Abstract
Mammalian reoviruses contain a genome of 10 segments of double-stranded RNA (dsRNA). Reovirus replication and assembly occur within distinct structures called viral inclusions, which form in the cytoplasm of infected cells. Viral nonstructural proteins μNS and σNS and core protein μ2 play key roles in forming viral inclusions and recruiting other viral proteins and RNA to these structures for replication and assembly. However, the precise functions of these proteins in viral replication are poorly defined. Therefore, to better understand the functions of reovirus proteins associated with formation of viral inclusions, we used plasmid-based vectors to establish 293T cell lines stably expressing small interfering RNAs (siRNAs) specific for transcripts encoding the μ2, μNS, and σNS proteins of strain type 3 Dearing (T3D). Infectivity assays revealed that yields of T3D, but not those of strain type 1 Lang, were significantly decreased in 293T cells stably expressing μ2, μNS, or σNS siRNA. Stable expression of siRNAs specific for any one of these proteins substantially diminished viral dsRNA, protein synthesis, and inclusion formation, indicating that each is a critical component of the viral replication machinery. Using cell lines stably expressing μNS siRNA, we developed a complementation system to rescue viral replication by transient transfection with recombinant T3D μNS in which silent mutations were introduced into the sequence targeted by the μNS siRNA. Furthermore, we demonstrated that μNSC, which lacks the first 40 amino residues of μNS, is incapable of restoring reovirus growth in the complementation system. These results reveal interdependent functions for viral inclusion proteins and indicate that cell lines stably expressing reovirus siRNAs are useful tools for the study of viral protein structure-function relationships.
Viral replication and assembly often take place in intracellular compartments called viral inclusions, where viral components concentrate. For several viruses, formation of viral inclusions occurs at distinct cytoplasmic sites, such as the perinuclear area, and involves complex interactions between viral and cellular factors. Studies of the composition and organization of viral inclusions have provided insight into essential steps in viral replication.
Mammalian orthoreoviruses (reoviruses) are members of the family Reoviridae and contain a genome of 10 segments of double-stranded RNA (dsRNA) (43). The genome is encapsidated by two protein shells, termed outer capsid and core. Following internalization by receptor-mediated endocytosis (3, 6, 51, 59), the core is released into the cytoplasm and begins to synthesize capped, single-stranded RNA (ssRNA) copies of the 10 dsRNA genome segments (6, 19, 23, 56). These mRNAs are competent for translation and serve as templates for minus strand synthesis, resulting in formation of nascent genomic dsRNA (33, 52, 55). Synthesis of the complementary strand appears to be concomitant with assortment of the 10-genome segments into progeny particles (1). Reovirus assembly is completed by the addition of outer-capsid proteins, resulting in the formation of mature, double-shelled virions (40).
Reovirus replication and assembly are thought to occur within viral inclusions that form in the cytoplasm of infected cells (21). Viral inclusions contain dsRNA (57), viral proteins (21), and both complete and incomplete particles (21). These structures are devoid of ribosomes (54) and cellular membranes (27, 46), suggesting that they offer an environment uniquely suited to efficient viral replication and assembly. However, it is not known how viral inclusions are organized with respect to structure or function. Core protein μ2 and viral nonstructural proteins μNS and σNS play important roles in forming viral inclusions and recruiting other viral proteins and RNA to inclusion structures for replication and assembly (4, 5, 7, 9, 36, 39, 45, 65).
The reovirus μ2 protein is encoded by the M1 genome segment and forms a structurally minor component of the viral core (14, 37, 41). The M1 genome segment is associated with viral-strain-specific differences in the in vitro transcriptase and nucleoside triphosphatase (NTPase) activities of viral particles (44, 64). M1 is also the genetic determinant of strain-specific differences in the morphology—globular or filamentous—of viral inclusions (36, 45). The μ2 proteins of some reovirus strains interact with and stabilize microtubules, which are properties responsible for the filamentous inclusion morphology exhibited by prototype strains type 1 Lang (T1L) and type 2 Jones (45, 65). Differences in inclusion morphology are correlated with a single-amino-acid polymorphism in μ2 at position 208 (45).
The reovirus μNS protein is encoded by the M3 genome segment (37, 41) and associates with viral mRNAs (1) and viral cores (8) but does not inhibit viral transcription or capping activities (8). Transiently expressed μNS forms inclusion-like structures, which are similar in appearance and localization to the globular inclusions observed in cells infected with prototype strain type 3 Dearing (T3D) (9). In addition, μNS associates with μ2 and recruits viral core proteins λ1, λ2, and σ2 into viral inclusion-like structures when these proteins are coexpressed in transiently transfected cells (7, 9). These findings suggest that μ2 and μNS together are essential for the formation and morphology of viral inclusions. μNSC, also produced naturally during reovirus infection (32), lacks the 40 amino-terminal residues of μNS and has been proposed to be a product of translation initiation from an alternative site in M3 mRNA (63). The 40 amino-terminal residues of μNS contain interacting domains for viral proteins μ2 and σNS (9, 39). However, the role of μNSC in reovirus replication has not been elucidated.
The reovirus σNS protein, encoded by the S3 genome segment, also plays a role in viral inclusion formation. Temperature-sensitive (ts) reovirus mutant tsE320, which contains a mutation in the S3 genome segment responsible for the ts phenotype, does not form inclusions when grown at nonpermissive temperature (4). The σNS protein binds ssRNA, including reovirus mRNAs, and forms higher-order structures (24-26, 30, 47). Large complexes containing σNS prepared from infected cells are dissociated by treatment with RNase A (24, 30), suggesting that RNA stabilizes these structures. Furthermore, σNS colocalizes with μNS in viral inclusions in reovirus-infected cells (5, 39). Although the function of σNS in viral replication has not been determined, it is possible that σNS is involved in recruitment of viral RNAs into or retention of viral RNAs within inclusions.
RNA interference (RNAi) is a sequence-specific gene-silencing mechanism that employs dsRNA to produce short interfering RNAs (siRNAs) approximately 21 nucleotides (nt) in length (12, 18). siRNAs assemble into endoribonuclease-containing complexes known as RNA-induced silencing complexes and subsequently guide these complexes to degrade mRNAs containing complementary sequences (29, 53). Since its discovery, RNAi has been developed into a widely used gene-silencing technique for studies of gene function. Indeed, numerous studies in which 21-nt synthetic siRNAs (12, 18) or siRNAs transcribed from an RNA polymerase III promoter, such as U6 (60, 66) or H1 (10, 38), were used to specifically suppress the expression of endogenous genes in mammalian cells without activating nonspecific responses to dsRNA have been reported. Similarly, siRNAs have been used to interfere with the replication of a number of viruses, including rotaviruses (2, 11, 17, 28, 34, 35, 58), which are closely related to reoviruses. Thus, the technology of siRNA-mediated gene silencing has the capability to provide information necessary to understand mechanisms of viral replication and functions of individual viral genes.
To understand the function of reovirus proteins associated with the formation of viral inclusions, we established cell lines stably expressing siRNAs specific for transcripts encoding the μ2, μNS, and σNS proteins of T3D by using plasmid-based vectors. Our results reveal that the effects of siRNAs on reovirus infection are strain specific and that μ2, μNS, and σNS proteins are each involved in inclusion formation and maturation as indispensable components of viral replication. Furthermore, we have established a complementation system to study reovirus replication by using a cell line stably expressing reovirus M3 siRNA. This system was used to demonstrate that μNS but not μNSC is required for reovirus replication.
MATERIALS AND METHODS
Cells and viruses.
Murine L929 (L) cells were grown in Joklik's modified Eagle's minimal essential medium (Irvine Scientific, Santa Ana, Calif.) supplemented to contain 5% fetal calf serum (Gibco-BRL, Gaithersburg, Md.), 2 mM l-glutamine, 100 U of penicillin G per ml, 100 μg of streptomycin per ml, and 250 ng of amphotericin B per ml (Gibco-BRL). Human embryonic kidney 293T cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented to contain 10% fetal calf serum, 2 mM l-glutamine, 100 U of penicillin G per ml, 100 μg of streptomycin per ml, and 250 ng of amphotericin B per ml. To establish cell lines stably expressing reovirus siRNAs, 293T cells transfected with siRNA expression vectors to suppress reovirus M1, M3, or S3 gene expression were selected using 5 μg per ml of puromycin (CALBIOCHEM, San Diego, Calif.).
Reovirus prototype strains T1L and T3D and reassortant viruses G16, EB85, and EB113 derived from crosses of T1L and T3D (42) are laboratory stocks. Purified virion preparations of reovirus were made using second-passage L-cell lysate stocks of twice-plaque-purified reovirus as previously described (22). Virus titers were determined by plaque assay using L-cell monolayers as previously described (61).
Plasmid construction.
To generate plasmid-based vectors expressing reovirus M1, M3, or S3 siRNAs, the pSUPER RNAi system (OligoEngine, Seattle, Wash.) was used in concert with a pair of custom oligonucleotides that contain a unique 19-nt sequence derived from the T3D M1, M3, or S3 transcripts targeted for suppression (Table 1). Oligonucleotides containing unique sequences in the sense and antisense orientations were annealed and cloned into the pSUPER.puro vector between the BglII and HindIII restriction sites 3′ of the polymerase III H1-RNA promoter. Mammalian expression vectors pCMVM3wt and pCMVS3wt, containing the open reading frames of the T3D μNS and σNS proteins, respectively, were described previously (5). To generate a mammalian expression vector encoding the T3D μ2 protein (pcFM1T3) fused to a FLAG epitope tag at the amino terminus, an M1 cDNA fragment was amplified by reverse transcription and PCR using viral dsRNA extracted from purified reovirus virions as a template. Amplified cDNA fragments were cloned into the XhoI-KpnI site of the pcFLAG vector, which was constructed by insertion of the FLAG tag sequence into the multicloning site of the pcDNA3 vector (Invitrogen, San Diego, Calif). To construct the mammalian expression vector pCMVM31861m, in which three silent mutations were introduced into the M3 1861 siRNA sequence, a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used with the primer set 5′-CGTATTTCTAAGGAAGCAGCaGCgAAgTGTCAAACTGTTATTGATGAC-3′ and 5′-GTCATCAATAACAGTTTGACAcTTcGCtGCTGCTTCCTTAGAAATACG-3′ (lowercase letters indicate altered nucleotides in the T3D M3 gene at positions 1866, 1869, and 1871) and pCMVM3wt as a template. Fragments of M3 cDNA containing site-directed substitutions were PCR amplified using the primer set 5′-TAAGGTACCATGGCTTCATTCAAGGGATTC-3′ and 5′-TAACTCGAGTTACAACTCATCAGTTGGAAC-3′ and pCMVM31861m as a template. Amplicons were cloned into the KpnI-XhoI site of pcDNA3 to generate pcM31861m. The μNSC cDNA was amplified with the specific primer set 5′-TAAGGTACCATGTCTCAATCGCGTGAATTCC-3′ and 5′-TAACTCGAGTTACAACTCATCAGTTGGAAC-3′ and pcM31861m as a template. Amplified fragments were cloned into the KpnI-XhoI site of pcDNA3 to generate pcM3dN138. pcM31ATGm and pcM32ATGm, containing site-direct substitutions of ATG to GCG in initiation codons corresponding to μNS (nucleotide positions 19 to 21 of M3 RNA) and μNSC (nucleotide positions 139 to 141 of M3 RNA), respectively, were generated using a QuikChange site-directed mutagenesis kit with the primer sets 5′-GAGACCCAAGCTTGGTACCgcGGCTTCATTCAAGG-3′ and 5′-CCTTGAATGAAGCCgcGGTACCAAGCTTGGGTCTC-3′ (pcM31ATGm) and 5′-CTCCGTCTGTGGATgcGTCTCAATCGCGTGAATTC-3′ and 5′-GAATTCACGCGATTGAGACgcATCCACAGACGGAG-3′ (pcM32ATGm) (lowercase letters indicate altered nucleotides in the T3D M3 gene) and pcM31861m as a template.
TABLE 1.
Oligonucleotides for siRNA expression
| siRNAa | Sequenceb |
|---|---|
| M1 895 | 5′-gatccccGGTGGATGTTGTAGACATGttcaagagaCATGTCTACAACATCCACCttttta-3′ 3′-gggCCACCTACAACATCTGTACaagttctctGTACAGATGTTGTAGGTGGaaaaattcga-5′ |
| M1 969 | 5′-gatccccCTATGCATACCGTTCCTGTttcaagagaACAGGAACGGTATGCATAGttttta-3′ 3′-gggGATACGTATGGCAAGGACAaagttctctTGTCCTTGCCATACGTATCaaaaattcga-5′ |
| M3 399 | 5′-gatccccGTTTGCCATAAAGCCAGGTttcaagagaACCTGGCTTTATGGCAAACttttta-3′ 3′-gggCAAACGGTATTTCGGTCCAaagttctctTGGACCGAAATACCGTTTGaaaaattcga-5′ |
| M3 642 | 5′-gatccccAGGGATAATGAAGGCTGCTttcaagagaAGCAGCCTTCATTATCCCTttttta-3′ 3′-gggTCCCTATTACTTCCGACGAaagttctctTCGTCGGAAGTAATAGGGAaaaaattcga-5′ |
| M3 860 | 5′-gatccccAGCAGTCGGGATTGATACTttcaagagaAGTATCAATCCCGACTGCTttttta-3′ 3′-gggTCGTCAGCCCTAACTATGAaagttctctTCATAGTTAGGGCTGACGAaaaaattcga-5′ |
| M3 1686 | 5′-gatccccGTCAGCTCAATCATGTAGCttcaagagaGCTACATGATTGAGCTGACttttta-3′ 3′-gggCAGTCGAGTTAGTACATCGaagttctctCGATGTACTAACTCGACTGaaaaattcga-5′ |
| M3 1756 | 5′-gatccccGATGAATTGCTTGACGCTGttcaagagaCAGCGTCAAGCAATTCATCttttta-3′ 3′-gggCTACTTAACGAACTGCGACaagttctctGTCGCAGTTCGTTAAGTAGaaaaattcga-5′ |
| M3 1861 | 5′-gatccccGCAGCTGCCAAATGTCAAAttcaagagaTTTGACATTTGGCAGCTGCttttta-3′ 3′-gggCGTCGACGGTTTACAGTTTaagttctctAAACTGTAAACCGTCGACGaaaaattcga-5′ |
| M3 2045 | 5′-gatccccATGTGGAATTGGACGCGTTttcaagagaAACGCGTCCAATTCCACATttttta-3′ 3′-gggTACACCTTAACCTGCGCAAaagttctctTTGCGCAGGTTAAGGTGTAaaaaattcga-5′ |
| S3 333 | 5′-gatccccTCATCAAGCATCCACCATGttcaagagaCATGGTGGATGCTTGATGAttttta-3′ 3′-gggAGTAGTTCGTAGGTGGTACaagttctctGTACCACCTACGAACTACTaaaaattcga-5′ |
| S3 501 | 5′-gatccccGCGTGTTCCGATTATGCACttcaagagaGTGCATAATCGGAACACGCttttta-3′ 3′-gggCGCACAAGGCTAATACGTGaagttctctCACGTATTAGCCTTGTGCGaaaaattcga-5′ |
| S3 630 | 5′-gatccccCGACGGACTCAAAGGATTAttcaagagaTAATCCTTTGAGTCCGTCGttttta-3′ 3′-gggGCTGCCTGAGTTTCCTAATaagttctctATTAGGAAACTCAGGCAGCaaaaattcga-5′ |
| S3 766 | 5′-gatccccGCCATCTGTGTGCTTAAGAttcaagagaTCTTAAGCACACAGATGGCttttta-3′ 3′-gggCGGTAGACACACGAATTCTaagttctctAGAATTCGTGTGTCTACCGaaaaattcga-5′ |
| GFP | 5′-gatccccGAACGGCATCAAGGTGAACttcaagagaGTTCACCTTGATGCCGTTCttttta-3′ 3′-gggCTTGCCGTAGTTCCACTTGaagttctctCAAGTGGAACTACGGCAAGaaaaattcga-5′ |
Numbers indicate the first nucleotide of the target T3D reovirus mRNA nucleotide sequence.
Uppercase letters indicate the 19-nt sense and antisense sequences corresponding to reovirus mRNA.
Protein expression in mammalian cells.
293T cells were transfected using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. Transfected cells were subjected to immunofluorescence assay or immunoblot analysis.
Antibodies.
To generate polyclonal antiserum against μNS, the T3D M3 gene was cloned 3′ of sequences encoding glutathione S-transferase (GST) in the pGEX-4T-3 vector (Amersham Biosciences, Piscataway, NJ). The GST-μNS fusion protein expressed in BL21-DE3 cells (Novagen, Madison, WI) was purified from the soluble fraction according to the manufacturer's instructions by using GSTrap affinity chromatography (Amersham Biosciences). The GST tag was removed by treating eluted protein with 20 units of thrombin (Amersham Biosciences) at 25°C for 2 h. Rabbits were immunized and boosted with precipitated μNS protein to generate μNS-specific serum (Cocalico Biologicals Inc., Reamstown, PA). σ3-specific monoclonal antibody (MAb) 4F2 (62) and antisera specific for σNS (5) and μ2 (67) have previously been described.
Immunofluorescence staining.
Cells were infected with reovirus at a multiplicity of infection (MOI) of 10 PFU per cell and seeded onto collagen-coated glass coverslips (Fisher Scientific, Pittsburgh, PA). After incubation for 20 h, cells were fixed with 1:1 methanol-acetone, washed with phosphate-buffered saline (PBS), and incubated with antisera specific for σNS, μNS, or μ2. After two washes with PBS, cells were incubated with Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG) or Alexa Fluor 546 goat anti-guinea pig IgG (Molecular Probes, Inc., Eugene, Oreg.) at a dilution of 1:1,000. Cells also were incubated with TO-PRO3 (Molecular Probes, Inc.) to label nuclei and then washed twice with PBS. Infected cells were visualized using a Zeiss inverted LSM510 confocal microscope (Carl Zeiss, New York, N.Y.).
Immunoblotting.
Cells infected with reovirus were lysed in buffer consisting of 50 mM Tris-HCl (pH 7.6), 1% deoxycholic acid, 1% IGEPAL CA-630 (MP Biomedicals, LLC, Aurora, Ohio), 0.1% sodium dodecyl sulfate (SDS), and 150 mM NaCl. After centrifugation, proteins in the soluble fraction were size fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) and electroblotted onto nitrocellulose membranes. Viral proteins were detected using enhanced chemiluminescence (Amersham Biosciences) following incubation with σ3-specific MAb 4F2 or antisera specific for σNS, μNS, μ2, or T1L virions and appropriate secondary antibodies.
Electrophoretic analysis of cytoplasmic viral dsRNA.
Cells were infected with reovirus at an MOI of 10 PFU per cell, harvested at 18 h postinfection, and lysed in buffer consisting of 100 mM Tris-HCl (pH 7.4), 0.5% IGEPAL CA-630, 1.5 mM MgCl2, and 140 mM NaCl. After removal of nuclei by low-speed centrifugation, RNAs were collected by ethanol precipitation. RNA precipitates were analyzed by 10% SDS-PAGE, followed by ethidium bromide staining.
Reovirus mRNA transcription and translation.
Linearized pcM31861m, pcM31ATGm, pcM32ATGm, and pcM3dN138 were prepared by digestion with XhoI. For generation of capped reovirus mRNA, linearized plasmids were transcribed in vitro using a MEGAscript high-yield transcription T7 kit and cap analog (Ambion Inc., Austin, Texas). Reovirus mRNAs were translated in vitro using rabbit reticulocyte lysates (Flexi Rabbit; Promega, Madison, Wis.) and Easy Tag Express [35S]methionine protein-labeling mix (PerkinElmer, Boston, Mass.). Labeled translation products were subjected to 10% SDS-PAGE, followed by autoradiography.
Complementation of reovirus replication in cells expressing reovirus siRNA.
293T cells (4 × 104) stably expressing M3 siRNA were seeded into 12-well plates (Costar, Cambridge, Mass.) approximately 24 h prior to transfection. Cells were transfected with recombinant plasmid DNA expressing either wild-type μNS or μNS mutants. After 4 h of incubation, transfected cells were infected with T3D at an MOI of 10 PFU per cell. Viral cultures were harvested following 24 h of incubation, and viral titers in cell lysates were determined by plaque assay.
RESULTS
Viral protein expression in 293T cells transiently transfected with μ2, μNS, or σNS siRNA expression vectors.
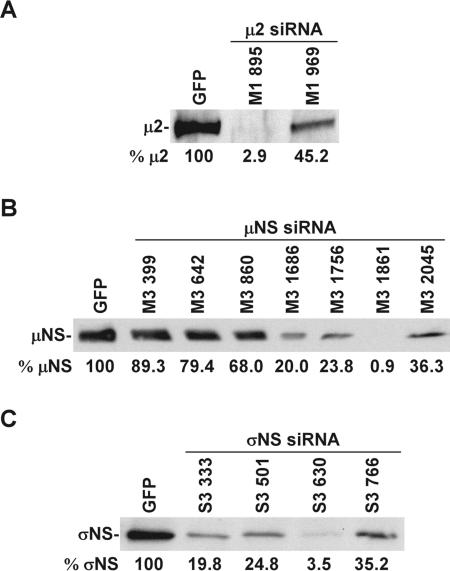
To better understand the function of reovirus core protein μ2 and nonstructural proteins μNS and σNS in viral replication, we selectively reduced the level of each protein by using RNAi (29, 53). We first designed several 19-nt siRNA sequences corresponding to different regions of the T3D M1 (μ2), M3 (μNS), and S3 (σNS) genes (Table 1) and cloned them into the siRNA-expressing vector pSUPER.puro under the control of the H1-RNA promoter. These constructs were cotransfected with T3D μ2 (pcFM1T3), μNS (pCMVM3wt), or σNS (pCMVS3wt) expression vectors into 293T cells. Viral protein expression in transfected cells was assayed by immunoblotting using anti-μ2, -μNS, or -σNS antibodies at 24 h posttransfection (Fig. 1A to C). In comparison to cells expressing green fluorescence protein (GFP) siRNA (Table 1), cells expressing M1, M3, and S3 siRNAs exhibited diminished synthesis of μ2, μNS, and σNS, respectively (Fig. 1A to C). The effects of specific siRNAs were variable, with some sequences exhibiting substantially more inhibition than others. Viral protein expression was not affected by mock transfection or transfection of cells with the empty pSUPER.puro vector (data not shown). These results demonstrate that siRNAs targeted to different regions of the T3D M1, M3, and S3 genes possess various capacities for suppression of reovirus protein synthesis and that M1 895, M3 1861, and S3 630 siRNAs, in particular, were highly effective at inhibiting the protein expression of the T3D μ2, μNS, and σNS proteins, respectively.
FIG. 1.
Viral protein expression in 293T cells transiently transfected with reovirus siRNA expression vectors. siRNA expression vectors encoding a 19-nt sequence from the genes encoding the T3D μ2 (A), μNS (B), or σNS (C) proteins were cotransfected with mammalian expression vector pcFM1T3 (μ2), pCMVM3wt (μNS), or pCMVS3wt (σNS) into 293T cells. Viral protein expression in transfected cells was assessed by immunoblotting with anti-μ2, -μNS, or -σNS antibodies at 24 h posttransfection. GFP siRNA-expressing vectors (GFP) were used as controls. Band intensities were quantitated using Scion Image software and are expressed relative to the GFP siRNA control at the bottom of each gel. The results shown are representative of one experiment out of at least two performed.
Kinetics of reovirus growth in cells stably expressing reovirus-specific siRNAs.
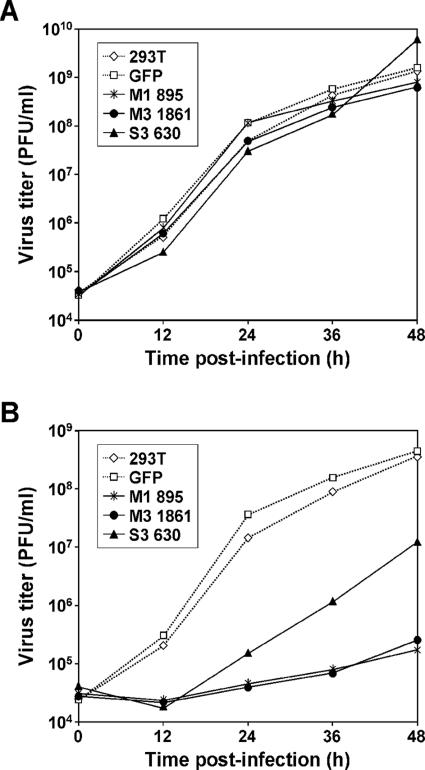
To test whether virus production is inhibited in cells expressing reovirus-specific siRNAs, we established 293T cells stably expressing siRNAs that inhibit M1, M3, or S3 gene expression. siRNA-expressing pSUPER.puro vectors encoding reovirus-specific siRNAs M1 895, M3 1861, and S3 630 were transfected into 293T cells. Transfected cells were selected using puromycin to establish stable lines. To test the effect of reovirus siRNAs on viral growth, these cells were infected with either T1L or T3D at an MOI of 2 PFU per cell. Virus production was assessed by plaque assay at 0, 12, 24, 36, and 48 h postinfection (Fig. 2). Growth of T1L was not diminished at any time point by siRNAs directed to the T3D M1, M3, or S3 transcripts (Fig. 2A). In contrast, growth of T3D was reduced 2,160-, 1,482-, or 97-fold in cells expressing M1 895, M3 1861, or S3 630 siRNAs, respectively, in comparison to growth in untransfected 293T cells (Fig. 2B). Titers of both T1L and T3D continued to rise through 48 h postinfection, consistent with the secondary and tertiary rounds of viral replication that would be expected for subtotal infection of cells at the MOI used (2 PFU per cell). Complete lysis of cultures was observed about 5 days after viral adsorption. To control for nonspecific effects of siRNA on T3D replication, growth of T3D in cells expressing GFP siRNA was not affected (Fig. 2B). The T1L M1, M3, and S3 genes differ from the T3D homologues at the siRNA target sites (Table 2). Thus, siRNAs directed to reovirus replication proteins cause marked reductions in viral growth. Furthermore, the effect appears to be specific, since growth of T1L was not altered by the T3D-based siRNAs.
FIG. 2.
Reovirus growth in cells stably expressing reovirus-specific siRNAs. 293T cells stably expressing M1 895, M3 1861, or S3 630 siRNAs were infected with either T1L (A) or T3D (B) at an MOI of 2 PFU per cell. Untransfected 293T cells (293T) and cell lines stably expressing GFP siRNA (GFP) were used as controls. Viral titers at the time points shown were determined by plaque assay. The results shown are representative of one experiment out of three performed.
TABLE 2.
Comparison of siRNA sequences in T1L and T3D
| siRNA | Gene (protein) | siRNA sequencea |
|---|---|---|
| M1 895 | M1 (μ2) | GGTGGATGTTGTAGACATG (895-913) |
| M3 1861 | M3 (μNS) | GCAGCTGCCAAATGTCAAA (1861-1879) |
| S3 630 | S3 (σNS) | CGACGGACTCAAAGGATTA (630-648) |
Numbers in parentheses correspond to reovirus mRNA nucleotide sequences. Nucleotides that differ between T1L and T3D are underlined. T3D sequences are shown.
Reovirus siRNAs are specific for T3D-derived targets.
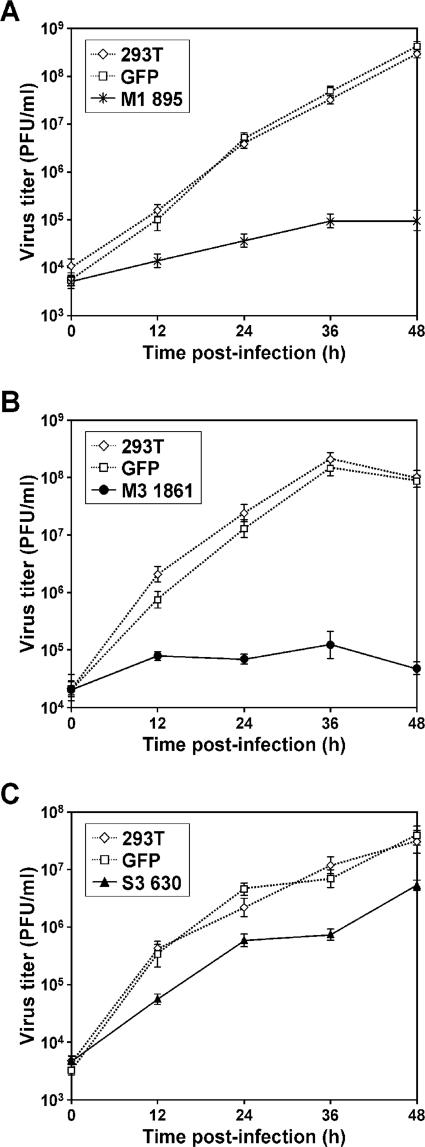
To confirm specific inhibition of T3D M1, M3, and S3 siRNAs on virus production, we investigated growth of selected T1L × T3D reassortant viruses G16, EB85, and EB113 in cells stably expressing reovirus siRNAs. G16 contains the T3D M1 and S2 genes, EB85 contains the T3D M3 and S2 genes, and EB113 contains the T3D M1 and S3 genes in an otherwise T1L background (42). Growth of G16 was inhibited up to 3,086-fold in cells expressing M1 895 siRNA (Fig. 3A), growth of EB85 was inhibited 2,058-fold in cells expressing M3 1861 siRNA (Fig. 3B), and growth of EB113 was inhibited 16-fold in cells expressing S3 630 siRNA (Fig. 3C), each in comparison to growth in untransfected 293T cells. In contrast, titers of G16, EB85, or EB113 in cells expressing GFP siRNA were not diminished at any time point (Fig. 3A to C). These results indicate that the effects of siRNAs on reovirus infection are strain and gene specific, and inhibition of virus growth caused by reovirus siRNAs does not result from nonspecific induction of antiviral responses.
FIG. 3.
Growth of reovirus reassortants following infection of cells stably expressing reovirus-specific siRNAs. 293T cells stably expressing M1 895, M3 1861, or S3 630 siRNAs were infected with reassortant virus G16 (A), EB85 (B), or EB113 (C), respectively, at an MOI of 2 PFU per cell. Untransfected 293T cells (293T) and cell lines stably expressing GFP siRNA (GFP) were used as controls. Virus titers at the time points shown were determined by plaque assay. The results shown are the means from two independent experiments. Error bars indicate standard deviations.
Viral protein expression in cells stably expressing reovirus-specific siRNAs.
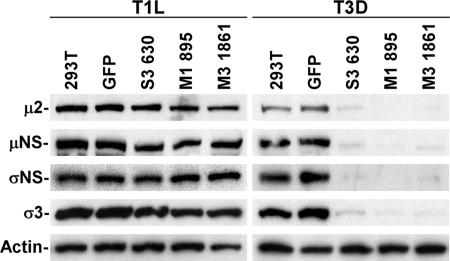
To investigate viral protein expression in 293T cells stably expressing reovirus-specific siRNAs, cells were infected with either T1L or T3D at an MOI of 5 PFU per cell and incubated for 20 h. Lysates prepared from infected cells were analyzed by immunoblotting using polyclonal antisera specific for the μ2, μNS, and σNS proteins, MAb 4F2 specific for the T3D σ3 protein, and an antiserum raised against T1L virions to detect T1L σ3 protein. Stable expression of siRNAs specific for T3D μ2 (M1 895), μNS (M3 1861), or σNS (S3 630) proteins substantially diminished viral protein expression in cells infected with T3D, in contrast to that in untransfected 293T cells or cells expressing GFP siRNA (Fig. 4). Expression of viral proteins in cells infected with T1L was not affected by siRNAs targeting replication proteins of T3D (Fig. 4). These results indicate that the effect of reovirus siRNAs on viral protein synthesis is strain specific and that μ2, μNS, and σNS proteins are each required for viral protein synthesis during reovirus replication.
FIG. 4.
Viral protein expression in cells stably expressing reovirus-specific siRNAs. 293T cells stably expressing S3 630, M1 895, or M3 1861 siRNAs were infected with either T1L or T3D at an MOI of 5 PFU per cell and incubated for 20 h. Lysates prepared from infected cells were analyzed by immunoblotting using polyclonal antisera specific for the μ2, μNS, and σNS proteins. Polyclonal antiserum raised against T1L virions and MAb 4F2 were used to detect the T1L and T3D σ3 proteins, respectively. An actin-specific antibody was used as a loading control. The results shown are representative of one experiment out of three performed.
Viral dsRNA synthesis in cells stably expressing reovirus-specific siRNAs.
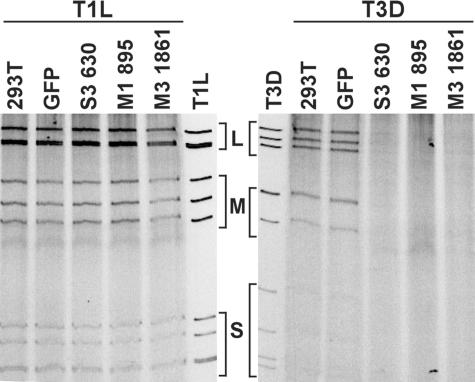
To define the importance of μ2, μNS, and σNS proteins in viral dsRNA synthesis, we assessed viral dsRNA production in cells expressing reovirus siRNAs. Cells were infected with either T1L or T3D at an MOI of 10 PFU per cell, harvested at 18 h postinfection, and lysed. After removal of nuclei by low-speed centrifugation, RNA precipitates were analyzed by SDS-PAGE and ethidium bromide staining (Fig. 5). Production of dsRNA by T1L in cells expressing T3D-specific M1, M3, or S3 siRNAs was not decreased in comparison to that in untransfected 293T cells or a cell line stably expressing GFP siRNA. In contrast, production of dsRNA by T3D was significantly decreased in cell lines expressing T3D-specific M1, M3, or S3 siRNAs (Fig. 5). These results indicate that μ2, μNS, and σNS proteins are each required for viral dsRNA synthesis.
FIG. 5.
Viral dsRNA expression in cells stably expressing reovirus-specific siRNAs. Untransfected 293T (293T) cells and cell lines stably expressing S3 630, M1 895, M3 1861, or GFP siRNA (GFP) were infected with either T1L or T3D at an MOI of 10 PFU per cell and incubated for 18 h. The cytoplasmic fraction containing viral dsRNA from infected cells was resolved by electrophoresis in 10% polyacrylamide gels and analyzed by ethidium bromide staining. Size classes of viral dsRNA segments (large [L], medium [M], and small [S]) are indicated. Viral dsRNA extracted from purified T1L and T3D virions was used as a control. The results shown are representative of one experiment out of two performed.
Subcellular localization and expression of reovirus proteins in cells expressing reovirus-specific siRNAs.
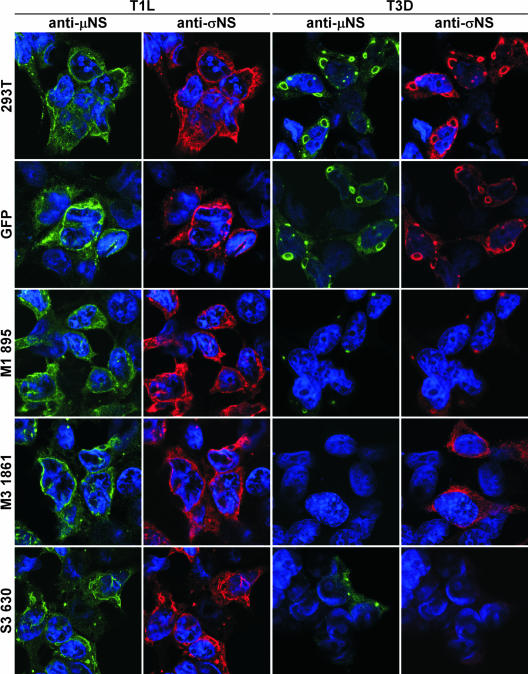
The results described thus far indicate that the μ2, μNS, and σNS proteins are required for viral protein and dsRNA synthesis. These proteins are also components of viral inclusions in reovirus-infected cells. Thus, we investigated the effect of siRNAs on viral inclusion formation in cells infected with either T1L or T3D. 293T cells infected with reovirus at an MOI of 10 PFU per cell were seeded onto collagen-coated glass coverslips, incubated for 20 h, and fixed and stained with primary antibodies specific for reovirus proteins. In previous studies, it was observed that T1L produces filamentous inclusions, whereas T3D forms globular inclusions in several cell lines (45). The M1 gene is the genetic determinant of this difference in inclusion morphology (36, 45, 65). In our experiments, μNS and σNS proteins were detected in globular inclusions in untransfected and GFP siRNA-expressing 293T cells infected with T3D. Both proteins were detected in filamentous inclusions in cells infected with T1L (Fig. 6). In T3D-infected cells expressing M3 1861 siRNA, μNS protein was not detected, and σNS protein was diffusely distributed in the cytoplasm. μNS was detected in small globular inclusions in T3D-infected cells not expressing σNS protein (S3 630 siRNA cells) (Fig. 6). Both μNS and σNS proteins were evident in filamentous inclusions in T1L-infected cells stably expressing M3 1861 or S3 630 siRNAs (Fig. 6).
FIG. 6.
Detection of protein expression in cells stably expressing M1, M3, or S3 siRNAs by immunofluorescence microscopy. 293T cells and cell lines stably expressing GFP, M1 895, M3 1861, or S3 630 siRNAs were infected with either T1L or T3D at an MOI of 10 PFU per cell and incubated for 20 h. Infected cells were fixed and stained using anti-μNS (rabbit) or anti-σNS (guinea pig) antiserum, followed by Alexa Fluor 488 goat anti-rabbit IgG (green) or Alexa Fluor 546 goat anti-guinea pig IgG (red), respectively. Cells were stained with TO-PRO3 (blue) to label nuclei.
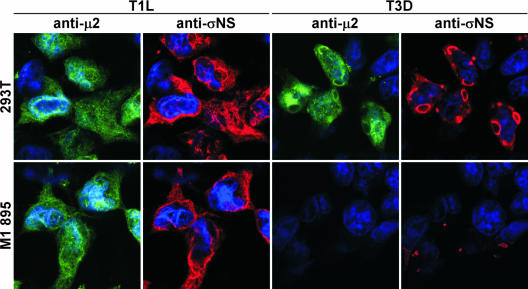
Following infection of cells expressing M1 895 siRNA with T3D, μNS and σNS were contained in small globular inclusions (Fig. 6). The μ2 protein was localized to cytoplasmic viral inclusions and nuclei of untransfected 293T cells infected with T3D (Fig. 7); μ2 was not detected in cells expressing M1 895 siRNA following infection with T3D (Fig. 7). The μ2, μNS, and σNS proteins were detected in filamentous cytoplasmic inclusions and nuclei of M1 895 siRNA-expressing cells infected with T1L (Fig. 6 and 7). These results indicate that the μNS protein plays an essential role in the formation of viral inclusions and recruitment of other viral proteins, including σNS, in reovirus-infected cells. Furthermore, the effects of reducing levels of μ2 and σNS proteins in infected cells reveal that both proteins serve essential functions in the formation and maturation of viral inclusions.
FIG. 7.
Detection of protein expression in cells stably expressing M1 siRNA by immunofluorescence microscopy. 293T cells and cell lines stably expressing M1 895 siRNA were infected with either T1L or T3D at an MOI of 10 PFU per cell and incubated for 20 h. Infected cells were fixed and stained using anti-μ2 (rabbit) or anti-σNS (guinea pig) antiserum, followed by Alexa Fluor 488 goat anti-rabbit IgG (green) or Alexa Fluor 546 goat anti-guinea pig IgG (red). Cells were stained with TO-PRO3 (blue) to label nuclei.
Complementation of reovirus replication defects in cells stably expressing T3D M3 siRNA.
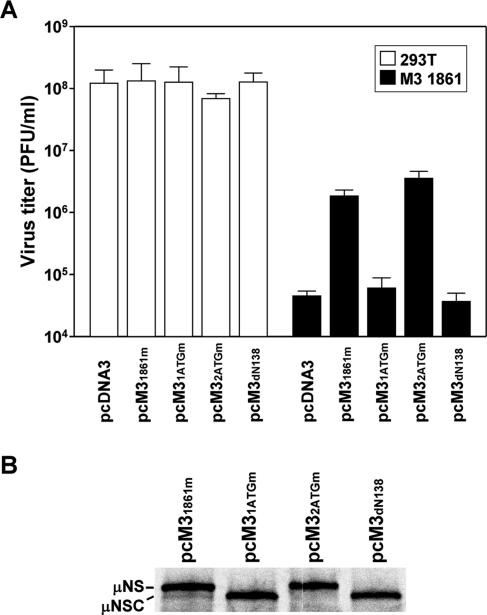
To develop a complementation system for studies of reovirus replication, we first constructed a T3D μNS expression plasmid containing three silent point mutations within the siRNA target sequence (pcM31861m). Cells stably expressing M3 1861 siRNA were transfected with pcM31861m followed by infection with T3D at an MOI of 10 PFU per cell at 4 h posttransfection. Viral titers were determined by plaque assay at 24 h postinfection. Yields of T3D were decreased approximately 3,000-fold in siRNA-expressing cells transfected with mock plasmid compared to an approximately 75-fold reduction in cells transfected with pcM31861m plasmid (Fig. 8A). Thus, provision of μNS in trans rescues a substantial level (∼40-fold) of reovirus growth in cells expressing M3 siRNA.
FIG. 8.
Complementation of reovirus replication in cells expressing M3 siRNA. (A) 293T cells or cells stably expressing M3 1861 siRNA were transfected with pcDNA3, pcM31861m, pcM31ATGm, pcM32ATGm, or pcM3dN138 and infected with T3D at an MOI of 10 PFU per cell. At 24 h postinfection, viral titers were determined by plaque assay. The results are means from three independent experiments. Error bars indicate standard deviations. (B) RNA transcripts of pcM31861m, pcM31ATGm, pcM32ATGm, and pcM3dN138 were translated in rabbit reticulocyte lysates containing [35S]methionine. Labeled products were resolved by 10% SDS-PAGE, followed by autoradiography.
To determine whether expression of μNSC, which lacks the 40 amino-terminal residues of μNS, can complement the viral growth defect in cells expressing M3 siRNA, pcM3dN138, which encodes μNSC but not μNS (Fig. 8B), and pcM31ATGm, which encodes μNS with a mutated start codon (19ATG21 → 19GCG21) such that only μNSC is expressed (Fig. 8B), were introduced into M3 1861 cells prior to infection with T3D. In these experiments, neither construct was capable of restoring reovirus replication (Fig. 8A). In contrast, transfection of pcM32ATGm, which expresses μNS but is incapable of expressing μNSC due to a mutated μNSC start codon (139ATG141 → 139GCG141), mediated the rescue of viral replication in cells stably expressing M3 siRNA. Complementation efficiency obtained using pcM32ATGm was equivalent to that effected with pcM31861m (Fig. 8A), which expresses both μNS and μNSC (Fig. 8B). These results demonstrate that the 40 amino-terminal residues of μNS are indispensable for its native function in reovirus-infected cells and that μNSC is not required for reovirus replication.
DISCUSSION
Reovirus mutant strains obtained by selection, screening, or passage have served as powerful tools to facilitate an understanding of a broad range of viral processes (15). However, there are some reovirus genes and proteins for which informative mutant viruses have not been isolated. Notably, a reovirus mutant bearing a lesion in the M3 gene has not been described. Although a reverse genetics system that allows incorporation of nonviral sequences into infectious reovirus particles has been established, current techniques are limited to modifications of the S2 gene (encoding core protein σ2) (48-50). Rescue of engineered changes in native reovirus RNAs has not been reported. Therefore, stable cell lines expressing reovirus-specific siRNAs, such as those established in this study, offer an opportunity to study functions of viral proteins for which mutant strains are unavailable.
Reovirus replication and assembly are thought to occur within cytoplasmic viral inclusions where viral and cellular proteins (21, 54), viral RNAs (57), and immature and mature viral particles (21) are concentrated. The μ2, μNS, and σNS proteins play key roles in forming viral inclusions (4, 5, 7, 9, 36, 39, 45, 65); however, little is known about their individual and corporate functions in viral RNA replication and particle assembly. In this study, we investigated the functions of μ2, μNS, and σNS proteins in viral replication by using RNAi, a process by which dsRNA directs sequence-specific degradation of mRNA. To overcome the barrier of low transfection efficiency problematic to the use of chemically synthesized siRNAs, we established cell lines stably expressing M1-, M3-, and S3-RNA-specific siRNAs, which were highly effective in inhibiting the expression of the T3D μ2, μNS, and σNS proteins, respectively (Fig. 1). Viral yields of T3D and selected T1L × T3D reassortant viruses containing T3D genome segments with siRNA-targeted sequences were significantly decreased in siRNA-expressing cell lines (Fig. 2 and 3). In contrast, viral yields of T1L, which differs from T3D in the siRNA-targeted sequences (Table 2), were not altered by T3D-based siRNAs (Fig. 2). Thus, inhibition of virus production by reovirus siRNAs is allele specific and, therefore, unlikely to result from induction of innate antiviral responses in cell lines constitutively producing siRNAs.
Stable expression of siRNA specific for the μ2 protein substantially diminished viral protein and dsRNA synthesis in cells infected with T3D (Fig. 4 and 5), demonstrating a tight functional association of μ2 protein with the viral replication machinery. In a previous study, reovirus ts mutant tsH11.2, which contains a defect mapped to the M1 genome segment, produced neither detectable viral proteins nor dsRNA late in infection at the nonpermissive temperature (13). These findings are consistent with our results for stable cell lines expressing M1 siRNA. The μ2 protein is genetically associated with viral-strain-specific differences in the in vitro transcriptase and NTPase activities of viral particles (44, 64), and purified μ2 possesses in vitro NTPase activity stimulated in the presence of λ3 (31). Thus, inhibition of reovirus replication by stable expression of M1 siRNA may reflect suppression of transcriptase or NTPase activities of the viral core. It is also possible that the effects of M1 siRNA are attributable to the inhibition of another μ2 function. For example, μ2 determines strain-specific differences in rate of viral inclusion formation in reovirus-infected cells (36). In cells infected with T3D, μNS and σNS are detected in very small inclusion-like structures in the absence of μ2 (Fig. 7). These results suggest a critical function for μ2 in the maturation of viral inclusions but not in their genesis.
Stable expression of S3 siRNA substantially diminished viral dsRNA and protein synthesis in cells infected with T3D (Fig. 4 and 5). These results suggest that functional σNS is essential for reovirus replication, consistent with a previous report that reovirus mutant tsE320, in which the ts phenotype maps to the S3 genome segment, produced reduced levels of viral proteins and dsRNA (16, 20) and exhibited diminished inclusion formation at the nonpermissive temperature (4). Expression of σNS was not detectable by immunofluorescence in T3D-infected cells stably expressing S3 siRNA (Fig. 6), although μNS was detected in small inclusion-like structures (Fig. 6). These findings suggest that σNS functions in the maturation of functional viral inclusions but does not initiate inclusion formation in the absence of other viral proteins. The σNS protein associates with ssRNA in a non-sequence-specific manner and forms higher-order complexes stabilized by RNAs (24-26, 30, 47). Thus, possible functions for σNS consistent with the results of this and other studies include recruitment of viral ssRNA to inclusions; sequestration of viral RNA in inclusions through a tripartite μNS-σNS-RNA complex; organization of viral RNA for replication, assortment, and packaging; scaffolding of viral inclusions; and regulation of viral translation. These or other possible σNS functions may be mediated by interactions between σNS and cellular proteins yet to be identified.
Stable expression of siRNAs specific for the μNS protein diminished viral protein and dsRNA synthesis in cells infected with T3D (Fig. 4 and 5). This is the first report that μNS plays an essential role in reovirus replication in infected cells. In previous studies, μNS protein was shown to bind core particles (8) and, when expressed from plasmid vectors, formed structures with morphologies similar to those of viral inclusions in infected cells and specifically recruited viral proteins λ1, λ2, μ2, σNS, and σ2 into inclusion-like structures (5, 7, 39). In T3D-infected cells stably expressing M3 siRNA, we found that σNS protein was diffusely distributed in the cytoplasm and μNS was not detectable (Fig. 6). In contrast, σNS and μNS were colocalized in small viral inclusions in infected cells stably expressing M1 siRNA (Fig. 7). Therefore, our results and previous findings suggest that μNS plays a central role in viral inclusion formation and recruits other viral proteins and viral RNA into the inclusions where replication and particle assembly occur.
We developed a complementation system for functional studies of μNS in cells that stably express M3-specific siRNAs. The complementing construct pcM31861m contains three nucleotide substitutions in the siRNA-targeted T3D μNS-encoding sequence, resulting in resistance to siRNA-mediated degradation. The efficiency with which this construct restored viral replication in infected cells was less than anticipated, even when correcting for potentially low transfection frequencies. It is possible that the low complementation efficiency is related to the process of inclusion formation or maturation during reovirus infection. In our complementation system, exogenous μNS was expressed in M3 siRNA-expressing cells prior to viral infection and, therefore, viral inclusion-like structures were likely nucleated by μNS prior to the expression of other inclusion-associated proteins, such as μ2 and σNS. Perhaps concurrent production of μNS and other replication proteins, such as μ2 and σNS, is required for maturation of fully functional inclusions. In support of this idea, coexpression of μ2 or σNS with exogenous μNS enhances viral yields in cells expressing M3 siRNA (T. Kobayashi and T. S. Dermody, unpublished data).
Our complementation strategy afforded the opportunity to define the importance of μNS isoform μNSC in viral replication. We found that transient expression of μNSC was incapable of restoring viral replication in reovirus-infected cells made deficient in μNS expression by RNAi (Fig. 8). Conversely, expression of μNSC was not requisite to the complementation of viral replication by μNS (Fig. 8). μNSC lacks the 40-residue amino-terminal segment of μNS, which contains interacting domains for μ2 and σNS (9, 39). Therefore, these results indicate that the association of μNS with μ2 or σNS (or both proteins) is required for viral replication. Although μNSC did not restore viral replication in the complementation system, this protein may nevertheless exercise functions beneficial to viral growth, consistent with preservation of the μNSC open reading frame. Additionally, it is possible that μNSC contributes to viral growth or spread in vivo.
Our study demonstrates the remarkable utility of stable cell lines expressing reovirus-specific siRNAs as tools to investigate the replication machinery of reovirus in infected cells. Targeting of other reovirus genes should lead to additional insights about the contribution of individual viral proteins to viral replication and assembly. The RNAi approach, combined with individual viral replication-complementation assays, opens new opportunities to understand biological activities of reovirus replication proteins.
Acknowledgments
We thank Earl Brown for providing μ2-specific antiserum, members of our laboratory for many useful discussions, and Denise Wetzel, Kristen Guglielmi, and Tim Peters for review of the manuscript.
This work was supported by a fellowship from the Naito Foundation (T.K.), Public Health Service awards K08 AI062862 (J.D.C.) and R01 AI32539 from the National Institute of Allergy and Infectious Diseases, and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards CA68485 for the Vanderbilt-Ingram Cancer Center and DK20593 for the Vanderbilt Diabetes Research and Training Center.
REFERENCES
- 1.Antczak, J. B., and W. K. Joklik. 1992. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology 187:760-776. [DOI] [PubMed] [Google Scholar]
- 2.Arias, C. F., M. A. Dector, L. Segovia, T. Lopez, M. Camacho, P. Isa, R. Espinosa, and S. Lopez. 2004. RNA silencing of rotavirus gene expression. Virus Res. 102:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, G. S., D. H. Ebert, C. J. Chung, A. H. Erickson, and T. S. Dermody. 1999. Mutant cells selected during persistent reovirus infection do not express mature cathepsin L and do not support reovirus disassembly. J. Virol. 73:9532-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, M. M., T. R. Peters, and T. S. Dermody. 2003. Reovirus σNS and μNS proteins form cytoplasmic inclusion structures in the absence of viral infection. J. Virol. 77:5948-5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borsa, J., M. D. Sargent, P. A. Lievaart, and T. P. Copps. 1981. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology 111:191-200. [DOI] [PubMed] [Google Scholar]
- 7.Broering, T. J., J. Kim, C. L. Miller, C. D. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. Parker. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broering, T. J., A. M. McCutcheon, V. E. Centonze, and M. L. Nibert. 2000. Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broering, T. J., J. S. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 11.Campagna, M., C. Eichwald, F. Vascotto, and O. R. Burrone. 2005. RNA interference of rotavirus segment 11 mRNA reveals the essential role of NSP5 in the virus replicative cycle. J. Gen. Virol. 86:1481-1487. [DOI] [PubMed] [Google Scholar]
- 12.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs, K. M. 1996. Identification and characterization of a double-stranded RNA− reovirus temperature-sensitive mutant defective in minor core protein μ2. J. Virol. 70:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombs, K. M. 1998. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 243:218-228. [DOI] [PubMed] [Google Scholar]
- 15.Coombs, K. M. 1998. Temperature-sensitive mutants of reovirus. Curr. Top. Microbiol. Immunol. 233:69-107. [DOI] [PubMed] [Google Scholar]
- 16.Cross, R. K., and B. N. Fields. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of viral RNA. J. Virol. 50:799-809. [DOI] [PubMed] [Google Scholar]
- 17.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 19.Faust, M., K. E. M. Hastings, and S. Millward. 1975. M7 G5′-ppp5′ Gm pCpUp at the 5′ terminus of reovirus messenger RNA. Nucleic Acids Res. 2:1329-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields, B. N., R. Laskov, and M. D. Scharff. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of peptides. Virololgy 50:209-215. [DOI] [PubMed] [Google Scholar]
- 21.Fields, B. N., C. S. Raine, and S. G. Baum. 1971. Temperature-sensitive mutants of reovirus type 3: defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology 43:569-578. [DOI] [PubMed] [Google Scholar]
- 22.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuichi, Y., M. Morgan, S. Muthukrishnan, and A. J. Shatkin. 1975. Reovirus messenger RNA contains a methylated blocked 5′-terminal structure M7G(5′)ppp(5′)GmpCp-. Proc. Natl. Acad. Sci. USA 72:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillian, A. L., and M. L. Nibert. 1998. Amino terminus of reovirus nonstructural protein σNS is important for ssRNA binding and nucleoprotein complex formation. Virology 240:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Gillian, A. L., S. C. Schmechel, J. Livny, L. A. Schiff, and M. L. Nibert. 2000. Reovirus protein σNS binds in multiple copies to single-stranded RNA and shares properties with single-stranded DNA binding proteins. J. Virol. 74:5939-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomatos, P. J., O. Prakash, and N. M. Stamatos. 1981. Small reovirus particles composed solely of sigma NS with specificity for binding different nucleic acids. J. Virol. 39:115-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomatos, P. J., I. Tamm, S. Dales, and R. M. Franklin. 1962. Reovirus type 3: physical characteristics and interactions with L cells. Virology 17:441-454. [DOI] [PubMed] [Google Scholar]
- 28.Haasnoot, P. C., D. Cupac, and B. Berkhout. 2003. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 29.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 30.Huismans, H., and W. K. Joklik. 1976. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with high affinity for single-stranded and double-stranded RNA, respectively. Virology 70:411-424. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J., J. S. Parker, K. E. Murray, and M. L. Nibert. 2004. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. J. Biol. Chem. 279:4394-4403. [DOI] [PubMed] [Google Scholar]
- 32.Lee, P. W. K., E. C. Hayes, and W. K. Joklik. 1981. Characterization of anti-reovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology 108:134-146. [DOI] [PubMed] [Google Scholar]
- 33.Li, J. K.-K., P. P. Scheible, J. D. Keene, and W. K. Joklik. 1980. The plus strand of reovirus gene S2 is identical with its in vitro transcript. Virology 105:282-286. [DOI] [PubMed] [Google Scholar]
- 34.Lopez, T., M. Camacho, M. Zayas, R. Najera, R. Sanchez, C. F. Arias, and S. Lopez. 2005. Silencing the morphogenesis of rotavirus. J. Virol. 79:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez, T., M. Rojas, C. Ayala-Breton, S. Lopez, and C. F. Arias. 2005. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 86:1609-1617. [DOI] [PubMed] [Google Scholar]
- 36.Mbisa, J. L., M. M. Becker, S. Zou, T. S. Dermody, and E. G. Brown. 2000. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology 272:16-26. [DOI] [PubMed] [Google Scholar]
- 37.McCrae, M. A., and W. K. Joklik. 1978. The nature of the polypeptide encoded by each of the ten double-stranded RNA segments of reovirus type 3. Virology 89:578-593. [DOI] [PubMed] [Google Scholar]
- 38.McManus, M. T., C. P. Petersen, B. B. Haines, J. Chen, and P. A. Sharp. 2002. Gene silencing using micro-RNA designed hairpins. RNA 8:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, C. L., T. J. Broering, J. S. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino terminus. J. Virol. 77:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan, E. M., and H. J. Zweerink. 1975. Characterization of transcriptase and replicase particles isolated from reovirus infected cells. Virology 68:455-466. [DOI] [PubMed] [Google Scholar]
- 41.Mustoe, T. A., R. F. Ramig, A. H. Sharpe, and B. N. Fields. 1978. Genetics of reovirus: identification of the dsRNA segments encoding the polypeptides of the μ and σ size classes. Virology 89:594-604. [DOI] [PubMed] [Google Scholar]
- 42.Nibert, M. L., R. L. Margraf, and K. M. Coombs. 1996. Nonrandom segregation of parental alleles in reovirus reassortants. J. Virol. 70:7295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 44.Noble, S., and M. L. Nibert. 1997. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 71:7728-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhim, J. S., L. E. Jordan, and H. D. Mayor. 1962. Cytochemical, fluorescent-antibody and electron microscopic studies on the growth of reovirus (ECHO 10) in tissue culture. Virology 17:342-355. [DOI] [PubMed] [Google Scholar]
- 47.Richardson, M. A., and Y. Furuichi. 1985. Synthesis in Escherichia coli of the reovirus nonstructural protein σNS. J. Virol. 56:527-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roner, M. R., K. Bassett, and J. Roehr. 2004. Identification of the 5′ sequences required for incorporation of an engineered ssRNA into the reovirus genome. Virology 329:348-360. [DOI] [PubMed] [Google Scholar]
- 49.Roner, M. R., and W. K. Joklik. 2001. Reovirus reverse genetics: incorporation of the CAT gene into the reovirus genome. Proc. Natl. Acad. Sci. USA 98:8036-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roner, M. R., and J. Roehr. 2006. The 3′ sequences required for incorporation of an engineered ssRNA into the Reovirus genome. Virol. J. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubin, D. H., D. B. Weiner, C. Dworkin, M. I. Greene, G. G. Maul, and W. V. Williams. 1992. Receptor utilization by reovirus type 3: distinct binding sites on thymoma and fibroblast cell lines result in differential compartmentalization of virions. Microb. Pathog. 12:351-365. [DOI] [PubMed] [Google Scholar]
- 52.Schonberg, M., S. C. Silverstein, D. H. Levin, and G. Acs. 1971. Asynchronous synthesis of the complementary strands of the reovirus genome. Proc. Natl. Acad. Sci. USA 68:505-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 54.Sharpe, A. H., L. B. Chen, and B. N. Fields. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399-411. [DOI] [PubMed] [Google Scholar]
- 55.Shatkin, A. J., and M. Kozak. 1983. Biochemical aspects of reovirus transcription and translation, p. 79-106. In W. K. Joklik (ed.), The Reoviridae. Plenum Press, New York, N.Y.
- 56.Silverstein, S. C., C. Astell, D. H. Levin, M. Schonberg, and G. Acs. 1972. The mechanism of reovirus uncoating and gene activation in vivo. Virology 47:797-806. [DOI] [PubMed] [Google Scholar]
- 57.Silverstein, S. C., and P. H. Schur. 1970. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology 41:564-566. [DOI] [PubMed] [Google Scholar]
- 58.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sturzenbecker, L. J., M. L. Nibert, D. B. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virgin, H. W., IV, R. Bassel-Duby, B. N. Fields, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 62:4594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virgin, H. W., IV, M. A. Mann, B. N. Fields, and K. L. Tyler. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65:6772-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiener, J. R., J. A. Bartlett, and W. K. Joklik. 1989. The sequences of reovirus serotype 3 genome segments M1 and M3 encoding the minor protein μ2 and the major nonstructural protein μNS, respectively. Virology 169:293-304. [DOI] [PubMed] [Google Scholar]
- 64.Yin, P., M. Cheang, and K. M. Coombs. 1996. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J. Virol. 70:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin, P., N. D. Keirstead, T. J. Broering, M. M. Arnold, J. S. Parker, M. L. Nibert, and K. M. Coombs. 2004. Comparisons of the M1 genome segments and encoded μ2 proteins of different reovirus isolates. Virol. J. 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, J. Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou, S., and E. G. Brown. 1996. Stable expression of the reovirus μ2 protein in mouse L cells complements the growth of a reovirus ts mutant with a defect in its M1 gene. Virology 217:42-48. [DOI] [PubMed] [Google Scholar]