 20 μl was used to infect the next well. The newly infected well was inspected 18 h later, and the experiment continued if 100 to several hundred green cells or small green plaques were observed. If the number of infected green cells was too large or too small, then the infecting dose from the previous pass was adjusted and used to infect a new well. The intent was to infect with an MOI of 0.01 to 0.001.
Growth assay.
D2F2/E2 or SKBR3 cells were plated in a six-well tray (Corning, Inc.) at 4 × 105 cells/well. One day later, individual wells were infected with rrVSV at an MOI of 0.001 in 1 ml of growth media with 2.5% fetal calf serum (FCS). After a 2-h viral incubation, the medium was replaced with 2 ml of growth medium with 10% FCS. One hundred microliters of media was harvested from each well at various time points and frozen at −70°C for later titer determinations. One hundred microliters of fresh media was added to the wells to replace each withdrawal. Titers of the yield were determined as described above on either D2F2/E2 or SKBR3 cells. The effect of antiserum to mouse IFN on rrVSV growth was assayed by adding various volumes of antiserum at the time of infection and harvesting and determining the titer of the yield 48 h after infection.
Burst size.
D2F2/E2 cells were plated in a 96-well tray (Corning, Inc.) at various concentrations. One day later, wells that were 80% confluent were infected. The number of cells in one representative well was counted. Individual wells were infected with rrVSV at an MOI of 10 in 50 μl of growth media with 2.5% FCS. After a 2-h viral incubation, the media were replaced with 100 μl of growth medium with 10% FCS. At various time points, all of the supernatant from individual wells was harvested and frozen at −70°C for later titer determination. Complete infection of all cells in the wells was confirmed visually by finding that 100% of the cells expressed EGFP. Titers of the yield were determined as described above.
Virus stability.
D2F2/E2 cells were infected for the burst size assay as noted above, and supernatant was harvested at 12 h. The supernatant was placed in empty individual wells of a 96-well tray and incubated in a humidified incubator at 37°C with 5% CO2. At various times, 5-μl aliquots were removed and frozen at −70°C for later titer determination.
IFN assay.
Interferon concentrations were measured using a previously described biological assay (40). Briefly, mouse L cells in 96-well trays were treated for 24 h with serial twofold dilutions of test samples. The cells were then challenged with wild-type (wt) VSV at an MOI of 0.1. At 72 h after infection, the monolayers were examined for virus-induced cytopathic effect using an inverted light microscope. In this assay, one unit of IFN is defined as that concentration that can protect 50% of the monolayer from virus-mediated destruction.
Confocal microscopy.
BHK-21 cells transfected with a vector expressing either the initial or the adapted gp were grown on coverslips, fixed in 2% paraformaldehyde for 20 min, and permeabilized in 0.1% Triton X-100 in phosphate-buffered saline. The cells were then washed three times in phosphate-buffered saline containing 0.5% bovine serum albumin and 0.15% glycine, pH 7.4 (buffer A), followed by a 30-min incubation with purified goat IgG (50 μg/ml) at 25°C and three additional washes with buffer A. All of the preceding steps are designed to ensure minimal nonspecific reaction to the antibodies used. Sections were then incubated for 60 min with primary antibody VR 1248, followed by three washes in buffer A and a 60-min incubation in fluorescently labeled secondary antibody, a 1- to 2-μg/ml mix of Alexa 488 phalloidin and Cy3-conjugated goat anti-mouse antibody (Molecular Probes, Eugene, OR). The sections were washed six times in buffer A and then mounted in gelvatol and coverslipped for confocal microscopy. Confocal imaging was performed with an Olympus FV500 microscope using a 60× plan apo objective. All imaging conditions were kept constant for each treatment. X, Y, and Z sections were collected at the optimal Nyquist sampling for each axis.
RESULTS
Growth of initial and adapted virus on D2F2/E2 cells.
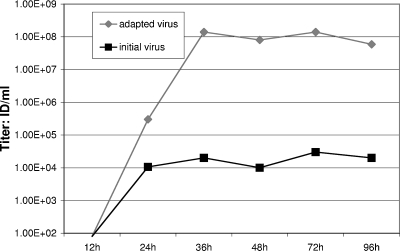
After construction, the Her2/neu-specific rrVSV was successfully grown on the Her2/neu-overexpressing human breast cancer cell line SKBR3. This initial virus stock had a titer measured by counting green cells on SKBR3 cells of 3.1 × 107 compared with a titer of 7.3 × 105 on 143 cells, a human osteosarcoma cell line that does not express Her2/neu. We intended to test the therapeutic efficacy of the virus against implants of D2F2/E2, a mouse mammary cell line that was stably transfected to express Her2/neu. However, the rrVSV was not well adapted to this line. The titer of the rrVSV stock on D2F2/E2 cells was low at 6 × 105/ml, and the virus grew very poorly on these cells. Viral yield following growth on D2F2/E2 cells reached a plateau after 24 h of only about 104 infectious doses (ID)/ml (Fig. 2).
FIG. 2.
Growth assay. Yield of virus at various times following infection of D2F2/E2 cells at an MOI of 0.001. The titer of the yield was determined on D2F2/E2 cells. Titers were <100 for both viruses at 12 h. Results are shown for one representative experiment of 5 replicates.
The initial rrVSV was serially passed for 15 passages on D2F2/E2 cells and evolved an adapted virus that had higher infectivity of D2F2/E2 cells and much better yield following low-MOI infection of D2F2/E2 cells. This adapted virus had no loss of infectivity or decreased growth on SKBR3 cells. The final adapted virus had a titer on D2F2/E2 cells of 2.2 × 107/ml; the initial virus had a titer of 6 × 105/ml. The titer on parent D2F2 cells that did not express Her2/neu was 9.8 × 105/ml for the final virus compared with 2.6 × 104 for the initial virus. The ratio of infection on Her2/neu-positive to Her2/neu-negative cells was 22.4 for the adapted virus and 23 for the initial virus. Thus, the increase in titer was not accompanied by any change in receptor specificity. The yield from a low-MOI infection of D2F2/E2 cells was 1.4 × 108 ID/ml at 36 h compared with a yield of 2 × 104 ID/ml for the initial virus (Fig. 2). In fact, the yield of the adapted virus was similar to that achieved by wild-type VSV, 2.2 × 108 ID/ml, in this cell line. We then conducted a series of experiments to determine the changes in the adapted virus accounting for the improved infection and growth in D2F2/E2 cells.
Genotype of initial and adapted viruses.
The genotypic changes were identified by sequencing the entire genomes of the initial and adapted viruses. The sequences of the initial and adapted viruses were identical except for two point mutations which were located in the SCA gene coding for the heavy chain resulting in the following amino acid substitutions: pro52aleu and ala78thr (18). The pro52aleu mutation is located in an antibody complementarity-determining region, CDR2, and the ala78thr mutation is in a framework region, FR3 (18). We proved that these mutations accounted for the adapted phenotype by creating a new rrVSV from vector components incorporating these mutations into the viral genome. As previously described (3), the virus was created in COS7 cells, amplified on BHK-21 cells, and then grown once on D2F2/E2 cells. The titer of this virus on D2F2/E2 cells was 2.24 × 107/ml and the titer on D2F2 cells was 6.21 × 105/ml, which is almost identical to the titer of the adapted virus noted above.
Serial passage of the virus on D2F2/E2 applied strong selective pressure for fixation of beneficial mutations in the gp gene. This chimeric foreign protein is not optimized for VSV but is now essential for its life cycle. Therefore, beneficial mutations in this gene will confer the greatest selective advantage to the progeny virus and will be fixed first. Sequence analysis showed that not only was there no change in the gene sequence of the 4 essential VSV genes, N, P, M, and L, but there was also no change in the gene sequence of the nonessential additional genes coding for GM-CSF and EGFP. We confirmed that all virus particles retained EGFP expression by finding that the titer of adapted virus measured by counting green cells was identical to the titer measured by the tissue culture infectious dose method. Similar stable expression of an extra gene in recombinant VSV has been noted previously (36). We next sought to determine how the genotypic changes in the chimeric gp improved infection of D2F2/E2 cells.
Properties of the initial and the adapted glycoproteins.
Analysis of 35S-labeled viral proteins by gel electrophoresis demonstrated that the adapted gp incorporated better into the viral particle and showed a change in migration of the E2 protein (Fig. 3). These studies were performed using pseudotype viruses to be absolutely certain that all genes were identical except for the gp gene and because the initial virus grew poorly on D2F2/E2 cells. The pseudotype viruses were constructed by transfecting BHK-21 cells with vectors expressing either the initial or adapted gp and then infecting them with rrVSV, whose genome did not contain any gp gene. The first and second lanes comparing the virus particles show that the densities of the N nucleocapsid proteins are virtually identical (relative density of the N bands on this gel are 0.97 and 1 and those on a much less exposed gel are 1.15 and 1), indicating that equal numbers of particles are being loaded on the gel. The concentration of E2, however, is fivefold greater in the adapted than the initial gp (relative densities of the E2 bands are 4.9 and 1). Incorporation of E1 was also improved in the adapted gp, probably because E1 and E2 are incorporated together into the virus (30). The fivefold increase in membrane gp was associated with a fivefold increase in infectivity of the pseudotypes. Titers of 35S-radiolabeled pseudotype viruses were determined on D2F2/E2 cells using equal numbers of radioactive CPM and, therefore, equal numbers of virus particles. The titer per 10,000 counts was 1.10 × 105 using the initial gp and 5.94 × 105 using the adapted gp (means of 4 separate constructions of pseudotypes). Improved incorporation of gp was not due to greater cellular production. Analysis of gp immunoprecipitates from intracellular proteins did not show greater production of the adapted gp (Fig. 3). Improved incorporation was apparently also not due to improved intracellular trafficking because flow cytometry showed no difference in surface expression following transfection of the different gp (Fig. 4). This result was confirmed by confocal fluorescent immunohistochemistry which showed equal staining of the cell surface following transfection of BHK-21 cells with either the initial or the adapted gp (Fig. 5). In addition, both gp appeared uniform on the cell surface, with no apparent difference in the extent of protein aggregation. Flow cytometry could not be used to measure surface expression of infected, rather than transfected, cells because infection killed the cells and dead cells are not accurately assessed by flow cytometry.
FIG. 3.
SDS-polyacrylamide gel electrophoresis analysis of pseudotype viruses and immunoprecipitates of Sindbis gp from cell extracts. Pseudotype viruses were prepared in BHK-21 cells metabolically labeled for 48 h with [35S]methionine and incubated at 32°C. Immunoprecipitates were prepared using VR1248, a mouse polyclonal antibody directed to the SV. The VSV proteins are used as molecular weight markers. The labeled lanes express either the initial or the adapted glycoprotein. The immunoprecipitates show 3 bands in the region of E2. The middle band represents the fully processed E2 found on the viral surface. The top band represents precursor E2 (PE2), and the bottom band represents either nonglycosylated E2 or an E2 breakdown product.
FIG. 4.
Flow cytometric analysis of D2F2/E2 cells transfected with an expression vector expressing either the initial (A) or the adapted (B) chimeric Sindbis gp. Cells were stained with the mouse polyclonal anti-Sindbis antibody, VR1248, followed by a fluorescein isothiocyanate-conjugated goat anti-mouse IgG antibody. Control plots from cells stained only with the secondary antibody are outlined on the left of each figure. Results are shown for one representative experiment of 6 replicates.
FIG. 5.
Confocal fluorescent immunohistochemistry. BHK-21 cells were transfected with either the initial gp (A and B) or the adapted gp (C and D). The gp was stained red, and the cytoplasm was counterstained green using VR 1248 and Alexa 488 phalloidin as detailed in Materials and Methods. Panels A and C are images of a single plane, and panels B and D are stacked images of multiple planes through the entire cell. Images of the initial virus in panels A and C compared with images of the adapted virus in panels B and D demonstrate no apparent difference in the extent of protein accumulation or aggregation on the cell surface.
The differences found between the initial and adapted E2 protein in pseudotype viruses was confirmed in replicating viruses (Fig. 6, lanes 2 and 3). wt VSV proteins labeled at the same time are shown for comparison (Fig. 6, lane 1). Metabolic labeling was technically difficult and harder to interpret because the titer of the initial virus was very low on D2F2/E2 cells and there was difficulty obtaining a synchronized infection. The background signal from host proteins is high in lane 2 derived from the initial virus and different from that seen in lane 3 derived from the adapted virus. In addition, the yield of virus following infection with the initial virus was low, requiring loading of more material, 35 μl, into lane 2 than in lane 3, 10 μl, and lane 1, 5 μl. Nonetheless, it remains clear that the adapted gp incorporated better into the viral particle and showed a change in migration of the E2 protein (Fig. 6). The relative density of the N bands in lanes 2 and 3 are 1 and 1.49, indicating that about 1.5 times more adapted virus particles have been loaded than initial virus particles, but the relative densities of the E2 bands are 1 and 7.27. The concentration of E2, per particle, is therefore 4.9-fold greater in the adapted than the initial replicating virus, just as it was in the pseudotype virus. Lane 3 has 1.5-fold more virus than lane 2 and was loaded with 3.5-fold less supernatant, indicating that the initial concentration of virus particles was 5.25 greater for the adapted than the initial virus. We did not measure the titers of these radiolabeled viruses, but we know that the yields of adapted virus on D2F2/E2 cells are 3 to 4 logs greater than the yields of initial virus, suggesting again that infectivity per virus particle was greater for the adapted than the initial virus.
FIG. 6.
SDS-polyacrylamide gel electrophoresis analysis of wt VSV (lane 1), initial virus (lane 2), and adapted virus (lane 3). Viruses were prepared in D2F2/E2 cells metabolically labeled for 24 h with [35S]methionine and incubated at 37°C.
Analysis of the mutations in the adapted gp indicated two possible causes for the slower electrophoretic mobility of the adapted gp. The ala78thr mutation produced a potential O-glycosylation site on the new threonine and also changed the amino acid sequence from Asn-Thr-Ala to Asn-Thr-Thr, thereby creating an N-glycosylation consensus sequence (Asn-X-Ser/Thr). The difference in electrophoretic mobility of the adapted gp was completely eliminated by treatment with PNGase, an enzyme that cleaves N-glycosylated sugars, indicating that the new N-glycosylation site in the adapted gp accounted for the difference in size of the glycoproteins (Fig. 7, lanes 5 and 7 without PNG show a difference in E2 mobility, but lanes 6 and 8 with PNG show no difference). An additional O-glycosylation site was excluded by demonstrating that creating the different pseudotypes in ldlD cells had no effect on the difference in size between the initial and the adapted gp (Fig. 7, lanes 1 and 2 using ldlD cells without Gal show the same degree of difference in mobility of E2 as lanes 3 and 4 using ldlD cells with Gal). ldlD cells are unable to O-glycosylate in the absence of supplemental GalNAc and Gal (43). If the adapted gp had an additional O-glycosylation, growth in ldlD cells would have reduced the difference in size between the two gp.
FIG. 7.
SDS-polyacrylamide gel electrophoresis analysis of immunoprecipitates of Sindbis gp from cell extracts. Vectors expressing either the initial or the adapted Sindbis SCA gp (labeled at the bottom of the figure) were transfected into BHK-21 or ldlD cells and incubated at 32°C. After 24 h, cells were metabolically labeled for 2 h with [35S]methionine. After a 1-h chase, cells were harvested and Sindbis gp was immunoprecipitated using VR1248, a mouse polyclonal antibody directed to the Sindbis virus. One set of ldlD cells were treated during the labeling and chase with GalNAc and Gal and are labeled Gal. One set of immunoprecipitates from the BHK-21 cells were treated with PNGase and are labeled PNG.
Biologic phenotypes of initial and adapted viruses.
As noted above, higher incorporation of gp into the envelope of the adapted virus was associated with improved infectivity of D2F2/E2 cells. As shown in the following results, this change was also associated with much better viral stability (Fig. 8), higher burst size (Fig. 9), and reduced IFN induction in low-multiplicity infections (Table 1).
FIG. 8.
Comparison of viral stability at 37°C between the initial and adapted viruses. The initial virus was tested after one passage through D2F2/E2 cells to compare viruses that were both made in D2F2/E2 cells. Loss of titer over time was fit to a one-phase exponential (exp) decay (goodness of fit R2 = 0.9948 [initial virus] and 0.9684 [adapted virus]). Data are means of results from 2 experiments with standard error bars (the standard error for the initial virus was almost zero at all time points).
FIG. 9.
Comparison of titers at various times following high-MOI (MOI = 10) infection of D2F2/E2 cells with either the initial or adapted virus. Data are means of results from 3 experiments with standard error bars.
TABLE 1.
Interferon assaya
| Time (h) | IFN titer of virus
|
|
|---|---|---|
| Adapted | Initial | |
| 12 | <4 | <4 |
| 24 | <4 | <4 |
| 36 | <4 | 16 |
| 48 | <4 | 32 |
| 72 | 16 | 32 |
D2F2/E2 cells were infected with the initial or adapted virus at an MOI of 0.001. At various times, supernatant was harvested and assayed for IFN titer. Results are from one representative experiment.
Viral stability.
Viruses expressing the adapted gp were much more stable at physiologic conditions than viruses expressing the initial gp (Fig. 8). The initial virus was tested after one passage through D2F2/E2 cells to compare viruses that were both made in D2F2/E2 cells. The half-life (t1/2) at 37°C for the adapted virus was 15.9 h compared with 2.0 h for the initial virus (P = 0.0095). The extremely short t1/2 of the initial virus meant that the virus lost 3 logs of infectivity over 24 h. To be certain that this dramatic difference in t1/2 was not due to differences in adsorption to the wells during incubation, the particle number in the supernatant from both viruses was assessed by qRT-PCR prior to and after 12 h of incubation. All values were within one cycle number, indicating that there was less than a twofold difference in particle number between any of the samples and there was no significant loss of particles to adsorption for either virus.
Virus production.
Burst size was assessed in single-step growth curve infections (Fig. 9). D2F2/E2 cells were infected with either the initial or the adapted virus at an MOI of 10. Almost all the cells were green by 12 h, indicating complete infection. Supernatant was harvested at 4, 8, 12, and 24 h for determination of viral yield (ID/ml). The burst size ID at 12 h as determined by titer on D2F2/E2 cells was 769 per infected cell for the adapted virus compared with 29 per infected cell for the initial virus. The rapid decrease in titer of the initial virus between 12 and 24 h was due to the poor stability of the virus. The burst size of particle production was simultaneously assessed by qRT-PCR (Fig. 10). At 12 h, the adapted virus yielded 1.4 times as many particles as the initial virus. The small difference in particle number compared to the large difference in ID titer on D2F2/E2 cells was due to the greater infectivity of the adapted virus on D2F2/E2 cells.
FIG. 10.
Particle production in single-step growth curve infections. qRT-PCR comparing the genome number from the viral yield at 12 h and 24 h in D2F2/E2 cells infected with either the initial or the adapted virus. Ct represents the cycle number required to achieve a threshold concentration of fluorescence. Each unit reduction of Ct indicates a doubling of the genome number.
Interferon induction.
We still required an explanation for the extremely low viral yield of ∼1 × 104 ID/ml following low-MOI infection of 4 × 105 D2F2/E2 cells. The cause was unlikely to be defective interfering particles because the infection was initiated at low MOI and because the same virus produced excellent viral yield on SKBR3 cells (1.2 × 107 ID/ml). We suspected that the initial virus induced an antiviral state in the D2F2/E2 cells mediated by IFN because viral production abruptly ceased after 24 h (Fig. 2), and usually, many cells in the monolayer remained uninfected. IFN was assayed in the supernatant at various times following infection of D2F2/E2 cells with either initial or adapted virus at an MOI of 0.001 (Table 1). The results show earlier and higher induction of IFN following infection with the initial virus. The physiologic importance of this IFN production was confirmed by growing virus in the presence of anti-IFN antibody. The yield of the initial virus from D2F2/E2 cells was increased from 8 × 103 ID/ml to 2.1 × 105 ID/ml by growth in medium containing 1,500 IFN neutralizing titer per ml (Table 2; one of 3 representative experiments). The yield of the adapted virus was completely unchanged by growth in media containing antibody to IFN. We concluded that the adapted virus was able to complete the final round of replication prior to the induction of an IFN-mediated antiviral state, but the initial virus produced infectious progeny virus more slowly and replication was ultimately terminated by an IFN response.
TABLE 2.
Yield of virus following low-MOI infection of D2F2/E2 cells incubated with various quantities of anti-IFN antibodya
| Amt of antibody (U/ml) | Yield (ID/ml) |
|---|---|
| 0 | 8.00E+03 |
| 1,500 | 2.11E+05 |
| 750 | 1.11E+05 |
| 375 | 4.90E+04 |
| 188 | 5.30E+04 |
| 94 | 4.10E+04 |
| 47 | 2.50E+04 |
| 24 | 9.00E+03 |
| 12 | 1.10E+04 |
Results are from one representative experiment of 3.
DISCUSSION
We have demonstrated the ability to use the genetic plasticity of RNA viruses to create an improved engineered VSV by serial passage on an appropriate cell line. The rrVSV was designed to preferentially infect cells that expressed the Her2/neu receptor. The initial rrVSV, which was made in human SKBR3 breast cancer cells, succeeded in this task (3). However, the rrVSV infected and replicated poorly in the mouse cell line, D2F2/E2, that was the intended target for animal studies. Serial passage for 15 cycles yielded an adapted replicating oncolytic virus with an improved titer on targeted D2F2/E2 cells of 2.2 × 107/ml and the ability to generate a productive infection in these cells that approximated the infection generated by wt VSV (Fig. 2). Previous reports using SCA to target retroviruses, adeno-associated virus, and attenuated measles virus (MV) have not achieved these high titers. For retroviruses and adeno-associated virus, titers at best were only about 1 × 105/ml (16, 19, 22, 23, 42). Failure of a chimeric retroviral gp to participate in the fusion process has been identified as a cause of poor infectivity (45). For MV, the titers against specific cells were 6 × 104 to 6 × 105/ml, but native MV binding was not altered (15).
We postulate that the essential phenotypic improvement was greater expression of the gp on the viral surface, which resulted in higher infectivity and greater stability. Higher infectivity enabled a productive low-MOI infection because the adapted virus progressed quickly through multiple rounds of replication within the infected D2F2/E2 monolayer and outpaced the development of an antiviral state in the uninfected cells in the culture. In contrast, the initial virus grew slowly in a low-multiplicity infection, and the progression of the infection was curtailed. This inhibition could be partially reversed by the addition of anti-IFN antibody. Neither virus produced detectable levels of IFN in the single-step growth assay (data not shown), suggesting that these viruses may initially induce very low levels of IFN which can prime the remaining cells to induce protective levels of IFN upon infection (1, 39).
It is not surprising that the initial gp incorporated poorly into the viral particle (3) because it is a novel construct consisting of an artificial SCA placed within a modified positive-strand RNA virus gp which is then incorporated into the envelope of a negative-strand RNA virus. Similar difficulties of incorporation into the viral envelope were reported for an SCA-retroviral surface protein chimera (23), and difficulties in gp folding, transport, and membrane fusion were reported for a VSV-G/human immunodeficiency virus chimera (35). It is possible that, in addition to greater surface expression, the higher infectivity might be due to better function of the adapted gp (45), such as tighter binding to the surface receptor or improved fusion with the endosomal membrane, but these properties were not directly tested.
The mechanism promoting improved incorporation of the adapted gp onto the viral surface remains unknown. We expected that the additional N-glycosylation site would allow the adapted gp to migrate more efficiently through the endoplasmic reticulum and Golgi apparatus and show higher expression on the cell plasma membrane (14). Flow cytometric analysis, however, repeatedly showed virtually identical expression of the different chimeric gp on the cell surface. Previous work has shown that the intracellular endodomain of glycoproteins can influence viral assembly either positively or negatively, but the mutations in our adapted rrVSV were all in the ectodomain (17). Possible mechanisms for improved incorporation without better cellular expression include conformational changes induced in the endodomain, allowing better association with the M protein, more efficient gp capping on the cell surface, more efficient processing of the precursor E2 (PE2) (12), or more efficient heterodimer and trimer association of E1 and E2. Analysis of intracellular proteins did not suggest more efficient processing of PE2 in cells infected with the adapted virus (Fig. 3), and confocal fluorescent immunohistochemistry did not suggest a difference in capping (Fig. 5).
Despite the fact that 2 mutations were in the SCA protein and one of these was in the complementarity-determining regions of the SCA, the gp mutations did not result in higher specificity of infection on cells expressing erbb2. The ratio of titer on erbb2 expressing D2F2/E2 cells to titer on erbb2-negative D2F2 cells was identical in the initial and the adapted virus. The increased expression of gp on the viral surface improved the residual nonspecific native binding of the Sindbis gp to the same extent as it improved the specific binding mediated by the SCA. Serial passage resulting in nonspecific increase in titer has been noted previously (33). Disabling nonspecific binding will probably require recombinant engineering to delete sites important for native E2 binding (25, 37, 44).
Gene therapy using rrVSV will require additional genes inserted into the genome, and an important finding in this study was that two nonessential genes added to rrVSV did not show any fixed mutations following 15 passages. Similar stable expression of an extra gene in recombinant VSV has been noted previously (36). Although VSV has a high mutation rate, averaging about 1 × 10−4 to 4 × 10−4 per base site, and exists as a mixture of genetic and phenotypic variants (11, 38), genes not placed under selection pressure retained sequence fidelity in the dominant species (32). On the other hand, when the same EGFP gene used in our rrVSV was added to the cytoplasmic end of the VSV-G gene, it was rapidly deleted because of strong selection against homotrimeric G protein bearing such a large cytoplasmic domain (6). Appropriately positioned additional genes in rrVSV are likely to retain expression in vivo for as long as the VSV continues replicating. Nonetheless, replicating VSV used clinically will have to be monitored for genetic changes that occur in response to unpredicted selective pressures in vivo.
MOI was a critical parameter for successful serial passage. MOI needed to be low to avoid creating defective interfering particles. In addition, it was best to dilute supernatant from an infected monolayer before passing it to a fresh monolayer to avoid passing a significant quantity of virally induced interferon. On the other hand, it was important not to pass too few virus which produces progressive loss of viral fitness, a phenomenon called Muller's ratchet. Because most mutations arising in well-adapted quasi-species will be deleterious, passing small numbers of a virus such as VSV with a high mutation rate causes fixation of these harmful mutations in the population (9). Five plaque passages of VSV can maintain fitness stability but are not optimal in sustaining gains of function (28). We used an MOI of ∼0.01 to 0.001 and passed 102 to 103 viral ID. An improved virus emerged after 15 passages which is similar to previous results showing nonlinear population dynamics during replicative competitions of RNA virus quasi-species with outgrowth of a superior species beginning after 8 to 11 passages (27).
These studies demonstrate that serial passage of rrVSV can be used to improve infection on specific cell lines based on mutations in the surface attachment glycoprotein. The rrVSV has been designed to allow easy substitution of genes expressing other SCA or binding proteins within the VSV genome. rrVSV with a wide variety of specificities can therefore be created and quickly adapted to specific cell types. VSV is an excellent candidate for development as an oncolytic virus because of its potency, safety and favorable combination of genomic adaptability and stability.
Acknowledgments
This study was supported in part by grants from the Department of the Army, award number DAMD17-00-1-0201, the PA Department of Health's Breast and Cervical Cancer Section, the Research Advisory Committee of Children's Hospital of Pittsburgh, and Public Health Service grant RO1 CA104404 from the National Cancer Institute.
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the granting institutions.
We thank Massimo Trucco, William A. Rudert, and Rebecca P. Hughey for invaluable intellectual and material assistance. Wei-Zen Wei, John K. Rose, Paul D. Robbins, Irvin S. Y. Chen, and the NIAID Reference Reagent Repository very generously supplied materials as noted in the text.
REFERENCES
- 1.Abreu, S. L., F. C. Bancroft, and W. E. Stewart. 1979. Interferon priming. Effects on interferon messenger RNA. J. Biol. Chem. 254:4114-4118. [PubMed] [Google Scholar]
- 2.Bergman, I., M. A. Barmada, J. A. Griffin, and D. J. Slamon. 2001. Treatment of meningeal breast cancer xenografts in the rat using an anti-p185/HER2 antibody. Clin. Cancer Res. 7:2050-2056. [PubMed] [Google Scholar]
- 3.Bergman, I., P. Whitaker-Dowling, Y. Gao, and J. A. Griffin. 2004. Preferential targeting of vesicular stomatitis virus to breast cancer cells. Virology 330:24-33. [DOI] [PubMed] [Google Scholar]
- 4.Bergman, I., P. Whitaker-Dowling, Y. Gao, J. A. Griffin, and S. C. Watkins. 2003. Vesicular stomatitis virus expressing a chimeric Sindbis glycoprotein containing an Fc antibody binding domain targets to Her2/neu overexpressing breast cancer cells. Virology 316:337-347. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, D. K., E. A. Duarte, S. F. Elena, A. Moya, E. Domingo, and J. Holland. 1994. The red queen reigns in the kingdom of RNA viruses. Proc. Natl. Acad. Sci. USA 91:4821-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton, K. P., and J. K. Rose. 2001. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology 279:414-421. [DOI] [PubMed] [Google Scholar]
- 7.de Mattos, C. A., C. C. de Mattos, and C. E. Rupprecht. 2001. Rhabdoviruses, p. 1245-1277. In D. Knipe and P. Howley (ed.), Fundamental virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 8.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 9.Duarte, E., D. Clarke, A. Moya, E. Domingo, and J. Holland. 1992. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc. Natl. Acad. Sci. USA 89:6015-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubuisson, J., and C. M. Rice. 1993. Sindbis virus attachment: isolation and characterization of mutants with impaired binding to vertebrate cells. J. Virol. 67:3363-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elena, S. F., F. Gonzalez-Candelas, I. S. Novella, E. A. Duarte, D. K. Clarke, E. Domingo, J. J. Holland, and A. Moya. 1996. Evolution of fitness in experimental populations of vesicular stomatitis virus. Genetics 142:673-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erwin, C., and D. T. Brown. 1980. Intracellular distribution of Sindbis virus membrane proteins in BHK-21 cells infected with wild-type virus and maturation-defective mutants. J. Virol. 36:775-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopalakrishna, Y., and J. Lenard. 1985. Sequence alterations in temperature-sensitive M-protein mutants (complementation group III) of vesicular stomatitis virus. J. Virol. 56:655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guibinga, G. H., F. L. Hall, E. M. Gordon, E. Ruoslahti, and T. Friedmann. 2004. Ligand-modified vesicular stomatitis virus glycoprotein displays a temperature-sensitive intracellular trafficking and virus assembly phenotype. Mol. Ther. 9:76-84. [DOI] [PubMed] [Google Scholar]
- 15.Hammond, A. L., R. K. Plemper, J. Zhang, U. Schneider, S. J. Russell, and R. Cattaneo. 2001. Single-chain antibody displayed on a recombinant measles virus confers entry through the tumor-associated carcinoembryonic antigen. J. Virol. 75:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, A., T. H. Chu, F. Nocken, K. Cichutek, and R. Dornburg. 1998. Cell-type-specific gene transfer into human cells with retroviral vectors that display single-chain antibodies. J. Virol. 72:10148-10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J. E., W. Rodgers, and J. K. Rose. 1998. A plasma membrane localization signal in the HIV-1 envelope cytoplasmic domain prevents localization at sites of vesicular stomatitis virus budding and incorporation into VSV virions. Virology 251:244-252. [DOI] [PubMed] [Google Scholar]
- 18.Jung, S., and A. Pluckthun. 1997. Improving in vivo folding and stability of a single-chain Fv antibody fragment by loop grafting. Protein Eng. 10:959-966. [DOI] [PubMed] [Google Scholar]
- 19.Khare, P. D., L. Shao-Xi, M. Kuroki, Y. Hirose, F. Arakawa, K. Nakamura, Y. Tomita, and M. Kuroki. 2001. Specifically targeted killing of carcinoembryonic antigen (CEA)-expressing cells by a retroviral vector displaying single-chain variable fragmented antibody to CEA and carrying the gene for inducible nitric oxide synthase. Cancer Res 61:370-375. [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marin, M., D. Noel, S. Valsesia-Wittman, F. Brockly, M. Etienne-Julan, S. Russell, F. L. Cosset, and M. Piechaczyk. 1996. Targeted infection of human cells via major histocompatibility complex class I molecules by Moloney murine leukemia virus-derived viruses displaying single-chain antibody fragment-envelope fusion proteins. J. Virol. 70:2957-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, F., S. Chowdhury, S. J. Neil, K. A. Chester, F. L. Cosset, and M. K. Collins. 2003. Targeted retroviral infection of tumor cells by receptor cooperation. J. Virol. 77:2753-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morizono, K., G. Bristol, Y. M. Xie, S. K. Kung, and I. S. Chen. 2001. Antibody-directed targeting of retroviral vectors via cell surface antigens. J. Virol. 75:8016-8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morizono, K., Y. Xie, G. E. Ringpis, M. Johnson, H. Nassanian, B. Lee, L. Wu, and I. S. Chen. 2005. Lentiviral vector retargeting to P-glycoprotein on metastatic melanoma through intravenous injection. Nat. Med. 11:346-352. [DOI] [PubMed] [Google Scholar]
- 26.Novella, I. S., E. A. Duarte, S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Exponential increases of RNA virus fitness during large population transmissions. Proc. Natl. Acad. Sci. USA 92:5841-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novella, I. S., and B. E. Ebendick-Corpus. 2004. Molecular basis of fitness loss and fitness recovery in vesicular stomatitis virus. J. Mol. Biol. 342:1423-1430. [DOI] [PubMed] [Google Scholar]
- 28.Novella, I. S., S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J. Virol. 69:2869-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno, K., K. Sawai, Y. Iijima, B. Levin, and D. Meruelo. 1997. Cell-specific targeting of Sindbis virus vectors displaying IgG-binding domains of protein A. Nat. Biotechnol. 15:763-767. [DOI] [PubMed] [Google Scholar]
- 30.Phinney, B. S., K. Blackburn, and D. T. Brown. 2000. The surface conformation of Sindbis virus glycoproteins E1 and E2 at neutral and low pH, as determined by mass spectrometry-based mapping. J. Virol. 74:5667-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ring, C. J. 2002. Cytolytic viruses as potential anti-cancer agents. J. Gen. Virol. 83:491-502. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez, L. L., W. M. Fitch, and S. T. Nichol. 1996. Ecological factors rather than temporal factors dominate the evolution of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 93:13030-13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Jarabo, C. M., N. Pariente, E. Baranowski, M. Davila, G. Gomez-Mariano, and E. Domingo. 2004. Expansion of host-cell tropism of foot-and-mouth disease virus despite replication in a constant environment. J. Gen. Virol. 85:2289-2297. [DOI] [PubMed] [Google Scholar]
- 34.Russell, S. J. 1994. Replicating vectors for cancer therapy: a question of strategy. Semin. Cancer Biol. 5:437-443. [PubMed] [Google Scholar]
- 35.Schlehuber, L. D., and J. K. Rose. 2004. Prediction and identification of a permissive epitope insertion site in the vesicular stomatitis virus glycoprotein. J. Virol. 78:5079-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnell, M. J., L. Buonocore, M. A. Whitt, and J. K. Rose. 1996. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 70:2318-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, T. J., R. H. Cheng, N. H. Olson, P. Peterson, E. Chase, R. J. Kuhn, and T. S. Baker. 1995. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 92:10648-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinhauer, D. A., and J. J. Holland. 1986. Direct method for quantitation of extreme polymerase error frequencies at selected single base sites in viral RNA. J. Virol. 57:219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taniguchi, T., and A. Takaoka. 2001. A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol. 2:378-386. [DOI] [PubMed] [Google Scholar]
- 40.Whitaker-Dowling, P., and J. S. Youngner. 1984. Characterization of a specific kinase inhibitory factor produced by vaccinia virus which inhibits the interferon-induced protein kinase. Virology 137:171-181. [DOI] [PubMed] [Google Scholar]
- 41.Wildner, O. 2001. Oncolytic viruses as therapeutic agents. Ann. Med. 33:291-304. [DOI] [PubMed] [Google Scholar]
- 42.Yang, Q., M. Mamounas, G. Yu, S. Kennedy, B. Leaker, J. Merson, F. Wong-Staal, M. Yu, and J. R. Barber. 1998. Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Hum. Gene Ther. 9:1929-1937. [DOI] [PubMed] [Google Scholar]
- 43.Zanni, E. E., A. Kouvatsi, M. Hadzopoulou-Cladaras, M. Krieger, and V. I. Zannis. 1989. Expression of ApoE gene in Chinese hamster cells with a reversible defect in O-glycosylation. Glycosylation is not required for apoE secretion. J. Biol. Chem. 264:9137-9140. [PubMed] [Google Scholar]
- 44.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, Y., L. Zhu, S. Lee, L. Li, E. Chang, N. W. Soong, D. Douer, and W. F. Anderson. 1999. Identification of the block in targeted retroviral-mediated gene transfer. Proc. Natl. Acad. Sci. USA 96:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zwiebel, J. A. 2001. Cancer gene and oncolytic virus therapy. Semin. Oncol. 28:336-343. [DOI] [PubMed] [Google Scholar]