Abstract
Neonatal infection of most mouse strains with lymphocytic choriomeningitis virus (LCMV) leads to a life-long persistent infection characterized by high virus loads in the central nervous system (CNS) in the absence of inflammation and tissue destruction. These mice, however, exhibit impaired learning and memory. The occurrence of cognitive defects in the absence of overt CNS pathology led us to the hypothesis that chronic virus infection may contribute to neuronal dysfunction by altering the host's gene expression profile. To test this hypothesis, we examined the impact of LCMV persistence on host gene expression in the CNS. To model the natural route of human congenital CNS infection observed with a variety of viruses, we established a persistently infected mouse colony where the virus was maintained via vertical transmission from infected mothers to offspring (LCMV-cgPi). LCMV-cgPi mice exhibited a lifelong persistent infection involving the CNS; the infection was associated with impaired spatial-temporal learning. Despite high viral loads in neurons of the brains of adult LCMV-cgPi mice, we detected changes in the host's CNS gene expression for only 75 genes, 56 and 19 being significantly induced and reduced, respectively. The majority of the genes induced in the brain of LCMV-cgPi mice were interferon (IFN)-stimulated genes (ISGs) and included the transcription factors STAT1 and IRF9, the ISG15 protease UBP43, and the glucocorticoid attenuated-response genes GARG16 and GARG49. Based on their crucial role in antiviral defense, these ISGs may play an important role in limiting viral spread and replication. However, since IFNs have also been implicated in adverse effects on neuronal function, the chronic induction of some ISGs may also contribute to the observed cognitive impairment.
Infection of the central nervous system (CNS) in humans by various DNA and RNA viruses can represent a severe health problem. Acute viral infections of the CNS frequently result in the destruction of specific neural cell populations as a direct consequence of virus multiplication or as a result of the host's antiviral immune response (23, 26). Many viruses, however, adopt a noncytolytic strategy of multiplication and can escape from the host immune surveillance by use of a plethora of mechanisms that may result in long-term persistent infections of the CNS. Despite the absence of overt signs of pathology such as cell lysis and inflammation, these infections can lead to severe alterations in neuronal function (45, 46) manifested by cognitive and behavioral impairment (20, 30, 39, 60). This, in turn, has led to the hypothesis that viruses may contribute to a variety of CNS disorders whose etiologies remain unknown (40, 44).
The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) provides an important model system for the investigation of the mechanisms and consequences of viral persistence in the CNS (8, 16, 18, 42, 43). Furthermore, increasing evidence indicates that LCMV might be a neglected human pathogen of clinical significance (3, 4, 64). LCMV has a noncytolytic strategy of multiplication, which enables the virus to persist both in vivo and in cultured cells. Neonatal infection of mice with LCMV leads to the establishment of a life-long persistent infection (LCMV-Pi). Brains of LCMV-Pi mice contain high virus load primarily in neurons in the neocortex, limbic system, and certain hypothalamic regions (19, 53). Neither inflammation nor cytolysis occurs within the brain parenchyma of LCMV-Pi mice. However, as adults LCMV-Pi mice exhibit an impaired learning ability and a reduced tendency to explore a novel environment (20, 25). Since cognitive defects occur in the absence of overt signs of pathology, we hypothesized that chronic virus infection might contribute to neuronal dysfunction by altering the host's gene expression profile. Accordingly, previous studies have shown altered levels of acetylcholine enzymes (46) as well as neurotransmitter mRNAs (31) in LCMV-infected mice. Moreover, we have documented that LCMV persistence causes a specific reduction in expression of the growth-associated protein 43 (GAP-43), a well-established marker of neuroplasticity (5) in hippocampus (15). LCMV persistent infection reduced GAP-43 mRNA levels by affecting both the rate of GAP-43 transcription and the posttranscriptional stabilization of GAP-43 mRNA (10).
In the present study, we aimed at the identification of additional host genes whose expression is changed in the CNS of mice persistently infected with LCMV. To recreate the natural route of congenital viral CNS infection (2), we established a model involving vertical transmission of LCMV from infected mothers to offspring (LCMV-cgPi). Despite high viral loads in neurons of the brains of adult LCMV-cgPi mice, our DNA array-based CNS gene expression profiling revealed remarkably subtle changes in the host's gene expression. The majority of genes with altered expression corresponded to interferon (IFN)-stimulated genes (ISGs), including some that are known to play a crucial role in antiviral defense but that have also been implicated in adverse effects on neuronal function. The chronic induction of these ISGs may be critical for the host's ability to contain viral infection and may also contribute to LCMV-cgPi-induced functional impairment of the CNS.
MATERIALS AND METHODS
Animals.
Female C57BL/6 mice were obtained from the rodent breeding colony at The Scripps Research Institute (La Jolla, CA) and were bred and maintained under specific pathogen-free conditions. For the establishment of a colony of mice persistently infected with LCMV, neonatal mice were inoculated intraperitoneally with serum-free Dulbecco's modified Eagle's medium containing 500 PFU of a plaque-purified Armstrong strain of LCMV (ARM53b). Mice that were persistently infected with LCMV, as determined by the presence of infectious virus in serum, were selected as breeders to establish a persistently infected colony. Prior to being used in experiments mice were verified to be persistently infected based on containing >4 logs of LCMV in their serum. Handling of mice and experimental procedures were conducted in accordance with the National Institute of Health guidelines for animal care and use and approved by the IACUC at the Scripps Research Institute.
Antibodies.
Polyclonal guinea pig antibody to LCMV has been described previously (9). Polyclonal rabbit antibody to glial acidic fibrillary protein (GFAP) and monoclonal antibody to NeuN were from Chemicon (Temecula, CA); rhodamine X-conjugated antibody to guinea pig immunoglobulin G (IgG), fluorescein isothiocyanate (FITC)-conjugated antibody to mouse IgG, and FITC-conjugated antibody to rabbit IgG were from Jackson ImmunoResearch (West Grove, PA).
Virus, virus purification, and virus quantification.
LCMV ARM53b is a triple-plaque-purified isolate of ARM CA 1371 (17). Seed stocks of ARM53b were prepared by growth in BHK-21 cells as described previously (17). Virus titers in serum and tissue were determined by plaque assays of Vero cells as described previously (15). For the analysis of viral RNA by Northern blot hybridization, tissue samples obtained from LCMV-cgPi mice and controls were homogenized in TRIreagent (Molecular Research Center, Inc., Cincinnati, OH). RNA was extracted according to the supplier's protocol and resuspended in Formazol (Nuclear Research Center, Inc.). RNA samples were analyzed by Northern blotting (12).
Detection of viral antigen by immunohistochemistry.
Brains from adult LCMV-cgPi and uninfected age- and sex-matched C57BL/6 mouse controls were collected, embedded in Tissue-Tek O.C.T. compound (Miles Diagnostic Division, Elkhardt, IN), and frozen on dry ice. Sections (6 μm) were cut, placed onto Fisher Superfrost Plus microscopic slides, dried, and fixed in 4% (wt/vol) paraformaldehyde in phosphate-buffered saline. Guinea pig anti-LCMV antibody (1:500), mouse monoclonal antibody anti-NeuN (1:200), and rabbit polyclonal antibody to GFAP (1:500) were incubated at room temperature for 1 h. As secondary antibodies, anti-guinea pig IgG conjugated to rhodamine X and anti-mouse or anti-rabbit IgG conjugated to FITC were used in a dilution of 1:100. Images were obtained using a Zeiss Axiovert S100 microscope (Carl Zeiss Inc. Thornwood, NY) fitted with a 5× objective, an AxioCam digital camera, and an automated stage. For digital image acquisition and processing, an Axiovision software package (Zeiss) and Adobe Photoshop were used. For a quantitative assessment of LCMV infection in specific neuronal populations, sets of three consecutive sagittal brain sections (6 μm) obtained from three different depths of the brain hemispheres were subjected to double staining with antibody to LCMV and an antibody against the neuronal marker NeuN. Two-color registered images were captured from four visual fields per section using a 20× objective, 490 and 570 nm excitation, and a narrow band-pass filter. The total number of NeuN-positive cells per visual field was determined, and the total number of LCMV antigen-positive cells was scored. Percentages of infected NeuN-positive cells were calculated from the averages of four visual fields obtained from three sections at three different depths of brain hemispheres.
For light microscopy, immunohistochemical staining for LCMV with diaminobenzidine as a chromogen was performed as described previously (6), using guinea pig anti-LCMV antibody in a dilution of 1:1,000 and horseradish peroxidase-conjugated anti-guinea pig IgG as a secondary antibody. Sections were counter-stained with Mayer's hematoxylin and eosin (H&E) stain (Sigma).
Nonconditional, spatial discrimination task.
LCMV-cgPi and age- and sex-matched control mice (n = 10/group) were subjected to a nonconditional, spatial discrimination task (7, 20). This test is based on measuring the ability of the mice to learn a Y-maze employing spatial discrimination to avoid the onset of a mild foot shock (0.43 mA). LCMV-cgPi and control mice were trained in five trials per day for 6 days. Errors were defined as entries into the wrong arm of the Y-maze or reentering the start compartment before or after the onset of shock. Both LCMV-cgPi and mock-infected control mice exhibited similar foot-shock thresholds; therefore, the reduced ability of LCMV-Pi mice to learn was not due to different sensitivities to the mild foot shock used to motivate performance.
DNA array analysis.
Total RNA was isolated from brain tissue by use of TRIreagent. DNA array analysis was performed at the DNA array core facility of the Scripps Research Institute as described previously (11). Briefly, 5 μg of total RNA from each sample was subjected to reverse transcription (RT) using a oligo(dT) primer containing a T7 promoter site (Genset, La Jolla, CA) and a cDNA synthesis kit (InVitrogen, San Diego, CA). The resulting double-stranded cDNA was in vitro transcribed into biotin-labeled cRNA by use of biotin-11-CTP and biotin-16-UTP (ENZO, Farmingdale, NY). After purification (RNeasy spin column; QIAGEN), 20 μg of labeled cRNA was fragmented by mild alkaline treatment and hybridized to 430 2.0 and U74Av2 Affymetrix murine genomic chips (Santa Clara, CA) according to the manufacturer's instructions. After washing and reacting with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR), hybridization signals were acquired using an Affymetrix laser scanner and the corresponding gene expression software provided by Affymetrix.
For data analysis, user-defined parameters were set to ensure confidence for detection of a transcript (defined as present) or its absence (http://www.affymetrix.com/products/. [Statistical algorithms reference guide]). For each experiment, four samples (two for each experimental group) were analyzed. Raw data were displayed in a Microsoft Excel worksheet; absent transcripts were excluded. For the detection of transcripts whose signals were significantly different between LCMV-cgPi and control samples, we considered only changes that were 1.41-fold or greater for total brain RNA and 3-fold or greater for the pooled hippocampus RNA samples. The data were combined from two independent experiments, and we considered changes that passed a statistical analysis with a Student's t test to ascertain significance at a P value of <0.01.
RNA analysis by RT-PCR.
Total RNA was isolated from brain tissue by using TRIreagent. Before the RT, contaminant DNA was removed by using a DNA-free kit (Ambion, Austin, TX). The RT reaction was performed using 5 μg RNA with SuperScript II and random hexamer primers (both from Invitrogen). The PCR was done by using Taq polymerase and the specific primer sets displayed in Table 1. The mRNA of the control housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified as described previously (56). For semiquantitative analysis we first determined a linear range of PCR product-template by serial dilution of the RT products obtained with the mock-infected samples. To validate quantitative differences in mRNA concentration of the candidate genes between LCMV-cgPi and control samples, we performed PCR on identical RT product dilutions within the linear range of PCR product-template. PCR products were separated on agarose gels and visualized by staining with ethidium bromide. Images were acquired using an Eagle-Eye digital camera.
TABLE 1.
Primers used for PCR amplification of candidate ISGs
| Gene | Forward primer | Reverse primer |
|---|---|---|
| STAT1 | 5′-GCT GAA TTC CAT CGA GCT CAC TCA-3′ | 5′-AAA CTC GAG CTC AGC TGC CAG ACT TCC-3′ |
| IRF9 | 5′-TTT CTC GAG AGG ACC AGG ATG CTG CCA TAT-3′ | 5′-AAA GAA TTC AGA GGA AGT TGA TGC TCC AGG-3′ |
| ISG15 | 5′-TTT GAA TTC CTG GTG TCC GTG ACT AAC-3′ | 5′-AAA CTC GAG CAC CCC TCA GGC GCA AAT GCT-3′ |
| UBP43 | 5′-GAA AGC CTT GCT GCC CAC GGA-3′ | 5′-TCC CTG GCA GAG AAT CTC ATG-3′ |
| GARG16 | 5′-ATG GAG AAT CTG CTT CAG CT-3′ | 5′-CTC CAC ACT TCA GCA AGG CC-3′ |
| GARG39 | 5′-AAT GCC ATT TCA CCT GGA AC-5′ | 5′-CAG AGG TGA ATT CTG GGT TC-3′ |
| GARG49 | 5′-AGG GAA GGA AGT ATG TCC AG-3′ | 5′-AGG TAC TGG TTC TGA GGA TT-3′ |
RESULTS
Characterization of vertically maintained LCMV persistence in mice.
Mice persistently infected with LCMV at birth via intracranial injection develop a life-long chronic infection (LCMV-Pi) in the absence of noticeable clinical symptoms (8). However, LCMV-Pi mice exhibit specific behavioral and learning deficits that are associated with defined neurochemical disturbances and altered CNS gene expression during adulthood (15, 20, 25, 31). Studies using the LCMV-Pi model have shed light on a variety of questions related to viral persistence and associated disorders as well as on the feasibility and benefits of immunocytotherapy to treat chronic viral infections (43). This model, however, has the limitation of not recreating accurately the natural route of transmission for the majority of congenital, either pre- or postnatal, CNS infections in humans; such infections involve vertical transmission from infected mothers to the fetus or neonate (1, 2).
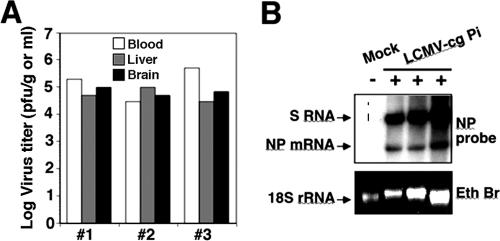
As vertical transmission plays an important role in arenavirus ecology, we reasoned that LCMV could provide us also with a valuable model system for the study of the consequences of congenital infections by noncytolytic persistent viruses. For this purpose, we established an LCMV persistently infected mouse colony where the virus is maintained via vertical transmission from infected mothers to offspring (LCMV-cgPi) instead of by intercranial inoculation at birth (LCMV-Pi). At 6 months of age, LCMV-cgPi mice and (sex- and age-matched) controls had similar gross appearances, body weights, and blood glucose levels, indicating no overt clinical abnormalities. Adult LCMV-cgPi mice had virus serum titers of 104 to 105 PFU/ml and 104 to 105 PFU per gram of tissue in liver and brain (Fig. 1A). High levels of viral S RNA and nucleoprotein (NP) mRNA were detected in the brain of LCMV-cgPi mice (Fig. 1B), indicating significant levels of viral replication and gene expression. Examination of H&E-stained brain sections from LCMV-cgPi mice failed to revealed anatomic abnormalities, significant changes in normal astroglia appearance, or the presence of immune cell infiltrates (data not shown). Hence, our LCMV-cgPi model represents a vertically maintained persistent infection in the absence of overt clinical signs or pathology.
FIG. 1.
Viral load in LCMV-cgPi mice. (A) Virus titers in brain (white), liver (gray), and blood (black) of LCMV-cgPi mice determined by plaque assay. (B) Levels of viral RNA in brain tissue. RNA was isolated from brain tissue of LCMV-cgPi and mock-infected control mice (6 months old). RNA (equal amounts of each sample) was analyzed by Northern blot hybridization using a DNA probe to LCMV nucleoprotein (NP) that recognizes the S genome RNA (replication) and the NP mRNA (transcription). Samples from three independent mice are shown.
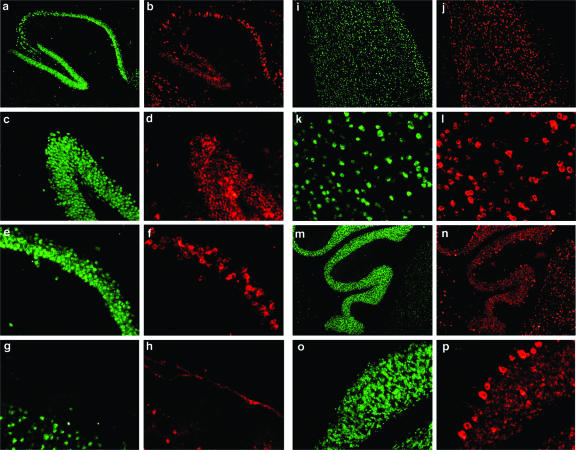
Next, we examined virus distribution in the brain of adult LCMV-cgPi mice. For this, we used immunofluorescence procedures with anti-LCMV hyperimmune guinea pig serum and a monoclonal antibody against the neuronal marker NeuN. We detected LCMV antigen throughout the brain, with a high abundance in neocortex, hippocampus, and cerebellum (Fig. 2) and, at lesser levels, in the brain stem, thalamus, and basal ganglia (data not shown). Within the brain parenchyma, the virus was primarily confined to neurons, and infection of astrocytes was negligible, as determined by the lack of detection of LCMV antigen in cells expressing the astrocytic marker GFAP (data not shown). In contrast to the infection patterns described for LCMV-Pi mice generated by LCMV intracranial inoculation at birth (19, 53), we detected significant amounts of LCMV antigen in cells of tissues lining the brain parenchyma, such as the meninges, the choroid plexus, and the ventricle walls (Fig. 2).
FIG. 2.
Virus distribution in the brain of LCMV-cgPi mice. Viral antigen in whole-brain sections of 6-month-old LCMV-cgPi mice was detected by immunofluorescence staining performed using frozen brain sections and hyperimmune guinea pig serum to LCMV and a rhodamine red X-labeled secondary antibody (red). Neurons were labeled with an antibody to NeuN and a FITC-labeled secondary antibody (green). a and b, hippocampus; c and d, dentate gyrus; e and f, C1 region; g and h, meninges; i to l, cerebral cortex; m to p, cerebellum.
Congenital CNS infections of humans with a number of viruses show a wide range of clinical and pathophysiological manifestations due to variation in the exact time point of infection, route of entry, and amount of pathogen involved in CNS invasion of the fetus (2). To address possible variability in the pattern of infection in the brain of LCMV-cgPi mice, we examined the distribution of viral antigen in a group (n = 7) of 6-month-old LCMV-cgPi mice with comparable serum viral loads. For this, sets of consecutive sagittal brain sections obtained from different depths of the brain were coimmunostained for LCMV antigen and NeuN. The extent of infection of a given neuronal population (Table 2) was assessed by determination of the percentage of LCMV antigen-positive cells normalized to the total number of NeuN-positive cells as described in Materials and Methods. The majority of mice (6 out of 7) showed high viral loads in neurons throughout the brain parenchyma as well as the meninges, choroid plexus, and ventricle walls. However, one mouse (mouse 4) showed limited infection of the brain parenchyma but high viral load in tissues lining the CNS. Notably, we observed significant variability in the extent and distribution of viral antigen expression within the hippocampus formation (Table 2). Three mice (mice 1 to 3) showed similar high viral loads in the granule cells of the dentate gyrus and the pyramidal cells of the CA1, CA2, and CA3 regions of the hippocampus as well as in the cortex and cerebellum and within the brain parenchyma. In contrast, three other mice (mice 5 to 7) exhibited significant region-specific differences in viral load. Together, these data indicated a significant degree of variability in the pattern of neuronal infection of virus in LCMV-cgPi mice, a situation reminiscent of human congenital CNS infection. We did not detect viral antigen in astrocytes, but some mice displayed a slight increase in the number of GFAP-positive astrocytes in the neocortex but not in the hippocampus or cerebellum (Table 2). The limited number of GFAP-positive astrocytes present in the brains of LCMV-cgPi mice is consistent with the absence of reactive astrogliosis as defined by the presence of GFAP-positive hypertrophic astrocytes and the overall normal aspect of the astroglia in routine H&E stains.
TABLE 2.
Heterogeneity in virus load and/or distribution in the brain of LCMV-cgPi micea
| Animal | Proportion of LCMVNP-positive neurons in different brain regions
|
Proportion of GFAP-positive astrocytes
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hippocampus
|
Cortex | Cerebellum | Meninges | Cortex | Hippocampus | Cerebellum | ||||
| DG | CA3 | CA2 | CA1 | |||||||
| 1 | +++ | +++ | +++ | +++ | +++ | +++ | ++ | + | − | − |
| 2 | +++ | +++ | +++ | +++ | +++ | +++ | ++ | + | − | − |
| 3 | +++ | +++ | +++ | +++ | +++ | +++ | ++ | + | − | − |
| 4 | + | + | + | + | + | ++ | ++ | − | − | − |
| 5 | ++ | +++ | +++ | +++ | +++ | +++ | ++ | + | − | − |
| 6 | ++ | +++ | ++ | ++ | +++ | ++ | ++ | + | − | − |
| 7 | + | ++ | ++ | ++ | +++ | ++ | ++ | − | − | − |
Immunohistochemical examination of LCMV-cgPi mice. Six-month-old female LCMV-cgPi mice (n = 7) were examined by immunohistochemistry (Fig 2). Virus loads in neurons of the hippocampus dentate gyrus (DG) and regions CA1, CA2, and CA3 as well as the cortex and cerebellum were determined as described in Materials and Methods and are indicated as follows: +++, >30% infected neurons; ++, 10% to 30% infected neurons; +, <10% infected neurons; −, no detectable infection. All mice exhibited significant infection of meninges, choroid plexus, and cells of the ventricle walls. The immunohistochemical analysis of brain sections of mouse 2 is shown in Fig. 2. Some mice showed mild increases in the numbers of GFAP-positive astrocytes in the cerebral cortex but not in the hippocampus or cerebellum.
Behavioral assessment of LCMV-cgPi mice.
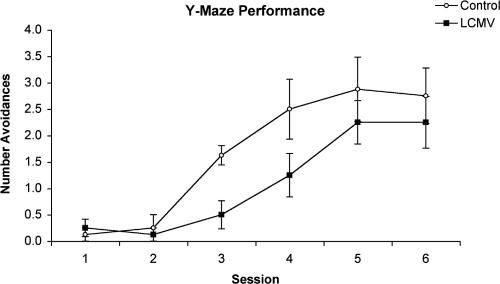
Mice persistently infected with LCMV as neonates exhibit impaired learning abilities and a reduced tendency to explore a novel environment during their adult life (20, 25). To investigate behavioral abnormalities associated with LCMV-cgPi mice we used a well-established nonconditional, spatial discrimination task (20). This test represents a short-term memory task involving trial-independent memory processes and is based on measuring the ability of the animals to learn a Y-maze spatial discrimination to avoid the onset of a mild foot shock (0.43 mA). Groups of 10 (age- and sex-matched) LCMV-cgPi and control mice were trained in five trials per day for 6 days. The number of errors (defined as entries into the wrong arm of the Y-maze or reentering the start compartment before or after the onset of shock) made by the mice provided us with a measure of acquisition performance. Compared to mock-infected controls, LCMV-cgPi mice exhibited impaired discrimination avoidance learning, reflected in a significantly lower number of correct avoidance responses on experimental days 3 and 4 with P values of <0.01 and <0.05, respectively, using unpaired t tests (Fig. 3). Following additional training sessions, LCMV-cgPi mice reached the same level of performance as the mock-infected control mice, suggesting retardation in acquisition of spatial discrimination in LCMV-cgPi mice.
FIG. 3.
Deficits in discriminated avoidance learning in LCMV-cgPi mice. Groups of 10 (age- and sex-matched) LCMV-cgPi mice (LCMV) and uninfected control mice (control) were trained in five trials per day for 6 days to choose the safe arm of a Y-shaped maze and hence to avoid a mild electric foot shock. The mean (± standard error of the mean) number of avoidance responses is shown.
Gene expression profiling of brain tissue from LCMV-cgPi mice.
Since the cognitive defects observed in LCMV-cgPi mice occurred in the absence of overt CNS pathology, we hypothesized that chronic virus infection may contribute to neuronal dysfunction by altering the host's gene expression profile. To test this hypothesis we evaluated the impact of persistent virus infection on host gene expression in the CNS by use of DNA array-based gene profiling. Since we observed significant variations in overall virus load and distribution in the LCMV-cgPi model (Table 2), we performed gene expression profiling using total RNA isolated from brain hemispheres of two selected individual LCMV-cgPi mice (mouse 2 and mouse 3) (Table 2) with similar patterns of CNS infection.
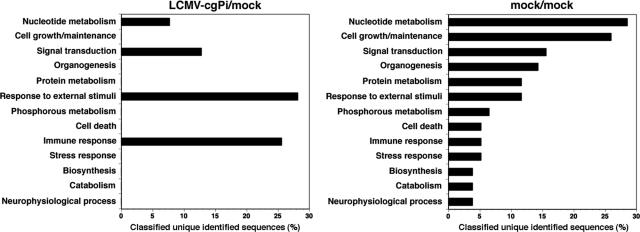
Comparison of all four permutations of LCMV-cgPi and control mouse datum sets revealed the consistent induction of 56 genes and reduction of 19 genes in all LCMV-cgPi versus control mice out of a total of 39,000 genes represented on the 430 2.0 array (for the complete gene list, see Table S1 in the supplemental material). Using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.niaid.nih.gov), we identified 21 out of the 56 and 12 out of the 19 genes with expression levels that had increased and decreased, respectively, in LCMV-cgPi compared to control mice (Table 3). Host genes with altered expression in LCMV-cgPi mice belonged to four main categories: those involved in (i) immune response, (ii) responses to external stimuli, (iii) signal transduction, and (iv) nucleotide metabolism (Fig. 4). We also examined individual variations of the gene expression profile between mock-infected mice. This analysis revealed significant differences in a number of genes (see Table S2 in the supplemental material). However, when genes with differential expression results between mock-infected mice were annotated, they fell into a broad range of categories and were essentially nonoverlapping with the group of genes found regulated in LCMV-cgPi versus control mice (Fig. 4). This finding underscores the specificity of changes in gene expression associated with LCMVcgPi. Moreover, our data indicate that the overall changes in host gene expression associated with LCMV-cgPi were very limited, affecting <0.2% of the 39,000 genes represented by the DNA assay. ISGs represented a major group among the genes with increased expression in LCMV-cgPi mice, suggesting that chronic activation of these genes may be involved in antiviral defense (Table 3). Genes down-regulated in LCMV-cgPi mice comprised a more heterogenous group that includes structural genes and genes involved in metabolic functions (Table 3).
TABLE 3.
Known host genes whose expression is changed the brain of LCMV-cgPi micea
| GenBank accession no. | Value for indicated gene
|
Gene identity | |||||||
|---|---|---|---|---|---|---|---|---|---|
| LCMV-cgPi2/ control 1
|
LCMV-cgPi2/ control 2
|
LCMV-cgPi3/ control 1
|
LCMV-cgPi3/ control 2
|
||||||
| Signal ratio | P | Signal ratio | P | Signal ratio | P | Signal ratio | P | ||
| U43084 | 2.2 | 0.000189 | 1.5 | 0.000023 | 2.6 | 0.000068 | 2 | 0.00002 | GARG-16 |
| AB067535 | 2 | 0.000307 | 0.9 | 0.000966 | 1.4 | 0.000774 | 0.3 | 0.001336 | 2′-5′ OAS 2 |
| AW047653 | 1.7 | 0.00002 | 1.9 | 0.00002 | 1.7 | 0.00002 | 1.8 | 0.00002 | UBP43 |
| BQ033138 | 1.7 | 0.00003 | 1.7 | 0.000023 | 1.4 | 0.000114 | 1.8 | 0.000078 | 2′-5′ OAS-like 2 |
| M74124 | 1.6 | 0.00006 | 2 | 0.00002 | 1.2 | 0.000241 | 1.6 | 0.000027 | Interferon-activated gene 205 |
| M29881 | 1.5 | 0.000147 | 1.6 | 0.000147 | 2 | 0.00006 | 2 | 0.000027 | MHCI Q region locus 7 |
| BG068242 | 1.4 | 0.000052 | 1.4 | 0.00003 | 1.2 | 0.000346 | 1.4 | 0.000346 | Tripartite motif protein 30-like |
| BB757349 | 1.2 | 0.00002 | 1.4 | 0.00002 | 1.2 | 0.00002 | 1 | 0.00002 | Zinc finger CCCH type, antiviral 1 |
| X67809 | 1.2 | 0.000046 | 1.1 | 0.000035 | 1.1 | 0.000078 | 1 | 0.000023 | Peptidylprolyl isomerase C-associated |
| U43086 | 1.1 | 0.000046 | 1.3 | 0.00002 | 1.4 | 0.000052 | 1.5 | 0.00002 | GARG-49 |
| U51992 | 0.9 | 0.000273 | 1.1 | 0.000052 | 0.8 | 0.000027 | 1.2 | 0.00002 | IRF9 |
| NM_008326 | 0.8 | 0.000052 | 0.6 | 0.000438 | 1.2 | 0.000035 | 0.8 | 0.000774 | Interferon inducible protein 1 |
| BM227980 | 0.8 | 0.000035 | 0.7 | 0.000078 | 0.7 | 0.00002 | 0.7 | 0.000147 | Zinc finger CCCH type domain-containing 1 |
| U06924 | 0.8 | 0.000214 | 0.6 | 0.00002 | 1 | 0.000167 | 0.9 | 0.00002 | STAT1 |
| NM_008330 | 0.7 | 0.00006 | 1.1 | 0.000346 | 1.5 | 0.000618 | 1.8 | 0.000692 | Olfactory receptor 56 |
| BC011306 | 0.7 | 0.000035 | 0.8 | 0.00003 | 0.7 | 0.000023 | 0.9 | 0.00002 | MHCI K region |
| M86502 | 0.7 | 0.00002 | 0.6 | 0.00002 | 0.5 | 0.00002 | 0.5 | 0.00002 | MHCIb D region locus 1 |
| NM_011579 | 0.7 | 0.000088 | 1 | 0.000189 | 0.8 | 0.00002 | 1.1 | 0.000052 | T-cell-specific GTPase |
| BF715219 | 0.7 | 0.00002 | 0.6 | 0.00002 | 0.7 | 0.00002 | 0.6 | 0.00002 | Beta-2 microglobulin |
| NM_010398 | 0.6 | 0.000023 | 0.5 | 0.000023 | 0.6 | 0.00002 | 0.6 | 0.00002 | MHCI T region locus 23 |
| BB418548 | 0.5 | 0.000068 | 0.5 | 0.00002 | 0.7 | 0.00002 | 0.9 | 0.00002 | Doublecortin |
| NM_010758 | -0.5 | 0.99998 | −0.5 | 0.99998 | −0.5 | 0.999977 | −0.5 | 0.00002 | Myelin-associated glycoprotein |
| NM_026731 | −0.6 | 0.999932 | −0.4 | 0.999922 | −0.7 | 0.999965 | −0.7 | 0.00004 | Protein phosphatase 1 regulatory subunit 14A |
| U64572 | −0.6 | 0.999977 | −0.8 | 0.99998 | −0.6 | 0.999922 | −0.7 | 0.00002 | Myelin oligodendrocyte glycoprotein |
| M58045 | −0.6 | 0.99998 | −0.5 | 0.99998 | −0.5 | 0.999693 | −0.3 | 0.00002 | Cyclic nucleotide phosphodiesterase 1 |
| BB353220 | −0.6 | 0.999899 | −0.6 | 0.99998 | −0.5 | 0.999899 | −0.6 | 0.00003 | LIM and senescent cell antigen-like domains 2 |
| NM_010354 | −0.7 | 0.99998 | −0.6 | 0.99998 | −0.6 | 0.99998 | −0.5 | 0.00002 | Gelsolin |
| NM_008614 | −0.7 | 0.99998 | −0.7 | 0.99998 | −0.4 | 0.999611 | −0.4 | 0.00002 | Myelin-associated oligodendrocytic basic protein |
| BQ175510 | −0.7 | 0.99998 | −0.5 | 0.99998 | −0.6 | 0.99998 | −0.4 | 0.00006 | G protein-coupled receptor 37 |
| BC024556 | −0.8 | 0.99998 | −0.6 | 0.999977 | −0.6 | 0.999911 | −0.3 | 0.00008 | PDZ and LIM domain 2 |
| AW550625 | −0.8 | 0.999727 | −0.8 | 0.999034 | −0.6 | 0.999611 | −0.8 | 0.007 | Procollagen, type III, alpha 1 |
| NM_026174 | −0.9 | 0.999977 | −0.8 | 0.99998 | −1 | 0.999922 | −0.9 | 0.00001 | Lysosomal apyrase-like 1 |
| NM_011674 | −1 | 0.99998 | −0.8 | 0.99998 | −0.6 | 0.999977 | −0.4 | 0.0003 | UDP-glucuronosyltransferase 8 |
Known host genes with altered expression in brains of LCMV-cgPi mice. Gene expression profiling was performed with total RNA isolated from brain hemispheres of LCMV-cgPi mice 1 and 2 (Table 2) and two controls (mock 1 and 2) by use of the Affymetrix murine genome oligonucleotide DNA array 430 2.0. All four permutations of comparisons between the two LCMV-cgPi mice and the two control animals were made. Genes were identified using the Annotation Tool of the Database for Annotation, Visualization, and Integrated Discovery (DAVID). Displayed are the known host genes that showed consistent regulation of >1.4-fold. The signal ratios for each permutation and the P values of changes are indicated.
MHCI, major histocompatibility complex class I.
FIG. 4.
Functional classification of genes with altered expression in LCMV-cgPi mice. Unique identified sequences up-regulated in all permutations between LCMV-cgPi and uninfected control mouse brains (LCMV-cgPi/mock) and between control mouse brains (mock/mock) were analyzed using the Annotation Tool of the Database for Annotation, Visualization, and Integrated Discovery (DAVID). Displayed are the percentages of classified unique identified sequences in the designated categories. Some genes may appear in two categories, and some remain unclassified.
Induction of a common set of IFN-regulated genes in total brain and hippocampus of LCMV-cgPi mice.
Based on the role of the hippocampus in spatial-temporal learning and memory, we next addressed the impact of LCMV persistent infection on hippocampal gene expression. Hippocampus tissue from LCMV-cgPi and control mice was obtained by manual dissection (21). This type of dissection provides tissue samples highly enriched in hippocampus material. However, because of variability between individual specimens, we used pools of 10 (age- and sex-matched) LCMV-cgPi mice and uninfected controls for each DNA array analysis of hippocampus tissue. For DNA array analysis, we used the less complex Affymetrix murine genomic DNA array U74Av2, which represents only a well-characterized subset of 12,000 messages, and considered only genes that exhibited a reproducible change in different experiments (n = 2) of threefold or higher between LCMV-cgPi and control mice. With this more stringent criterion we found 26 and 5 genes with expression that had increased and decreased, respectively, in LCMV-cgPi mice compared to controls. As with our findings using total brain RNA, the majority (13) of the 18 known genes induced in hippocampus tissue of LCMV-cgPi mice were ISGs. Comparison between whole-brain and hippocampal gene expression profiles revealed induction in LCMV-cgPi mice of an overlapping set of ISGs, including the transcription factors STAT1 and IRF9, the ISG15 protease UBP43, and the glucocorticoid attenuated response genes GARG16 and GARG49. Several ISGs, including the ubiquitin-like protein ISG15, GARG39, and the IFN-regulated GTPases LRG47 and GTPI, appeared to be specifically induced in the hippocampus (Table 4).
TABLE 4.
Known host genes whose expression is changed the hippocampus of LCMV-cgPi micea
| GenBank accession no. | LCMV-cgPi signalb | Mock signalb | Signal ratio | P | Gene identityc |
|---|---|---|---|---|---|
| AW047653 | 206.3 | 6.2 | 33.27 | 0.000002 | UBP43 |
| U43086 | 493.6 | 34.8 | 14.18 | 0.000001 | GARG-49 |
| U43084 | 454 | 39.6 | 11.46 | 0.000001 | GARG-16 |
| X56602 | 377.2 | 42.1 | 8.96 | 0.000001 | ISG15 |
| D90146 | 122 | 16.6 | 7.35 | 0.000533 | MHCI Q8/9d |
| M27134 | 176.3 | 24.9 | 7.08 | 0.000085 | MHCI K region locus 2 |
| U06924 | 330.8 | 49.3 | 6.71 | 0.000001 | STAT1 |
| X16202 | 892.1 | 195 | 4.57 | 0.000001 | MHCI Q4 |
| U55060 | 234.2 | 57.2 | 4.09 | 0.000054 | β-Gal binding lectin |
| X67809 | 892.2 | 233.1 | 3.83 | 0.000001 | Peptidylprolyl isomerase C-associated protein |
| D63902 | 119.6 | 31.6 | 3.78 | 0.000004 | Zinc finger protein 147 |
| U43085 | 121 | 33.1 | 3.66 | 0.000253 | GARG-39 |
| U19119 | 193 | 55.2 | 3.50 | 0.000001 | LRG-47 |
| AJ007972 | 265.7 | 76.3 | 3.48 | 0.000021 | GTPI |
| X01838 | 3369.6 | 989.1 | 3.41 | 0.000001 | Beta-2 microglobulin |
| U51992 | 502.5 | 157.2 | 3.20 | 0.000001 | IRF-9 |
| L12367 | 34.6 | 215.1 | −6.22 | 0.000074 | Adenylyl cyclase-associated protein |
| V00727 | 80.5 | 321.9 | −4.00 | 0.001304 | FBJ osteosarcoma oncogene |
Known host genes with altered expression in hippocampal tissue of LCMV-cgPi mice. Hippocampus tissue was pooled from age- and sex-matched LCMV-cgPi mice (n = 10) with virus titers of 104 to 105 PFU/ml and from control mice (Mock) (n = 10), and total RNAs were isolated and used for gene profiling studies using Affymetrix murine genomic DNA array U74Av2. Genes were identified as described for Table 3. Displayed are the known host genes that showed consistent regulation differences of >3-fold between LCMV-cgPi mice and controls. The mean fluorescence intensities for each probe set and the signal ratios with corresponding P values of changes are indicated.
Fluorescence units (intensity).
MHCI, major histocompatibility complex class I; β-Gal, β-galactosidase.
The overall high loads of viral antigen detected in hippocampal neurons (Table 2) compared to the absence of detectable changes in the expression of neuron-specific genes was surprising. This finding might be due, at least in part, to the limited sensitivity of the screening assay. However, it was also possible that LCMV persistence interfered specifically with neuronal genes, whose expression is induced only in direct response to certain environmental stimuli, such as a learning task. To test for this latter possibility we examined the hippocampal gene expression profile in LCMV-cgPi and control mice with tissue material collected immediately after the mice were subjected to the Y-maze discriminated avoidance task. This analysis revealed 21 known genes with >3-fold-increased expression in LCMV-cgPi compared to control mice (Table 5). Thirteen of these 21 genes were also up-regulated >3-fold in mice that were not exposed to the behavioral test. The regulation of only two genes was reduced >3-fold in hippocampal tissue of LCMV-cgPi collected immediately after the learning task. All ISGs induced in hippocampus of unchallenged LCMV-cgPi animals were also detected in the challenged group. Additional ISGs induced only in the challenged group were the IFN-regulated GTPases IIGP and mGBP2.
TABLE 5.
Known host genes whose expression is changed the hippocampus of LCMV-cgPi mice exposed to an avoidance-learning testa
| Genbank accession no. | Mock signalb | LCMV-cgPi signalb | Signal ratio | P | Gene identityc |
|---|---|---|---|---|---|
| M27134 | 20.7 | 225.1 | 10.87 | 0.000006 | MHCIb K region locus 2 |
| U43086 | 43.6 | 462.9 | 10.62 | 0.000001 | GARG-49 |
| M58156 | 23.3 | 236.8 | 10.16 | 0.000004 | MHCI H-2K-f |
| AW047653 | 21.7 | 190.8 | 8.79 | 0.000333 | UBP43 |
| U06924 | 32.7 | 258.7 | 7.91 | 0.000001 | STAT1 |
| U43084 | 40.5 | 318 | 7.85 | 0.000001 | GARG-16 |
| M18237 | 65 | 505.9 | 7.78 | 0.000001 | Ig kappa chain V-region |
| AB035174 | 47.7 | 341.9 | 7.17 | 0.001304 | GalNAc alpha 2,6-sialyltransferase |
| AF099973 | 31.3 | 191 | 6.10 | 0.000285 | Schlafen2 |
| AJ007971 | 24.4 | 147.4 | 6.04 | 0.000001 | IIGP |
| X56602 | 54 | 310.4 | 5.75 | 0.000002 | ISG15 |
| X67809 | 177.7 | 1001.7 | 5.64 | 0.000001 | Peptidylprolyl isomerase C-associated protein |
| X16202 | 215.8 | 976.7 | 4.53 | 0.000001 | MHCI Q4 |
| M69069 | 351.3 | 1271 | 3.62 | 0.000002 | MHCI D region locus 1 |
| X01838 | 827.7 | 2918.6 | 3.53 | 0.000001 | Beta-2 microglobulin |
| V00746 | 490.5 | 1603.2 | 3.27 | 0.000001 | MHCI H-2K |
| U43085 | 24.8 | 78.1 | 3.15 | 0.000007 | GARG-39 |
| U51992 | 157.5 | 494.2 | 3.14 | 0.000001 | IRF9 |
| U60020 | 58.3 | 175.7 | 3.01 | 0.000004 | Transporter 1, ABC |
| AJ007972 | 86.7 | 204.6 | 2.36 | 0.001767 | GTPI |
| U19119 | 72.7 | 162.6 | 2.24 | 0.000021 | LRG-47 |
| U66918 | 113.1 | 25.3 | −4.47 | 0.00021 | Short stature homeobox 2 |
| AF035684 | 238.5 | 27.3 | −8.74 | 0.00002 | Mus musculus beta chemokine TCA4 |
Known host genes whose expression is changed in the hippocampus of LCMV-cgPi mice exposed to an avoidance-learning test. Groups of 10 LCMV-cgPi mice with virus titers of 104 to 105 PFU/ml and 10 age- and sex-matched uninfected controls were subjected to the Y-maze discriminated avoidance task as described for Fig. 3. After the last session on day 6, animals were immediately euthanized and hippocampus tissue was dissected. Hippocampus tissue was pooled, and total RNAs were isolated and used for gene profiling studies with Affymetrix murine genomic DNA array U74Av2. Genes were identified as described for Table 3. Displayed are the known host genes that showed consistent regulation differences of >3-fold between LCMV-cgPi mice and controls. The mean fluorescence intensities for each probe set and the signal ratios with corresponding P values of changes are indicated.
Fluorescence units (intensity).
MHCI, major histocompatibility complex class I.
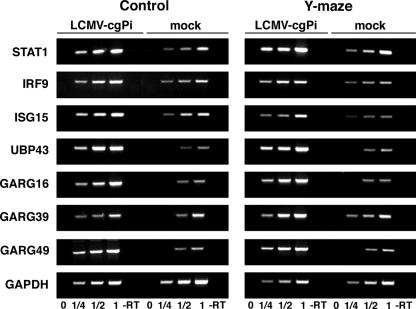
To validate the changes in mRNA levels identified by DNA array studies, we used semi-quantitative RT-PCR to determine the expression levels of the candidate ISGs STAT1, IRF9, ISG15, UBP43, GARG49, GARG39, and GARG16 in hippocampus tissue. For each of the candidate genes a segment of 400 to 500 bp within the open reading frame was amplified using specific sets of primers. As a control, we amplified a 200-bp fragment of the housekeeping gene GAPDH. The linear range of template/PCR product ratios for each candidate gene was determined by performing PCR on serial dilutions of the corresponding RT reaction product (for details, see Materials and Methods). Consistent with the DNA array data (Table 4 and Table 5), we detected significantly higher levels of STAT1, IRF9, ISG15, UBP43, GARG49, and GARG16 mRNA in total hippocampus RNA from LCMV-cgPi mice than from mock-infected animals (Fig. 5). However, we were unable to detect significant changes of GARG39 by RT-PCR, most probably due to its relatively weak induction and low signal intensity (Tables 4 and 5).
FIG. 5.
Validation of changes in gene expression of selected candidate genes in LCMV-cgPi mice. Total RNA was isolated from hippocampus tissue, and contaminant DNA was removed. A reverse transcription (RT) reaction was performed using 5 μg RNA and random hexamer primers, and specific PCR fragments were amplified using specific primers for the candidate genes indicated and the housekeeping gene GAPDH. For semiquantitative analysis the linear range of PCR product-template was determined by serial dilution of the RT products obtained with the mock-infected samples. To detect quantitative differences in mRNA concentration of the candidate genes between LCMV-cgPi and control samples, PCR was performed on identical RT product dilutions within the linear range of PCR product-template. Results for mock-infected controls and LCMV-cgPi animals either from the control group (control) or exposed to the Y-maze learning paradigm (Y-maze) are shown. A control reaction without RT (−RT) is indicated.
DISCUSSION
Congenitally acquired viral chronic infection of the CNS can frequently result in CNS abnormalities, including cognitive defects in the absence of overt pathology. We therefore hypothesized that viral persistence may contribute to neuronal dysfunction by altering the host's gene expression profile in the absence of cytolysis and inflammation. To test this hypothesis, we established a congenitally acquired LCMV persistent infection that recreates the natural route of viral transmission in its natural host, the mouse, and examined its impact on the host's CNS gene expression profile with DNA arrays. Our data show that congenital LCMV infection by vertical transmission (LCMV-cgPi) results in a lifelong persistent infection involving the CNS and that this condition is associated with impaired cognitive function and specific changes in CNS gene expression.
Previous studies aimed at examining the effect of LCMV persistence on CNS function were based on the use of intracranial inoculation of virus into the brain of neonate mice (LCMV-Pi). This model has been instrumental in addressing a variety of important questions, but it does not reflect the natural routes of transmission implicated in most congenital viral infections of the CNS, both pre- and postnatal, which involve vertical transmission from infected mothers to offspring (1, 2). To overcome this limitation, we established the LCMV-cgPi model, which allows for vertical transmission of LCMV from infected mothers to offspring (LCMV-cgPi). This resulted in a lifelong persistent infection in the absence of signs of inflammation. Viral titers in serum and tissues of LCMV-cgPi mice were found to be comparable to those in LCMV-Pi mice. As reported for LCMV-Pi mice (19, 53), the majority of LCMV-cgPi mice had high viral loads in neurons throughout the brain, including the cerebral cortex, hippocampus, and cerebellum. Although within the brain parenchyma, virus was primarily confined to neurons in both models, LCMV-cgPi mice had high virus loads in cells of meninges, choroid plexus, and the linings of the ventricle walls, which are largely free of virus in LCMV-Pi mice. Furthermore, we observed significant variability in viral distribution in the brains of LCMV-cgPi mice with similar serum titers, a situation reminiscent of human congenital CNS infection by rubella virus, varicella-zoster virus, and LCMV, viruses for which the pattern and consequences of infection relate directly to the timing of maternal infection and subsequent invasion of fetal tissue (2).
We and others have shown that LCMV-Pi mice exhibit impaired learning abilities and a reduced tendency to explore a novel environment (20, 25). Using the same nonconditional spatial discrimination task previously applied to the LCMV-Pi model (20), we found similar defects in spatial and temporal learning in LCMV-cgPi mice. This suggests that the cognitive defects occur independently of the route and time of CNS infection.
Despite high viral loads in CNS neurons of LCMV-cgPi mice, our gene expression profiling studies revealed remarkably limited changes in the host's gene expression profile. Out of 39,000 host genes probed by our DNA array analysis only 56 and 19 genes showed significant induction and reduction, respectively, in LCMV-cgPi and control mice, corresponding to less than 0.2% of the host genome. Notably, the majority of the known genes with increased expression in LCMV-cgPi mice belonged to the group of ISGs and included members known to play important roles in the host's innate and adaptive immune responses that contribute to the control of virus multiplication and spread. In addition, chronic upregulation of ISGs may also contribute to altered CNS function.
Changes in acetylcholine enzymes, GAP-43, and neurotransmitters have been previously reported for LCMV-Pi, and LCMV-Pi and LCMV-cgPi mice exhibit similar behavioral deficiencies. Our cDNA array data did not detect changes in these genes. Several reasons could account for this apparent lack of consistency. Thus, in the case of GAP-43, previous work reported a decrease of less than 30% in levels of GAP-43 immunoreactivity within the hippocampus molecular layer of LCMV-Pi mice compared to control mouse results. Since GAP-43 is a presynaptic protein, its deficiency in the molecular layer of the hippocampus likely reflects altered GAP-43 transcription in entorhinal cortex and septal region neurons that originally produce GAP-43. This modest and localized reduction in GAP-43 expression may have a significant impact in brain function, but it would be very difficult to detect via cDNA arrays. Likewise, only very modest changes have been reported for neurotransmitters and acetylcholine enzymes in LCMV-Pi compared to control mice, which could explain the difficulties in reliably detecting such alterations using cDNA arrays.
Since the hippocampus is implicated in learning and memory, we performed DNA array analysis on dissected hippocampus tissue of LCMV-cgPi mice and uninfected controls. As with our findings for total brain, only a small fraction of host genes examined (<0.5%) showed altered expression in LCMV-cgPi mice; these genes again included a large proportion of IFN-regulated genes. Considering the high viral loads found in hippocampal neurons of the LCMV-cgPi mice examined, these remarkably few changes in the host's gene expression profile are rather surprising. The absence of changes in the expression of neuron-specific genes was unexpected, but it could be explained, at least in part, by the limited sensitivity of the screening assay. It is possible that only infected neurons would exhibit altered expression of specific neuronal genes, and histological analysis showed that generally <50% of hippocampus neurons are infected in LCMV-cgPi mice. Thus, using a cutoff of >3-fold changes, our gene expression profiling of hippocampus tissue would be expected to detect reliably only changes in the range of 6-fold, or higher, for host genes whose expression is specifically affected in LCMV-infected neurons, thereby limiting our detection of some potentially important genes with altered CNS expression in LCMV-cgPi mice. The paucity of detectable changes in neuronal gene expression could be due to other factors. It is plausible that LCMV persistence would interfere only with genes whose expression is induced in response to certain environmental stimuli, such as a learning task. However, DNA array analysis of hippocampus RNA isolated from mice immediately after they were subjected to the Y-maze testing revealed quantitative and qualitative changes in gene expression similar to those seen with mice that were not exposed to the behavioral test. Initial synaptic changes involved in memory formation do not require gene transcription (35, 36), so it is conceivable that the learning deficits observed in LCMV-cgPi mice may be due to a direct or indirect interference of the virus with the signal transduction events underlying these early steps of synaptic plasticity.
The most consistent changes in CNS gene expression in LCMV-cgPi mice involved genes implicated in type I alpha/beta IFN (IFNα/β) response. Type I IFNs represent a first line of defense against virus infection by generating an intracellular environment that restricts viral replication and influences the magnitude and quality of the subsequent antimicrobial adaptive immune response (22, 55). Type I IFNs induced in response to viral infections can function in an autocrine, paracrine, or endocrine fashion by binding to a common receptor (IFNAR). IFNα/β binding to IFNAR activates receptor-associated JAK kinases, which phosphorylate the signal transducer and activator of transcription STAT1 and STAT2. Activated STAT1 and STAT2 associate with IFN regulatory factor 9 (IRF-9) to form ISG factor 3 (ISGF3), which translocates to the nucleus and activates gene transcription via the IFN-stimulated response element sequence present within the promoters of ISG. While a number of studies have established a role for type I IFNs in acute LCMV infection (24, 37, 41, 47, 58), their role in persistent LCMV infection is less well known. Notably, levels of type I IFN-induced genes with antiviral activity like those coding for double-stranded RNA-activated kinase (PKR) and the Mx proteins, frequently induced in response to acute viral infection, were not elevated in the hippocampus of LCMV-cgPi mice. In contrast, we found long-term expression changes in a distinct subset of ISGs, including STAT1, IRF9, ISG15, UBP43, GARG49, and GARG16. The IFN-regulated transcription factors STAT1 and IRF9 are expressed at low levels in the normal CNS, but perhaps their expression can be induced under pathological conditions, including viral infection (27, 33, 48, 49, 63). ISG15 is a small ubiquitin-like protein strongly induced upon IFN stimulation. ISG15 covalently modifies target proteins following activation by the E1 enzyme UBE1L5. Recent studies demonstrated the induction of ISG15 in the CNS and an increase in ISG15 conjugation (ISGylation) in brain proteins after acute LCMV infection (50). The protease UBP43 is involved in removal of ISG15 from ISGylated proteins (32, 34, 57), and evidence indicates that UBP43 plays a key role in the regulation of ISGylation (51) and innate immunity to viral infection (50). Mice deficient in UBP43 were found to be resistant to lethal LCM or myeloencephalitis after intracerebral inoculation with LCMV or vesicular stomatitis virus, respectively. The protection against LCMV-induced lethal LCM in UBP43 (−/−) mice correlated with enhanced protein ISGylation and a concomitant reduction in viral replication. However, increased ISGylation may not be directly responsible for these findings (29).
Interestingly, gene expression profiling in a monkey model of neuroAIDS revealed a strong, concomitant up-regulation of the IFN-regulated genes ISG15, STAT1, and IRF9, as with our LCMV-cgPi model (52). However, while the induction of ISG15, STAT1, and IRF9 in our LCMV-cgPi model occurs in the absence of detectable signs of inflammation and adaptive immune response, the neuroAIDS model exhibits T-cell infiltration (62).
The glucocorticoid attenuated response genes GARG16 (interferon-induced protein with tetratricopeptide repeats; IFIT-1), GARG39 (IFIT-2), and GARG49 (IFIT-3) define a highly conserved family of IFN-induced genes, which contain multiple tetratricopeptide domains (59) involved in protein-protein interactions. Up-regulation of GARG39 and GARG49 has been found in the CNS after infection with Sindbis virus, rabies virus, and Japanese encephalitis virus (27, 49, 54), suggesting a role of these proteins in antiviral defense in the CNS.
Various neurological side effects have been found associated with the therapeutic use of IFN, including impaired learning and memory (13), and type I IFNs interfere with hippocampal synaptic plasticity (14, 38). In addition, STAT1-dependent signaling pathways have been shown to induce dendritic atrophy and perturb synaptogenesis and synaptic maturation in cultured hippocampus neurons (28, 61). Our findings may reflect a balance between the host responses aimed at limiting virus replication while minimizing damage and functional impairment of infected neurons. Chronic regulation of specific genes of the IFN system likely contributes to curtail the effects of virus replication and gene expression in LCMV-cgPi mice and therefore provides the host with a certain degree of protection. However, since elevated levels of IFNs found in chronic inflammation and persistent viral infections in the CNS are frequently accompanied by behavioral side effects, the up-regulation of IFN-induced genes may act as a double-edged sword and contribute to CNS disturbances in the host.
Supplementary Material
Acknowledgments
We thank Jennifer Hammond and other personnel of the Scripps DNA array core facility for their help with data acquisition and analysis and Debra Rosario for technical help.
This research was supported by U.S. Public Health Service grants AG04342 (J.C.T.) and AI009484 (M.B.A.O., S.K.), grant P30 MH062261-05 from Scripps NeuroAIDS Prelinical Studies (SNAPS) (S.K. and A.J.R.), and National Institutes of Health grant NS048866-01 and a grant from the Dana Foundation (D.B.M.).
Footnotes
This is publication no. 18119 from the Molecular and Integrative Neurosciences Department (MIND) of the Scripps Research Institute.
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Arvin, A. N. 1997. Viral infections of the fetus and neonate, p. 801-814. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven, Philadelphia, Pa.
- 2.Bale, J. F. 2005. Congenital and perinatal viral infections, p. 37-54. In R. K. L. Ross (ed.), Principles of neurologic infectious diseases. McGraw-Hill, New York, N.Y.
- 3.Barton, L. L., and M. B. Mets. 2001. Congenital lymphocytic choriomeningitis virus infection: decade of rediscovery. Clin. Infect. Dis. 33:370-374. [DOI] [PubMed] [Google Scholar]
- 4.Barton, L. L., M. B. Mets, and C. L. Beauchamp. 2002. Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am. J. Obstet. Gynecol. 187:1715-1716. [DOI] [PubMed] [Google Scholar]
- 5.Benowitz, L. I., and A. Routtenberg. 1997. GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci. 20:84-91. [DOI] [PubMed] [Google Scholar]
- 6.Borrow, P., C. F. Evans, and M. B. Oldstone. 1995. Virus-induced immunosuppression: immune system-mediated destruction of virus-infected dendritic cells results in generalized immune suppression. J. Virol. 69:1059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brot, M. D., G. F. Rall, M. B. Oldstone, G. F. Koob, and L. H. Gold. 1997. Deficits in discriminated learning remain despite clearance of long-term persistent viral infection in mice. J. Neurovirol. 3:265-273. [DOI] [PubMed] [Google Scholar]
- 8.Buchmeier, M. J., M. D. Bowen, and C. J. Peters. 2001. Arenaviridae: the viruses and their replication, p. 1635-1668. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 9.Buchmeier, M. J., H. A. Lewicki, O. Tomori, and M. B. Oldstone. 1981. Monoclonal antibodies to lymphocytic choriomeningitis and pichinde viruses: generation, characterization, and cross-reactivity with other arenaviruses. Virology 113:73-85. [DOI] [PubMed] [Google Scholar]
- 10.Cao, W., M. B. Oldstone, and J. C. De La Torre. 1997. Viral persistent infection affects both transcriptional and posttranscriptional regulation of neuron-specific molecule GAP43. Virology 230:147-154. [DOI] [PubMed] [Google Scholar]
- 11.Chismar, J. D., T. Mondala, H. S. Fox, E. Roberts, D. Langford, E. Masliah, D. R. Salomon, and S. R. Head. 2002. Analysis of result variability from high-density oligonucleotide arrays comparing same-species and cross-species hybridizations. BioTechniques 33:516-518, 520, 522 passim. [DOI] [PubMed] [Google Scholar]
- 12.Cornu, T. I., and J. C. de la Torre. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dafny, N. 1998. Is interferon-alpha a neuromodulator? Brain Res. Brain Res. Rev. 26:1-15. [DOI] [PubMed] [Google Scholar]
- 14.D'Arcangelo, G., F. Grassi, D. Ragozzino, A. Santoni, V. Tancredi, and F. Eusebi. 1991. Interferon inhibits synaptic potentiation in rat hippocampus. Brain Res. 564:245-248. [DOI] [PubMed] [Google Scholar]
- 15.de la Torre, J. C., M. Mallory, M. Brot, L. Gold, G. Koob, M. B. Oldstone, and E. Masliah. 1996. Viral persistence in neurons alters synaptic plasticity and cognitive functions without destruction of brain cells. Virology 220:508-515. [DOI] [PubMed] [Google Scholar]
- 16.de la Torre, J. C., and M. B. Oldstone. 1996. Anatomy of viral persistence: mechanisms of persistence and associated disease. Adv. Virus Res. 46:311-343. [DOI] [PubMed] [Google Scholar]
- 17.Dutko, F. J., and M. B. Oldstone. 1983. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J. Gen. Virol. 64:1689-1698. [DOI] [PubMed] [Google Scholar]
- 18.Evans, C. F., J. M. Redwine, C. E. Patterson, S. Askovic, and G. F. Rall. 2002. LCMV and the central nervous system: uncovering basic principles of CNS physiology and virus-induced disease. Curr. Top. Microbiol. Immunol. 263:177-195. [DOI] [PubMed] [Google Scholar]
- 19.Fazakerley, J. K., P. Southern, F. Bloom, and M. J. Buchmeier. 1991. High resolution in situ hybridization to determine the cellular distribution of lymphocytic choriomeningitis virus RNA in the tissues of persistently infected mice: relevance to arenavirus disease and mechanisms of viral persistence. J. Gen. Virol. 72:1611-1625. [DOI] [PubMed] [Google Scholar]
- 20.Gold, L. H., M. D. Brot, I. Polis, R. Schroeder, A. Tishon, J. C. de la Torre, M. B. Oldstone, and G. F. Koob. 1994. Behavioral effects of persistent lymphocytic choriomeningitis virus infection in mice. Behav. Neural. Biol. 62:100-109. [DOI] [PubMed] [Google Scholar]
- 21.Goslin, K., H. Asmussen, and G. Banker. 1998. Rat hippocampal neurons in low-density culture, p. 339-370. In G. Banker and K. Gioslin (ed.), Culturing nerve cells. MIT Press, Cambridge, MA.
- 22.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15:259-267. [DOI] [PubMed] [Google Scholar]
- 23.Griffin, D. E. 2003. Immune responses to RNA-virus infections of the CNS. Nat. Rev. Immunol. 3:493-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hahm, B., M. J. Trifilo, E. I. Zuniga, and M. B. Oldstone. 2005. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity 22:247-257. [DOI] [PubMed] [Google Scholar]
- 25.Hotchin, J., and R. Seegal. 1976. Virus-induced behavioral alteration of mice. Science 196:671-674. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, R. T. 1998. Viral infections of the nervous system. Lippincott-Raven, Philadelphia, Pa.
- 27.Johnston, C., W. Jiang, T. Chu, and B. Levine. 2001. Identification of genes involved in the host response to neurovirulent alphavirus infection. J. Virol. 75:10431-10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, I. J., H. N. Beck, P. J. Lein, and D. Higgins. 2002. Interferon gamma induces retrograde dendritic retraction and inhibits synapse formation. J. Neurosci. 22:4530-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knobeloch, K. P., O. Utermohlen, A. Kisser, M. Prinz, and I. Horak. 2005. Reexamination of the role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol. Cell. Biol. 25:11030-11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristensson, K., and E. Norrby. 1986. Persistence of RNA viruses in the central nervous system. Annu. Rev. Microbiol. 40:159-184. [DOI] [PubMed] [Google Scholar]
- 31.Lipkin, W. I., E. L. F. Battenberg, F. E. Bloom, and M. B. A. Oldstone. 1988. Viral infection of neurons can depress neurotransmitter mRNA levels without histologic injury. Brain Res. 451:333-339. [DOI] [PubMed] [Google Scholar]
- 32.Liu, L. Q., R. Ilaria, Jr., P. D. Kingsley, A. Iwama, R. A. van Etten, J. Palis, and D. E. Zhang. 1999. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol. Cell. Biol. 19:3029-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier, J., C. Kincaid, A. Pagenstecher, and I. L. Campbell. 2002. Regulation of signal transducer and activator of transcription and suppressor of cytokine-signaling gene expression in the brain of mice with astrocyte-targeted production of interleukin-12 or experimental autoimmune encephalomyelitis. Am. J. Pathol. 160:271-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malakhov, M. P., O. A. Malakhova, K. I. Kim, K. J. Ritchie, and D. E. Zhang. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277:9976-9981. [DOI] [PubMed] [Google Scholar]
- 35.Malenka, R. C. 2003. The long-term potential of LTP. Nat. Rev. Neurosci. 4:923-926. [DOI] [PubMed] [Google Scholar]
- 36.Malenka, R. C., and M. F. Bear. 2004. LTP and LTD: an embarrassment of riches. Neuron 44:5-21. [DOI] [PubMed] [Google Scholar]
- 37.Malmgaard, L., T. P. Salazar-Mather, C. A. Lewis, and C. A. Biron. 2002. Promotion of alpha/beta interferon induction during in vivo viral infection through alpha/beta interferon receptor/STAT1 system-dependent and -independent pathways. J. Virol. 76:4520-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendoza-Fernandez, V., R. D. Andrew, and C. Barajas-Lopez. 2000. Interferon-alpha inhibits long-term potentiation and unmasks a long-term depression in the rat hippocampus. Brain Res. 885:14-24. [DOI] [PubMed] [Google Scholar]
- 39.Mohammed, A. M., E. Norrby, and K. Kristensson. 1993. Viruses and behavioral changes: a review of clinical and experimental findings. Rev. Neurosci. 4:267-280. [DOI] [PubMed] [Google Scholar]
- 40.Morozov, P. V. (ed). 1983. Research on the viral hypothesis of mental disorders. Advances in biological psychiatry, vol. 12. Karger, New York, N.Y.
- 41.Moskophidis, D., M. Battegay, M. A. Bruendler, E. Laine, I. Gresser, and R. M. Zinkernagel. 1994. Resistance of lymphocytic choriomeningitis virus to alpha/beta interferon and to gamma interferon. J. Virol. 68:1951-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldstone, M. B. 2002. Biology and pathogenesis of lymphocytic choriomeningitis virus infection. Curr. Top. Microbiol. Immunol. 263:83-118. [DOI] [PubMed] [Google Scholar]
- 43.Oldstone, M. B. 2004. Future trends in neurovirology: neuronal survival during virus infection and analysis of virus-specific T cells in central nervous system tissues. J. Neurovirol. 10:207-215. [DOI] [PubMed] [Google Scholar]
- 44.Oldstone, M. B. 1993. Viruses and diseases of the twenty-first century. Am. J. Pathol. 143:1241-1249. [PMC free article] [PubMed] [Google Scholar]
- 45.Oldstone, M. B., and F. J. Dixon. 1974. Aging and chronic virus infection: is there a relationship? Fed. Proc. 33:2057-2059. [PubMed] [Google Scholar]
- 46.Oldstone, M. B., J. Holmstoen, and R. M. Welsh, Jr. 1977. Alterations of acetylcholine enzymes in neuroblastoma cells persistently infected with lymphocytic choriomeningitis virus. J. Cell Physiol. 91:459-472. [DOI] [PubMed] [Google Scholar]
- 47.Ou, R., S. Zhou, L. Huang, and D. Moskophidis. 2001. Critical role for alpha/beta and gamma interferons in persistence of lymphocytic choriomeningitis virus by clonal exhaustion of cytotoxic T cells. J. Virol. 75:8407-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ousman, S. S., J. Wang, and I. L. Campbell. 2005. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J. Virol. 79:7514-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prosniak, M., D. C. Hooper, B. Dietzschold, and H. Koprowski. 2001. Effect of rabies virus infection on gene expression in mouse brain. Proc. Natl. Acad. Sci. USA 98:2758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ritchie, K. J., C. S. Hahn, K. I. Kim, M. Yan, D. Rosario, L. Li, J. C. de la Torre, and D. E. Zhang. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10:1374-1378. (First published 7 Nov. 2004; doi: 10.1038/nm1133.) [DOI] [PubMed] [Google Scholar]
- 51.Ritchie, K. J., M. P. Malakhov, C. J. Hetherington, L. Zhou, M. T. Little, O. A. Malakhova, J. C. Sipe, S. H. Orkin, and D. E. Zhang. 2002. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 16:2207-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, E. S., M. A. Zandonatti, D. D. Watry, L. J. Madden, S. J. Henriksen, M. A. Taffe, and H. S. Fox. 2003. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am. J. Pathol. 162:2041-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez, M., M. J. Buchmeier, M. B. Oldstone, and P. W. Lampert. 1983. Ultrastructural localization of viral antigens in the CNS of mice persistently infected with lymphocytic choriomeningitis virus (LCMV). Am. J. Pathol. 110:95-100. [PMC free article] [PubMed] [Google Scholar]
- 54.Saha, S., and P. N. Rangarajan. 2003. Common host genes are activated in mouse brain by Japanese encephalitis and rabies viruses. J. Gen. Virol. 84:1729-1735. [DOI] [PubMed] [Google Scholar]
- 55.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanchez, A. B., M. Perez, T. Cornu, and J. C. de la Torre. 2005. RNA interference-mediated virus clearance from cells both acutely and chronically infected with the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 79:11071-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwer, H., L. Q. Liu, L. Zhou, M. T. Little, Z. Pan, C. J. Hetherington, and D. E. Zhang. 2000. Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (Usp18). Genomics 65:44-52. [DOI] [PubMed] [Google Scholar]
- 58.Sevilla, N., D. B. McGavern, C. Teng, S. Kunz, and M. B. Oldstone. 2004. Viral targeting of hematopoietic progenitors and inhibition of DC maturation as a dual strategy for immune subversion. J. Clin. Investig. 113:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, J. B., and H. R. Herschman. 1996. The glucocorticoid attenuated response genes GARG-16, GARG-39, and GARG-49/IRG2 encode inducible proteins containing multiple tetratricopeptide repeat domains. Arch. Biochem. Biophys. 330:290-300. [DOI] [PubMed] [Google Scholar]
- 60.ter Meulen, V. 1991. Virus-cell interactions in the nervous system. Semin. Neurosci. 3:81-173. [Google Scholar]
- 61.Vikman, K. S., B. Owe-Larsson, J. Brask, K. S. Kristensson, and R. H. Hill. 2001. Interferon-gamma-induced changes in synaptic activity and AMPA receptor clustering in hippocampal cultures. Brain Res. 896:18-29. [DOI] [PubMed] [Google Scholar]
- 62.von Herrath, M., M. B. Oldstone, and H. S. Fox. 1995. Simian immunodeficiency virus (SIV)-specific CTL in cerebrospinal fluid and brains of SIV-infected rhesus macaques. J. Immunol. 154:5582-5589. [PubMed] [Google Scholar]
- 63.Wang, J., and I. L. Campbell. 2005. Innate STAT1-dependent genomic response of neurons to the antiviral cytokine alpha interferon. J. Virol. 79:8295-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright, R., D. Johnson, M. Neumann, T. G. Ksiazek, P. Rollin, R. V. Keech, D. J. Bonthius, P. Hitchon, C. F. Grose, W. E. Bell, and J. F. Bale, Jr. 1997. Congenital lymphocytic choriomeningitis virus syndrome: a disease that mimics congenital toxoplasmosis or cytomegalovirus infection. Pediatrics 100:E9. [Online.] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.