Abstract
Using a novel cDNA microarray prepared from sources of actively responding immune system cells, we have investigated the changes in gene expression in the target tissue during the early stages of infection of neonatal chickens with infectious bursal disease virus. Infections of two lines of chickens previously documented as genetically resistant and sensitive to infection were compared in order to ascertain early differences in the response to infection that might provide clues to the mechanism of differential genetic resistance. In addition to major changes that could be explained by previously described changes in infected tissue, some differences in gene expression on infection, and differences between the two chicken lines, were observed that led to a model for resistance in which a more rapid inflammatory response and more-extensive p53-related induction of apoptosis in the target B cells might limit viral replication and consequent pathology. Ironically, the effect in the asymptomatic neonatal infection is that more-severe B-cell depletion is seen in the more genetically resistant chicken. Changes of expression of many chicken genes of unknown function, indicating possible roles in the response to infection, may aid in the functional annotation of these genes.
While the poultry industry is now the most important source of animal proteins for the human diet, it is facing increasing problems from progressively more virulent forms of pathogenic viruses. Many studies have documented the existence of genetically determined differences in resistance to infectious disease in chickens (16, 19, 25, 35), but there is little knowledge of the mechanisms responsible. Recent outbreaks of avian influenza and severe acute respiratory syndrome emphasize the threat from emerging disease carried by domestic animals. Improved control of infectious disease in domestic populations by selection for genetic resistance can make a significant contribution to the reduction of this threat. The recent development of genomic resources for the chicken (4, 22) presents new opportunities for rapid advances in our knowledge of the genetic mechanisms of resistance to infection in chickens.
Transcriptional profiling has provided a great amount of information about many host-pathogen interactions (40, 46, 54). Most of these studies monitored the changes in gene expression in host cells after contact with a specific pathogen in vitro (11, 20, 24, 27, 57). While these studies can provide information concerning cell-autonomous differences in the response to infection, in vivo analyses are required to detect differences in the complex multifactorial interactions between the pathogen and host in a real infection, especially in the coordinated behavior of the cells comprising the host immune system. In order to undertake this kind of study of the chicken, we developed a cDNA array enriched in genes expressed in active immune responses and applied it in the study of a well-defined infection of a single major target organ with infectious bursal disease virus (IBDV) in lines of chickens previously shown to differ in susceptibility (6).
IBDV is a small RNA virus (Birnaviridae) which specifically targets early B cells, especially those in a gut-associated lymphoid organ, the bursa of Fabricius, in which chicken B cells undergo an obligatory stage of development. Infection results in rapid depletion of bursal B cells. B cells in other organs, such as spleen, thymus, and cecal tonsils, are also affected. In young chicks, with small bursa and few postbursal B cells, IBDV causes an acute, short-lived infection, substantially depleting the bursal B cells. The consequent immunosuppression is an important aspect of the disease. Virus peaks from day 2 to day 5 and is essentially cleared by day 7. This provides a model of an acute, local infection with innate response, cytokine induction, and infiltration of T cells (43). Live IBDV vaccines can control infection but are imperfect, and this control is threatened by the emergence of increasingly virulent IBDV strains (9, 15, 36). A study of IBDV infection in older birds provided evidence for genetic differences in susceptibility (16). The line BrL was highly susceptible to IBDV infection, with a mortality rate of 80% 8 days after infection, despite being one of the most robust lines when facing many other diseases (5, 6). On the contrary, line 6 showed only 8% mortality 8 days after infection.
We analyzed gene expression levels in bursas of young chicks from the lines described as resistant, line 6, and susceptible, line BrL. The results provide a global view of the early changes in response to IBDV infection, from which we were able to extract components pointing to possible mechanisms accounting for differences in susceptibility.
MATERIALS AND METHODS
Construction of cDNA arrays.
The primary sources of cDNA clones for arraying were four libraries, each in pre- and postnormalized forms: from inflamed cecal tonsils of chickens infected with Eimeria tenella, an intracellular protozoan parasite, from spleens of birds infected with Marek's disease virus, from normal splenocytes activated in vitro with phytohemagglutinin for various times, and from a lipopolysaccharide-activated HD11 retrovirus-transformed macrophage cell line (3). The preparation of these libraries and a detailed analysis of the expressed sequence tag (EST) data obtained will be presented elsewhere. After filtering the EST data obtained from these libraries and clustering them with other EST data to reduce redundancy, we chose 3,456 clones for the array. These were amplified by two rounds of PCR from bacteria by using vector primers, and the products were purified (Millipore, Massachusetts) and dried under vacuum. They were supplemented with some cDNA clones from a bursal cDNA library (1) and with some cDNAs amplified with gene-specific primers, as detailed in Table S1 in the supplemental material.
Spotting of arrays was carried out by Eurogentec (Lieges, Belgium). Purified PCR products were resuspended in EGT Bioprint buffer, an SSC-sodium dodecyl sulfate-based buffer (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate) to a concentration of approximately 250 ng/μl. These were spotted in adjacent duplicates onto GAPS II-coated glass slides (Corning Inc.) by using the robotic arrayer ChipWriterPro (Bio-Rad) and SMP3 stealth quill spotting pins (Telechem Int.). Amersham Scorecard artificial cDNA and luciferase cDNA were also deposited.
Birds and IBDV infection.
One- to 2-day-old chicks were obtained from the unvaccinated flocks of inbred BrL and White Leghorn line 6 chickens maintained in isolation accommodation at the Institute for Animal Health (Compton, United Kingdom). The parent flocks were free of specified pathogens, including IBDV, chicken infectious anemia, and other immunosuppressive pathogens. An infectious dose of the virulent strain F52/70 of IBDV was administered which had been determined by titration (59) to cause severe depletion of bursal lymphocytes without evident morbidity or mortality. Chicks were infected with 25 μl of purified virus in phosphate-buffered saline (PBS) per nares and 25 μl of virus per eye. All procedures were conducted in accordance with the 1986 United Kingdom Animals (Scientific Procedures) Act and local ethical review procedures.
Experimental design.
For each of the chicken lines, BrL and 6, four chicks, 1 to 2 days old, were randomly assigned to infected and uninfected groups. Twelve birds from each line were infected with F52/70 stock by the oral-nasal route as described above. Uninfected control groups, 12 birds from each line, were kept in a separate room and mock infected with PBS. Four infected and four uninfected chickens from each line were killed at 1, 2, and 4 days after infection for recovery of the bursa. Half of each bursa was used individually for RNA isolation, and half was fixed in formol saline for immunohistology. Microarray analyses were carried out with RNAs from three infected and two uninfected birds from each line at each time point. Each of these 30 RNAs was compared to a common reference RNA in two microarrays where the fluorochromes were inverted, using a total of 60 arrays. The same reference RNA mixture was used in all analyses. It was prepared from samples of uninfected small intestine from lines CB12 (30 samples) and 15I (29 samples).
RNA isolation.
Tissues removed immediately, from chicks killed 1, 2, or 4 days after infection, and cut to less than 5-mm thickness were immersed in at least 10 volumes of RNAlater (QIAGEN). These samples were stored at 20°C for 16 to 20 h and at 4°C for up to 30 days. For longer-term storage, they were stored at −80°C after the removal of excess RNAlater. Up to 30 mg tissue was transferred to RLT lysis buffer (QIAGEN) and homogenized with a 5-mm stainless steel bead for 4 min at 20 Hz using a bead mill (MM300; Retch), and the lysate was passed through a QIAshredder column (QIAGEN). The resulting filtrate was used to prepare RNA using the RNeasy system (QIAGEN), including on-column DNase digestion, according to the manufacturer's instructions. All RNA preparations were analyzed with the lab-on-a-chip Agilent bioanalyzer (RNA 6000 LabChip kit) to confirm RNA integrity and purity (28S:18S ratio > 2.0).
Probe labeling and hybridization.
Fluorochrome-labeled cDNAs were prepared using the direct labeling method. Twenty-five micrograms of total RNA was mixed in a total volume of 40 μl containing Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT) 1× reaction buffer (Promega), 1.5 μg oligo(dT)18-21, 0.5 mM each dATP, dGTP and dTTP, 25 μM dCTP, 40 μM labeled (cyanine 3, Cy3, or cyanine 5, Cy5) dCTP (Amersham Biosciences), 10 mM dithiothreitol and 40 units of RNasin (Promega), and diethyl pyrocarbonate-treated water. The mixture was incubated for 5 min at 65°C, followed by 5 min at 42°C. Reverse transcription was initiated by the addition of another 40 units of RNasin and 200 units of M-MLV reverse transcriptase RNase H minus (Promega), and incubation continued at 42°C for 1 h. A further 200 units of M-MLV reverse transcriptase was then added, followed by another incubation of 1 h at 42°C. The reaction was stopped by incubation for 20 min at 65°C with 5 μl 50 mM EDTA, pH 8, and 2 μl 10 N NaOH.Four microliters of 5 M acetic acid was then added. The labeled probes were cleaned using the QIAquick PCR purification kit (QIAGEN), concentrated in a Speed Vac for 1 h, and rehydrated in 5 μl H2O. Prior to hybridization, Cy3- and Cy5-labeled cDNA probes were mixed, together with 50 μg of heat-denatured salmon sperm DNA, 1 μg sonicated genomic DNA, and 1.5 μg A80 oligo(A) (Sigma), in a total volume of 12.5 μl and incubated for 2 min at 95°C. Forty microliters of Eurogentec hybridization buffer was then added. A LifterSlips, 25 by 44 mm (Erie Scientific Co.), was added onto the printed area of the microarray, and the probes were injected in between. Hybridization was achieved by incubation overnight at 37°C in a humid chamber (Corning hybridization chamber). The coverslip was removed by washing the array in 0.2× SSC and 0.1% sodium dodecyl sulfate for 10 min at room temperature, with occasional agitation. After another wash in 0.2× SSC for 10 min, arrays were dried by a 5-min, 500-rpm centrifugation. Each hybridization was conducted twice, inverting the labeling of the experimental and reference RNAs with Cy3 and Cy5.
Data analysis.
Arrays were scanned using a 4100A scanner (Axon Instruments, California), and raw data were extracted using GenePixPro 3.1 software. The NIT limma (linear model of microarray analysis) version 1.1.5 from the Marray package (12, 44, 61) was used to perform background subtraction and normalize the data. All elements were present in duplicate on each array, and all analyzed RNA preparations were used on two arrays, with the fluorochrome labeling inverted between reference and experimental RNAs. Thus, each RNA preparation yielded four measurements for each array element. Preliminary examination confirmed that variation between duplicate spots and between fluorochrome-inverted arrays was much lower than that between biological replicate RNAs (always from different birds). To avoid overestimation of statistical significance, the mean of the four technical replicates for each RNA preparation was used as a single data point in subsequent analysis using the SAM (significance of microarray analysis) v1.0 program (53). This level of analysis yielded a single measurement of expression level, relative to the reference RNA, for each gene in each biological replicate. SAM was used to assess the significance of the differences between the measured expression levels between infected (n = 3) and uninfected (n = 2) birds at each time point. Two-class response analyses were used to identify differentially expressed genes with a false-positive rate (false discovery rate) below 5%. Two criteria were used to select array elements that had yielded useful data. First, it was required that the geometric mean of the fluorescence signal intensity (calculated after background subtraction) exceed a threshold for each experimental condition (mean log2(A) > 5; A = median fluorescence intensity). Secondly, it was required that significant changes of expression level between infected and uninfected birds be observed at at least one time point in each line (false discovery rate < 5%).
Quantitative real-time PCR.
RNA samples used for the microarrays were analyzed by quantitative RT-PCR (qRT-PCR) experiments to validate microarray data. Table 1 and Fig. 1 and 3 show a correlation between the data derived from microarray analysis and from qRT-PCR of three genes explicitly discussed in Results. Single-stranded cDNA was synthesized from 2 μg of total RNA using M-MLV reverse transcriptase and primer dT23 VN from the ProtoScript first-strand cDNA synthesis kit (Biolabs). Gene-specific primers were designed for the immunoglobulin J (IgJ) chain, interleukin-8 (IL-8), LITAF, IL-6, IL-2, alpha interferon (IFN-α), IFN-γ, VP2, and the maternal G10 transcript (see Table S1 in the supplemental material). The last gene was chosen as an internal control because it had the lowest standard deviation calculated from all of the different conditions analyzed in the microarray experiment. We therefore used it to correct the differences in cDNA concentration of the different samples.
TABLE 1.
Comparison between microarray and qRT-PCR results
| Gene | Method | Line BrL ratio (infected/uninfected)a
|
Line 6 ratio (infected/uninfected)a
|
||||
|---|---|---|---|---|---|---|---|
| 1 dpi | 2 dpi | 4 dpi | 1 dpi | 2 dpi | 4 dpi | ||
| IL-6 gene | Microarray | 0.1 | 2.0 | 1.1b | 2.5b | 2.7b | 1.5b |
| qRT-PCR | 0.8 | 3.9 | 0.8 | 1.8 | 2.5 | 0.2 | |
| IgJ gene | Microarray | −0.1 | 0.0 | 0.7 | 0.2 | −3.2b | −1.9b |
| qRT-PCR | 0.0 | −0.4 | 0.3 | 0.3 | −6.0 | −3.2 | |
| IL-8 gene | Microarray | −0.8 | 4.9b | 1.6b | 2.6b | 3.8b | 2.8b |
| qRT-PCR | 0.5 | 4.6 | 2.5 | 3.3 | 5.8 | 0.2 | |
Numbers correspond to the log2 values of the ratio calculated between infected and uninfected birds at each time point. dpi, days postinfection.
Significant differences in expression between infected and uninfected birds (false discovery rate < 0.05).
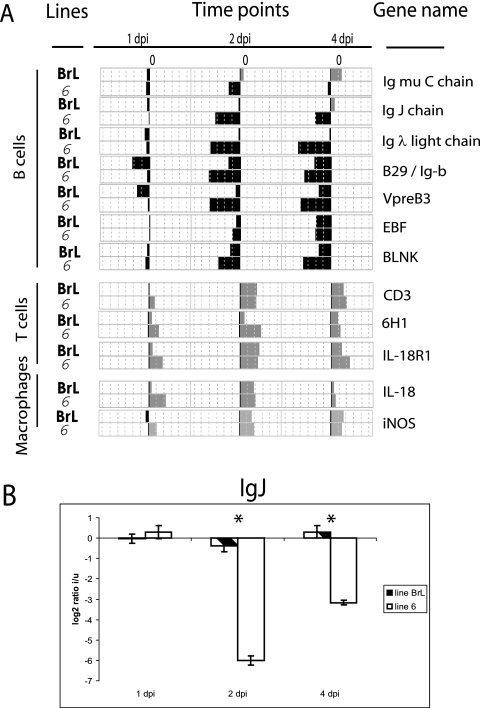
FIG. 1.
Changes in the cellular composition of the bursa after infection. (A) Comparison of changes in gene expression at 1, 2, and 4 days postinfection (dpi) with IBDV in the BrL and 6 lines. At each time point, the bars represent the ratios calculated between infected and uninfected birds. The length of each bar is proportional to the log2 value of the ratio. The positive and negative ratios correspond to a light-gray bar and a dark bar, respectively. The name of each gene is indicated. (B) Quantitative RT-PCR analysis of IgJ gene expression at 1, 2, and 4 days after infection (dpi). The log2 values of the ratios calculated between infected (i) and uninfected (u) birds are shown for each time point. BrL and 6 lines are represented by streaked and white bars, respectively. Significant differences in expression between chicken lines at a particular time point (P ≤ 0.05) are indicated by an asterisk. Errors bars indicate standard errors of the means.
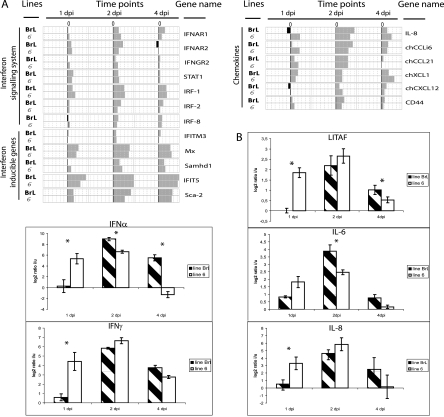
FIG. 3.
Transcriptional patterns of cytokine genes. (A) Comparison of changes in gene expression at 1, 2, and 4 days postinfection (dpi) with IBDV in the BrL and 6 lines. (B) Quantitative RT-PCR analysis of the IFN-α, IFN-γ, IL-6, IL-8, and LITAF gene expression at 1, 2, and 4 days after infection (dpi). The log2 values of the ratios calculated between infected (i) and uninfected (u) birds are shown for each time point. BrL and 6 lines are represented by streaked and white bars, respectively. Significant differences in expression between chicken lines at a particular time point (P ≤ 0.05) are indicated by an asterisk. Errors bars indicate standard errors of the means.
Prior to quantification, a step of amplification (Applera) was carried out using each pair of primers. The PCR products were purified using the High Pure PCR product purification kit (Roche). A series of five 10-fold dilutions of this product was used as a template to generate a standard curve for each gene in the quantification experiments. Quantification was performed on a LightCycler (Roche) using a LightCycler FastStart DNA Master Plus SYBR green I kit (Roche). The cycling conditions were 10 min at 95°C, 40 cycles of 10 s at 95°C, 30 s at annealing temperature (see Table S1 in the supplemental material), and 30 s at 72°C, followed by a final step of melting curve analysis wherein the annealing temperature increases up to 95°C. All samples were assayed in duplicate. The cycle threshold values were used to plot a standard curve in which the cycle threshold decreased in linear proportion with the log of the template concentration. Each experimental sample was compared to the standard curve in order to determine the starting amount of each particular transcript. Results were adjusted for total cDNA content by using the G10 internal standard. Melting curves were examined to verify the amplification of a specific target sequence.
Immunohistochemistry.
The monoclonal antibody (MAb) AV20 recognizes a monomorphic determinant on the Bu-1 molecule, expressed by B cells (42, 52). The R63 MAb (45) recognizing the VP2 protein was used to identify IBDV-infected cells. Sections of tissue frozen in Tissue-Tek OCT (Miles Inc., Indiana) were fixed in acetone, treated with 0.03% H2O2 in PBS to inactivate endogenous peroxidase, and washed three times in PBS before incubation with primary Abs. Bound Ab was detected using the Vectastain Elite ABC kit for mouse IgG and NovaRED substrate 5 (Vector Laboratories, United Kingdom), as described in detail elsewhere (59). Sections were counterstained with hematoxylin and permanently mounted for microscopy.
Microarray data accession number.
This microarray experiment has been assigned accession number E-MEXP-756 in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress).
RESULTS
Gene expression during infection by IBDV.
Immunologically focused cDNA arrays were used to measure the relative expression levels of genes involved in active immune responses after IBDV infection of two lines, 6 and BrL, the former described as resistant to the virus and the latter as susceptible (6). Birds were infected between 1 and 2 days after hatch, and the bursas were removed at days 1, 2, and 4 after infection. Since this tissue undergoes developmental changes during the period of infection, control samples were taken from uninfected birds at each time point. A total of 30 RNA preparations and a pool of RNA extracted from various tissues as a reference were used for microarray hybridizations.
The analysis after the microarray hybridizations (see Materials and Methods) revealed that at 2 and 4 days after infection, significant changes were evident in the expression of many genes. In general, the patterns at 2 and 4 days are very similar, with the levels of expression of the same genes being up or down at both time points. One day after infection, significant changes in gene expression were few.
Changes in the cellular composition of the bursa after infection.
Some changes in gene expression were the consequences of known changes in the cellular composition of the infected tissue, including the destruction of B cells and the influx of T cells and macrophages (59). The destruction of infected B cells was reflected in the reduction of relative levels of Ig genes and other B-cell-specific genes, B29, VpreB3, EBF, and BLNK, in both lines (Fig. 1A). The reduction was greater in line 6 than in line BrL birds. This difference between the lines was also evident in sections of bursas stained for the B-cell marker Bu-1 (Fig. 2, left panel) and was confirmed by quantitative PCR for the IgJ chain gene (Fig. 1B).
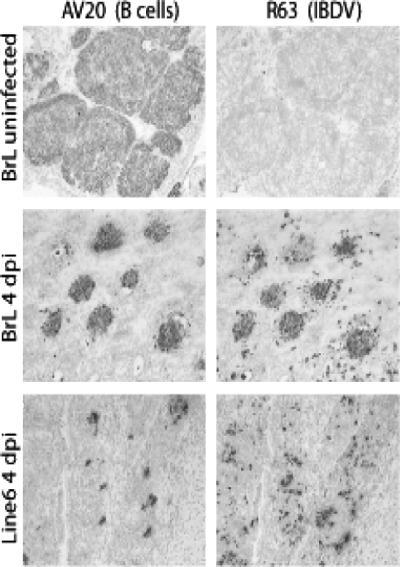
FIG. 2.
Immunohistological analysis of bursa at 4 days after infection. Paired left and right panels show serial sections stained with either AV20 MAb to reveal surviving B cells or R63 MAb to reveal viral antigen. The upper four panels are sections from BrL birds, uninfected at the top and infected below. The bottom panels are sections from an infected White Leghorn line 6 bird.
Infiltration of T lymphocytes and macrophages into the bursa is observed early after infection (59). These processes were also evident from the gene expression data, for example by the up-regulation of CD3, 6H1/P2Y5, a G protein-coupled receptor induced in activated T cells (29, 58), and IL-18R1 (Fig. 1A). Furthermore, the overexpression of IL-18R1 suggests the influx of NK cells, and increases in IL-18 and inducible nitric oxide synthase (iNOS) mRNAs reveal the appearance of activated macrophages (Fig. 1A).
Transcriptional pattern of immune response genes.
Many genes involved in the immune responses were up-regulated after infection, mostly with a high level of expression at days 2 and 4 after infection, and they can be described in several categories.
They included genes of the antiviral interferon system, IFN-α and IFN-γ, IFNAR1, IFNAR2, IFNGR2, and STAT1, involved in the IFN signaling pathway; interferon regulatory factors 1, 2, and 8; and the IFN-inducible genes Mx, Sca-2 (LY6E.2), IFITM3, Samhd1, and IFIT5. A minority of these, IFNAR2, STAT1, IRF-1, Mx, IFIT5, and Sca-2, were up-regulated as early as the first day after infection (Fig. 3A).
Also up-regulated were cytokines IL-18 and IL-6, which are involved in the switch from innate to acquired immunity (28), LITAF, a factor involved in the regulation of several inflammatory cytokines (51), and the chemokines IL-8, chCCLi6 (MIP-1β), chXCL1, chCXCL12, and chCCL21 (Fig. 3A).
Genes whose products are implicated in the innate immune response.
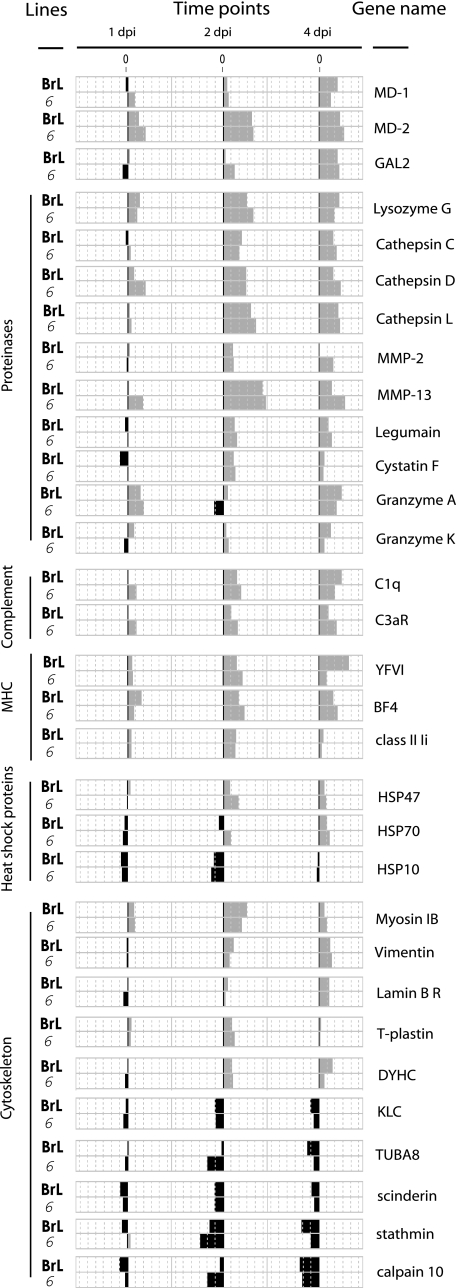
MD-1 and MD-2, associated with Toll-like receptors, and gallinacin 2 (GAL2), a member of the β-defensin family (21), were up-regulated, as were genes involved in the function of lymphocytes, the cell surface glycoprotein CD44, and several proteases, including lysozyme G, lysosomal cathepsins C, D, and L, matrix metalloproteinases MMP-2 and MMP-13, legumain, cystatin F, and granzymes A and K (Fig. 4).
FIG. 4.
Transcriptional profiles of immune response genes. Comparison of changes in gene expression at 1, 2, and 4 days postinfection (dpi) with IBDV in the BrL and 6 lines.
Increased expression was also seen for two complement components, opsonin C1q, which promotes phagocytosis, and the anaphylatoxin C3a receptor, which is involved in chemotaxis of leukocytes (18). The observed up-regulation (Fig. 4) of two MHC class I genes and the class II-associated invariant chain genes may correspond to the influx of immune cells into the bursa and/or to a regulation of their expression by type I (alpha/beta) and type II (gamma) IFNs.
Stress-response genes HSP70 and HSP47, key “danger” signals of the innate immune system (Fig. 4), were up-regulated, while the transcription of the gene for HSP10, which is involved in the modulation of inflammatory processes (26), showed a down-regulation with a minimum of expression 2 days after infection (Fig. 4).
Finally, genes encoding cytoskeleton elements myosin IB, vimentin, lamin B receptor, and T-plastin, as well as a component of the dynein motor, the dynein heavy chain, showed increased expression. In contrast, kinesin light chain, tubulin alpha 8 (TUBA8), and genes involved in the destabilization of cytoskeletal elements, scinderin, stathmin, and calpain 10 (13, 23), were down-regulated after infection (Fig. 4). Drastic effects of viral infection on cell morphology, along with with changes in the cytoskeleton, have been reported before (37).
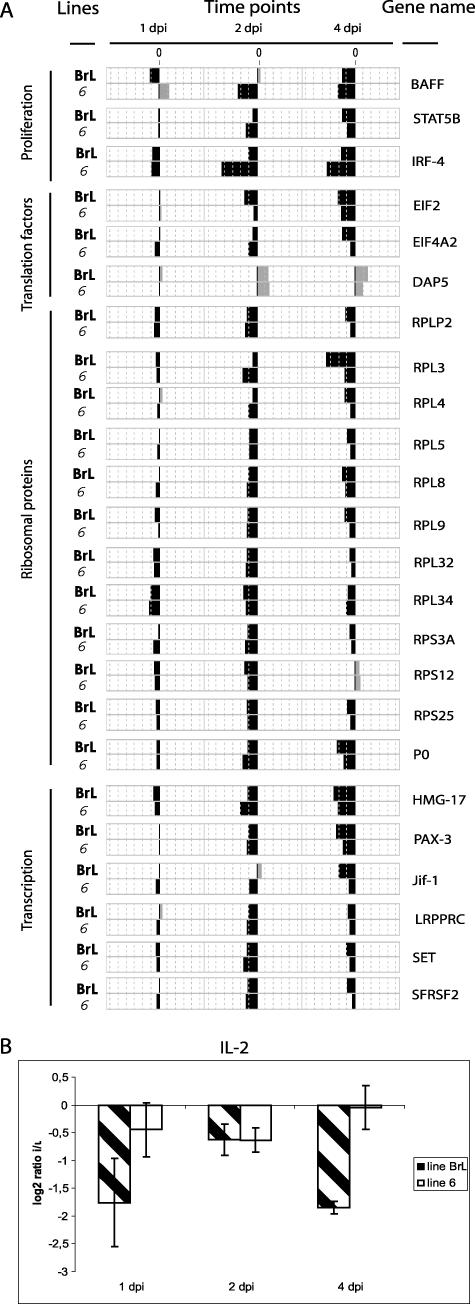
On the other hand, a number of genes involved in the proliferation of B and T cells were down-regulated in the bursa by the infection. Among these genes were those encoding the BAFF cytokine, a member of the tumor necrosis factor family that promotes B-cell survival and typically highly expressed by bursal B cells (Fig. 5A) (30), IL-2 (Fig. 5B), STAT5b, one of the components of the IL-2 signaling pathway (32), and IRF-4 (Fig. 5A) (34, 60).
FIG. 5.
Transcriptional profiles of down-regulated immune response genes. (A) Comparison of changes in gene expression at 1, 2, and 4 days postinfection (dpi) with IBDV in the BrL and 6 lines. (B) Quantitative RT-PCR analysis of the IL-2 gene expression at 1, 2, and 4 days after infection (dpi). The log2 values of the ratios calculated between infected (i) and uninfected (u) birds are shown for each time point. BrL and 6 lines are represented by streaked and white bars, respectively. Errors bars indicate standard errors of the means.
Down-regulation of EIF2 and EIF4A2, two factors involved in the early steps of protein synthesis, and of the genes encoding ribosomal proteins, as well as the up-regulation of DAP5 (EIF4G2) (Fig. 5A), a general repressor of translation, was expected since one of the major components of the antiviral response is to target the translational machinery and to disrupt the protein synthesis during the course of infection. The expression of genes involved in the transcription processes, those encoding HMG-17, PAX-3, Jif-1, LRPPRC, SET, and the splicing factor SFRS2, was also likely reduced by the virus after infection (Fig. 5A).
Differences in gene expression in resistant and susceptible lines.
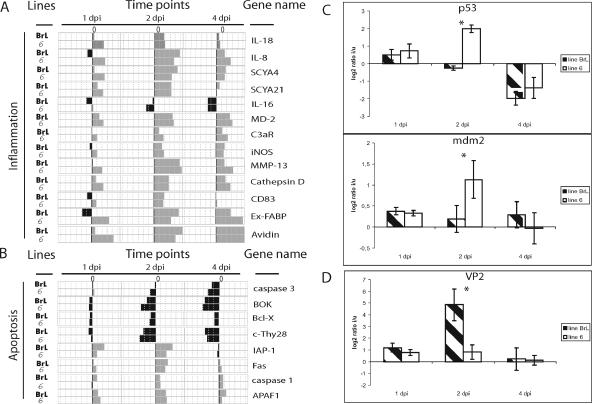
In a comparison of the relative infection-induced changes in gene expression levels in the two chicken lines at different times following infection, relatively few differences were detected at days 2 and 4. However, significant interline differences were detected (using SAM limma from the Marray package; see Materials and Methods) 1 day after infection in genes involved in the inflammatory response, which are up-regulated only in line 6, as shown in Fig. 6A.
FIG. 6.
Analysis of the difference of gene expression during response to infection by IBDV in inbred resistant and susceptible lines. (A) Transcriptional profiles of inflammatory response genes and comparison of changes in gene expression at 1, 2, and 4 days postinfection (dpi) with IBDV in the BrL and 6 lines. (B) Transcriptional profiles of apoptosis-related genes. (C) Quantitative RT-PCR analysis of p53 and mdm2 gene expression at 1, 2, and 4 days after infection (dpi). The log2 values of the ratios calculated between infected (i) and uninfected (u) birds are shown for each time point. BrL and 6 lines are represented by streaked and white bars, respectively. (D) Virus load all along the infection in lines 6 and BrL. Quantitative RT-PCR analysis of VP2 gene expression at 1 and 2 days after infection (dpi). Significant differences in expression between chicken lines at a particular time point (P ≤ 0.05) are indicated by an asterisk. Errors bars indicate standard errors of the means.
These were mostly indicators of the inflammatory process. They included the genes for IFN-α, IFN-γ, the proinflammatory cytokines IL-2, IL-18, and IL-6, the chemokines IL-8, chCCLi6, and chCCL21, and LITAF. IL-16, which is generated concomitantly or sequentially with proinflammatory factors to attenuate the inflammatory process (33), was also upregulated at day 1 in the line 6 birds. Other mediators of inflammation showing these line-restricted early changes included MD-2, which has been proposed to play a role in organ inflammation during sepsis (39), C3aR, the receptor of C3a, one of the most potent inflammatory molecules involved in chemotaxis (48), iNOS, and two proteases secreted at inflammatory sites, MMP-13 and cathepsin D. CD83, a marker of mature activated dendritic cells (7), Ex-FABP, and avidin, which show increased expression in response to inflammatory stimuli (14, 31), were also expressed at increased levels early in line 6 (Fig. 6A).
IBDV infection induces apoptosis in chicken embryo and tissue culture cells (17, 41, 55, 56). Therefore, we examined the transcriptional level of the genes which contribute to apoptosis in the two lines after infection. Proapoptotic factors, such as caspase 3, BOK (Bcl-2-related ovarian killer), Bcl-X, and cThy28, suggested to be implicated in spontaneous apoptotic cell death in thymus (10), were down-regulated (Fig. 6B). Furthermore, the apoptosis inhibitor IAP1, a suppressor of mammalian cell apoptosis (49), was up-regulated (Fig. 6B). However, conversely, the patterns of Fas, caspase-1, APAF1, involved in the initiation/amplification of apoptosis (8), and galectin-2, shown to induce apoptosis in activated human T cells (47), showed an increased expression, with a peak at day 2 (Fig. 6B).
These contradictory results prompted us to investigate whether p53 is activated in virally infected birds to induce an apoptotic response. An important role of p53 in innate antiviral host defenses, enhanced by IFN-α/β, has been reported, and the p53 gene is transcriptionally induced by these cytokines (38, 50).
Therefore, we examined the p53 gene expression pattern in uninfected birds and during the time course of infection in the two lines. There was a significant difference between the two lines at day 2 after infection, with overexpression of the p53 gene only in line 6. Four days after the infection, the gene was repressed in both lines. The expression level of p53 in uninfected birds was identical at each time point in both lines (data not shown).
To confirm a functional consequence of differential p53 expression, we looked at the transcriptional pattern of mdm2, a target gene of p53 (2). No changes were observed in the level of transcription of the mdm2 gene in line BrL at any of the three chosen time points, but up-regulation of this gene was evident in line 6 at day 2 (Fig. 6C), consistent with the observed increase of p53.
Virus load during infection in lines 6 and BrL.
Activation in line 6 of the inflammation processes at day 1 and of p53 and its proapoptotic target gene at day 2 after infection suggested that these events may limit the virus replication in this line at the early stage of infection. We used a qRT-PCR assay to test the amount of viral RNA in the two lines. A significant difference between the lines was noticeable 2 days after infection, with a higher VP2 RNA level in line BrL (Fig. 6D). The level was down at day 4 in both lines, although protein antigen was still detectable at levels that followed the relative amounts of RNA at day 2 (Fig. 2).
DISCUSSION
Naturally available genetic resistance to pathogens is a potent resource for improvement of the control and prevention of disease. Genetic resistance is generally a complex multigenic trait, usually involving the immune system and its interactions with physiologic and environmental factors. Transcriptome analysis, using microarray technologies, provides a global approach to the description of such complex biological phenomena. It offers a rapid and systematic means to generate data required for the understanding of responses to infection and for the identification of individual genes whose products may be responsible for genetically determined resistance in specific infection models.
The construction of an immune system-targeted gene array for the chicken, and its use in analysis of differences in gene expression in affected tissues from genetically resistant and susceptible animals infected with IBDV, has provided a large data set from which we have been able to derive some potentially valuable observations. Gene expression data, obtained from whole-bursa samples, infected and uninfected, were highly reproducible, yielding strong statistical support for differences in the expression levels of many genes. This reproducibility, supported by independent qPCR data for example genes, validated the methodology for detecting significant changes in gene expression, even in the analysis of changes in complex tissues.
The major differences in infected tissue were those expected from the depletion of B cells and the influx of T cells and macrophages that were known to occur. Indeed, the infection of chicks with IBDV caused B-lymphocyte depletion in both lines and, in line 6, resistant to IBDV-induced pathology in older birds (6), resulted in the loss of almost all B cells. The results also revealed the activation of a variety of antiviral host defenses early after infection: activation of innate immune responses, modulation of host transcription and protein processing, interferons, complement, natural killer cells, activation of macrophages, and inhibition of lymphocyte proliferation. These indicated the induction of an important local inflammatory response by the host after the infection in both lines of chickens.
Our ultimate aim was to determine whether differences in response to infection, detectable by differences of gene expression between the two lines of birds, could be used to make inferences concerning the mechanisms of the previously reported differences in disease resistance. The two array elements with the largest differences in response to infection between the lines were endogenous retroviral sequences. This probably reflects differences in the presence and locations of these genomic passengers in the two lines. Transcription is clearly enhanced in response to infection, raising the possibility that the products might have a role in immune system activation or regulation, although this remains highly speculative. While the levels of these sequences were very similar in all of the inbred line 6 chicks, the outbred line BrL birds fell into two groups with very different levels of transcribed retroviral sequences, suggesting that the highly transcribed endogenous retroviral elements may be segregating in these chickens (see Fig. S1 and Table S2 in the supplemental material).
There were other striking differences between the lines of birds, even at the earliest time point, 1 day after infection. The proinflammatory and inflammatory genes, those encoding IFNs, interleukins, chemokines, cathepsin D, MMP-13, and iNOS, were specifically up-regulated in the resistant, line 6 chicks compared with the line BrL chicks 1 day after infection. Thus, the virus induced a very early inflammatory response in the resistant line, which was delayed in the susceptible line. Following the observation of differences in apoptosis-related genes, analysis of p53 gene expression revealed increased expression in line 6 at 2 days after infection that was not observed in the susceptible line. This suggests that the earlier inflammatory response in the line 6 birds is followed by active p53-dependent apoptosis and thus the more rapid and extensive B-cell depletion that is clearly evident from the gene expression data as well as from immunohistology. It is possible that the increased apoptosis is the consequence of more-rapid inflammation, although this causal link is not established by these data. The more rapid removal of the target B cells could then serve to limit viral replication in the line 6 birds. Consistent with this hypothesis, we did detect less viral RNA in the line 6 birds. Using neonatal infections, we saw more-extensive B-cell depletion in the line 6 birds, although this line was previously reported to be less susceptible to the pathological effects of infection in older birds (6). At first sight, this appears to suggest a paradoxical age-dependent inversion of susceptibilities. However, neonatally infected birds do not suffer from the acute effects seen in older birds, which provided the parameters of susceptibility. Furthermore, our results with neonatal birds allow us to suggest a model for the difference in the outcome of infection in older birds: that a more rapid inflammatory response and more-rapid activation of apoptotic pathways in target B cells, as seen in our experiments, also limit the extent of viral replication and thus the pathological effects of IBDV infection in the resistant line 6 birds.
Many genes represented on the arrays are of tentatively identified or completely unknown function. This includes many of those which show differential expression, such as the anonymous gene mdvn119_a07_r2, which is up-regulated at days 1 and 2 but back to normal levels on day 4 (see Fig. S1 in the supplemental material). Thus, the data set we have produced may yield further insights as the accelerating effort to annotate the chicken genome gathers pace. Indeed, the observation of changes in expression during viral infection may contribute to the functional annotation of these genes.
Supplementary Material
Acknowledgments
This research was supported by grants from the European Commission to R.Z. (KA5-QLRT-CT99-1591 Chicken IMAGE). D.R.W. benefited from a BBSRC studentship.
Additional clones of the array were kindly provided by P. Kaiser, J. M. Buerstedde, and N. Bumstead. We thank B. Bed'Hom and F. Dautry for helpful discussion. The late Nat Bumstead was instrumental in the initiation of this work.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abdrakhmanov, I., D. Lodygin, P. Geroth, H. Arakawa, A. Law, J. Plachy, B. Korn, and J. M. Buerstedde. 2000. A large database of chicken bursal ESTs as a resource for the analysis of vertebrate gene function. Genome Res. 10:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beug, H., A. von Kirchbach, G. Doderlein, J. F. Conscience, and T. Graf. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375-390. [DOI] [PubMed] [Google Scholar]
- 4.Boardman, P. E., J. Sanz-Ezquerro, I. M. Overton, D. W. Burt, E. Bosch, W. T. Fong, C. Tickle, W. R. Brown, S. A. Wilson, and S. J. Hubbard. 2002. A comprehensive collection of chicken cDNAs. Curr. Biol. 12:1965-1969. [DOI] [PubMed] [Google Scholar]
- 5.Bumstead, N., and B. J. Millard. 1992. Variation in susceptibility of inbred lines of chickens to seven species of Eimeria. Parasitology 104:407-413. [DOI] [PubMed] [Google Scholar]
- 6.Bumstead, N., R. L. Reece, and J. K. Cook. 1993. Genetic differences in susceptibility of chicken lines to infection with infectious bursal disease virus. Poult. Sci. 72:403-410. [DOI] [PubMed] [Google Scholar]
- 7.Cao, W., S. H. Lee, and J. Lu. 2005. CD83 is preformed inside monocytes, macrophages and dendritic cells, but it is only stably expressed on activated dendritic cells. Biochem. J. 385:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecconi, F., G. Alvarez-Bolado, B. I. Meyer, K. A. Roth, and P. Gruss. 1998. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 94:727-737. [DOI] [PubMed] [Google Scholar]
- 9.Chettle, N., J. C. Stuart, and P. J. Wyeth. 1989. Outbreak of virulent infectious bursal disease in East Anglia. Vet. Rec. 125:271-272. [DOI] [PubMed] [Google Scholar]
- 10.Compton, M. M., J. M. Thomson, and A. H. Icard. 2001. The analysis of cThy28 expression in avian lymphocytes. Apoptosis 6:299-314. [DOI] [PubMed] [Google Scholar]
- 11.Coussens, P. M., N. Verman, M. A. Coussens, M. D. Elftman, and A. M. McNulty. 2004. Cytokine gene expression in peripheral blood mononuclear cells and tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis: evidence for an inherent proinflammatory gene expression pattern. Infect. Immun. 72:1409-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudoit, S., and Y. H. Yang. 2003. Bioconductor R packages for exploratory analysis and normalization of cDNA microarray data, p. 73-101. In G. Parmigiani, E. S. Garrett, R. A. Irizarry, and S. L. Zeger (ed.), The analysis of gene expression data: methods and software, Springer, New York, N.Y.
- 13.Dumitrescu Pene, T., S. D. Rose, T. Lejen, M. G. Marcu, and J. M. Trifaro. 2005. Expression of various scinderin domains in chromaffin cells indicates that this protein acts as a molecular switch in the control of actin filament dynamics and exocytosis. J. Neurochem. 92:780-789. [DOI] [PubMed] [Google Scholar]
- 14.Elo, H. A., M. S. Kulomaa, and P. J. Tuohimaa. 1979. Avidin induction by tissue injury and inflammation in male and female chickens. Comp. Biochem. Physiol. B 62:237-240. [DOI] [PubMed] [Google Scholar]
- 15.Eterradossi, N., C. Gauthier, I. Reda, S. Comte, G. Rivallan, D. Toquin, C. de Boisseson, J. Lamande, V. Jestin, Y. Morin, C. Cazaban, and P. M. Borne. 2004. Extensive antigenic changes in an atypical isolate of very virulent infectious bursal disease virus and experimental clinical control of this virus with an antigenically classical live vaccine. Avian Pathol. 33:423-431. [DOI] [PubMed] [Google Scholar]
- 16.Fadly, A. M., and R. L. Witter. 1986. Resistance of line 6(3) chickens to reticuloendotheliosis-virus-induced bursa-associated lymphomas. Int. J. Cancer 38:139-143. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Arias, A., S. Martinez, and J. F. Rodriguez. 1997. The major antigenic protein of infectious bursal disease virus, VP2, is an apoptotic inducer. J. Virol. 71:8014-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasque, P. 2004. Complement: a unique innate immune sensor for danger signals. Mol. Immunol. 41:1089-1098. [DOI] [PubMed] [Google Scholar]
- 19.Girard-Santosuosso, O., P. Menanteau, M. Duchet-Suchaux, F. Berthelot, F. Mompart, J. Protais, P. Colin, J. F. Guillot, C. Beaumont, and F. Lantier. 1998. Variability in the resistance of four chicken lines to experimental intravenous infection with Salmonella enteritidis phage type 4. Avian Dis. 42:462-469. [PubMed] [Google Scholar]
- 20.Granucci, F., P. R. Castagnoli, L. Rogge, and F. Sinigaglia. 2001. Gene expression profiling in immune cells using microarray. Int. Arch. Allergy Immunol. 126:257-266. [DOI] [PubMed] [Google Scholar]
- 21.Harwig, S. S., K. M. Swiderek, V. N. Kokryakov, L. Tan, T. D. Lee, E. A. Panyutich, G. M. Aleshina, O. V. Shamova, and R. I. Lehrer. 1994. Gallinacins: cysteine-rich antimicrobial peptides of chicken leukocytes. FEBS Lett. 342:281-285. [DOI] [PubMed] [Google Scholar]
- 22.Hillier, L. W., W. Miller, E. Birney, W. Warren, R. C. Hardison, C. P. Ponting, P. Bork, D. W. Burt, M. A. Groenen, M. E. Delany, J. B. Dodgson, A. T. Chinwalla, P. F. Cliften, S. W. Clifton, K. D. Delehaunty, C. Fronick, R. S. Fulton, T. A. Graves, C. Kremitzki, D. Layman, V. Magrini, J. D. McPherson, T. L. Miner, P. Minx, W. E. Nash, M. N. Nhan, J. O. Nelson, L. G. Oddy, C. S. Pohl, J. Randall-Maher, S. M. Smith, J. W. Wallis, S. P. Yang, M. N. Romanov, C. M. Rondelli, B. Paton, J. Smith, D. Morrice, L. Daniels, H. G. Tempest, L. Robertson, J. S. Masabanda, D. K. Griffin, A. Vignal, V. Fillon, L. Jacobbson, S. Kerje, L. Andersson, R. P. Crooijmans, J. Aerts, J. J. van der Poel, H. Ellegren, R. B. Caldwell, S. J. Hubbard, D. V. Grafham, A. M. Kierzek, S. R. McLaren, I. M. Overton, H. Arakawa, K. J. Beattie, Y. Bezzubov, P. E. Boardman, J. K. Bonfield, M. D. Croning, R. M. Davies, M. D. Francis, S. J. Humphray, C. E. Scott, R. G. Taylor, C. Tickle, W. R. Brown, J. Rogers, J. M. Buerstedde, S. A. Wilson, L. Stubbs, I. Ovcharenko, L. Gordon, S. Lucas, M. M. Miller, H. Inoko, T. Shiina, J. Kaufman, J. Salomonsen, K. Skjoedt, G. K. Wong, J. Wang, B. Liu, J. Wang, J. Yu, H. Yang, M. Nefedov, M. Koriabine, P. J. Dejong, L. Goodstadt, C. Webber, N. J. Dickens, I. Letunic, M. Suyama, D. Torrents, C. von Mering, et al. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695-716. [DOI] [PubMed] [Google Scholar]
- 23.Honnappa, S., B. Cutting, W. Jahnke, J. Seelig, and M. O. Steinmetz. 2003. Thermodynamics of the Op18/stathmin-tubulin interaction. J. Biol. Chem. 278:38926-38934. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Q., D. Liu, P. Majewski, L. C. Schulte, J. M. Korn, R. A. Young, E. S. Lander, and N. Hacohen. 2001. The plasticity of dendritic cell responses to pathogens and their components. Science 294:870-875. [DOI] [PubMed] [Google Scholar]
- 25.Ignjatovic, J., R. Reece, and F. Ashton. 2003. Susceptibility of three genetic lines of chicks to infection with a nephropathogenic T strain of avian infectious bronchitis virus. J. Comp. Pathol 128:92-98. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, B. J., T. T. Le, C. A. Dobbin, T. Banovic, C. B. Howard, M. Flores Fde, D. Vanags, D. J. Naylor, G. R. Hill, and A. Suhrbier. 2005. Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. J. Biol. Chem. 280:4037-4047. [DOI] [PubMed] [Google Scholar]
- 27.Jones, J. O., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77:1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, S. A. 2005. Directing transition from innate to acquired immunity: defining a role for IL-6. J. Immunol. 175:3463-3468. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan, M. H., D. I. Smith, and R. S. Sundick. 1993. Identification of a G protein coupled receptor induced in activated T cells. J. Immunol. 151:628-636. [PubMed] [Google Scholar]
- 30.Koskela, K., P. Nieminen, P. Kohonen, H. Salminen, and O. Lassila. 2004. Chicken B-cell-activating factor: regulator of B-cell survival in the bursa of Fabricius. Scand. J. Immunol. 59:449-457. [DOI] [PubMed] [Google Scholar]
- 31.Lechmann, M., E. Zinser, A. Golka, and A. Steinkasserer. 2002. Role of CD83 in the immunomodulation of dendritic cells. Int. Arch. Allergy Immunol. 129:113-118. [DOI] [PubMed] [Google Scholar]
- 32.Lin, J. X., and W. J. Leonard. 2000. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene 19:2566-2576. [DOI] [PubMed] [Google Scholar]
- 33.Lynch, E. L., F. F. Little, K. C. Wilson, D. M. Center, and W. W. Cruikshank. 2003. Immunomodulatory cytokines in asthmatic inflammation. Cytokine Growth Factor Rev. 14:489-502. [DOI] [PubMed] [Google Scholar]
- 34.Matsuyama, T., A. Grossman, H. W. Mittrucker, D. P. Siderovski, F. Kiefer, T. Kawakami, C. D. Richardson, T. Taniguchi, S. K. Yoshinaga, and T. W. Mak. 1995. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE). Nucleic Acids Res. 23:2127-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrisroe, L. S. 1976. Genetic resistance to Marek's disease. Aust. Vet. J. 52:215-219. [DOI] [PubMed] [Google Scholar]
- 36.Parede, L. H., S. Sapats, G. Gould, M. Rudd, S. Lowther, and J. Ignjatovic. 2003. Characterization of infectious bursal disease virus isolates from Indonesia indicates the existence of very virulent strains with unique genetic changes. Avian Pathol. 32:511-518. [DOI] [PubMed] [Google Scholar]
- 37.Ploubidou, A., and M. Way. 2001. Viral transport and the cytoskeleton. Curr. Opin. Cell Biol. 13:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porta, C., R. Hadj-Slimane, M. Nejmeddine, M. Pampin, M. G. Tovey, L. Espert, S. Alvarez, and M. K. Chelbi-Alix. 2005. Interferons alpha and gamma induce p53-dependent and p53-independent apoptosis, respectively. Oncogene 24:605-615. [DOI] [PubMed] [Google Scholar]
- 39.Pugin, J., S. Stern-Voeffray, B. Daubeuf, M. A. Matthay, G. Elson, and I. Dunn-Siegrist. 2004. Soluble MD-2 activity in plasma from patients with severe sepsis and septic shock. Blood 104:4071-4079. [DOI] [PubMed] [Google Scholar]
- 40.Ricciardi-Castagnoli, P., and F. Granucci. 2002. Interpretation of the complexity of innate immune responses by functional genomics. Nat. Rev. Immunol. 2:881-889. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Lecompte, J. C., R. Nino-Fong, A. Lopez, R. J. Frederick Markham, and F. S. Kibenge. 2005. Infectious bursal disease virus (IBDV) induces apoptosis in chicken B cells. Comp. Immunol. Microbiol. Infect. Dis. 28:321-337. [DOI] [PubMed] [Google Scholar]
- 42.Rothwell, C. J., L. Vervelde, and T. F. Davison. 1996. Identification of chicken Bu-1 alloantigens using the monoclonal antibody AV20. Vet. Immunol. Immunopathol. 55:225-234. [DOI] [PubMed] [Google Scholar]
- 43.Sharma, J. M., I. J. Kim, S. Rautenschlein, and H. Y. Yeh. 2000. Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Dev. Comp. Immunol. 24:223-235. [DOI] [PubMed] [Google Scholar]
- 44.Smyth, G. K., and T. Speed. 2003. Normalization of cDNA microarray data. Methods 31:265-273. [DOI] [PubMed] [Google Scholar]
- 45.Snyder, D. B., D. P. Lana, B. R. Cho, and W. W. Marquardt. 1988. Group and strain-specific neutralization sites of infectious bursal disease virus defined with monoclonal antibodies. Avian Dis. 32:527-534. [PubMed] [Google Scholar]
- 46.Staudt, L. M., and P. O. Brown. 2000. Genomic views of the immune system. Annu. Rev. Immunol. 18:829-859. [DOI] [PubMed] [Google Scholar]
- 47.Sturm, A., M. Lensch, S. Andre, H. Kaltner, B. Wiedenmann, S. Rosewicz, A. U. Dignass, and H. J. Gabius. 2004. Human galectin-2: novel inducer of T cell apoptosis with distinct profile of caspase activation. J. Immunol. 173:3825-3837. [DOI] [PubMed] [Google Scholar]
- 48.Takabayashi, T., S. Shimizu, B. D. Clark, M. Beinborn, J. F. Burke, and J. A. Gelfand. 2004. Interleukin-1 upregulates anaphylatoxin receptors on mononuclear cells. Surgery 135:544-554. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi, R., Q. Deveraux, I. Tamm, K. Welsh, N. Assa-Munt, G. S. Salvesen, and J. C. Reed. 1998. A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 273:7787-7790. [DOI] [PubMed] [Google Scholar]
- 50.Takaoka, A., S. Hayakawa, H. Yanai, D. Stoiber, H. Negishi, H. Kikuchi, S. Sasaki, K. Imai, T. Shibue, K. Honda, and T. Taniguchi. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424:516-523. [DOI] [PubMed] [Google Scholar]
- 51.Tang, X., D. L. Marciano, S. E. Leeman, and S. Amar. 2005. LPS induces the interaction of a transcription factor, LPS-induced TNF-alpha factor, and STAT6(B) with effects on multiple cytokines. Proc. Natl. Acad. Sci. USA 102:5132-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tregaskes, C. A., N. Bumstead, T. F. Davison, and J. R. Young. 1996. Chicken B-cell marker chB6 (Bu-1) is a highly glycosylated protein of unique structure. Immunogenetics 44:212-217. [DOI] [PubMed] [Google Scholar]
- 53.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Pouw Kraan, T. C., P. V. Kasperkovitz, N. Verbeet, and C. L. Verweij. 2004. Genomics in the immune system. Clin. Immunol. 111:175-185. [DOI] [PubMed] [Google Scholar]
- 55.Vasconcelos, A. C., and K. M. Lam. 1995. Apoptosis in chicken embryos induced by the infectious bursal disease virus. J. Comp. Pathol 112:327-338. [DOI] [PubMed] [Google Scholar]
- 56.Vasconcelos, A. C., and K. M. Lam. 1994. Apoptosis induced by infectious bursal disease virus. J. Gen. Virol. 75:1803-1806. [DOI] [PubMed] [Google Scholar]
- 57.Vos, J. B., M. A. van Sterkenburg, K. F. Rabe, J. Schalkwijk, P. S. Hiemstra, and N. A. Datson. 2005. Transcriptional response of bronchial epithelial cells to Pseudomonas aeruginosa: identification of early mediators of host defense. Physiol. Genomics 21:324-336. [DOI] [PubMed] [Google Scholar]
- 58.Webb, T. E., M. G. Kaplan, and E. A. Barnard. 1996. Identification of 6H1 as a P2Y purinoceptor: P2Y5. Biochem. Biophys. Res. Commun. 219:105-110. [DOI] [PubMed] [Google Scholar]
- 59.Withers, D. R., J. R. Young, and T. F. Davison. 2005. Infectious bursal disease virus-induced immunosuppression in the chick is associated with the presence of undifferentiated follicles in the recovering bursa. Viral Immunol. 18:127-137. [DOI] [PubMed] [Google Scholar]
- 60.Yamagata, T., J. Nishida, S. Tanaka, R. Sakai, K. Mitani, M. Yoshida, T. Taniguchi, Y. Yazaki, and H. Hirai. 1996. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol. Cell. Biol. 16:1283-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, Y. H., and A. C. Paquet. 2005. Preprocessing two-color spotted arrays, p. 49-69. In R. C. Gentleman, V. J. Carey, W. Huber, R. Irizarry, and S. Dudoit (ed.), Bioinformatics and computational biology solutions using R and bioconductor. Springer-Verlag, New York, N.Y.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.