Abstract
Whereas evolutionary inferences derived from present-day DNA sequences are by necessity indirect, ancient DNA sequences provide a direct view of past genetic variants. However, base lesions that accumulate in DNA over time may cause nucleotide misincorporations when ancient DNA sequences are replicated. By repeated amplifications of mitochondrial DNA sequences from a large number of ancient wolf remains, we show that C/G-to-T/A transitions are the predominant type of such misincorporations. Using a massively parallel sequencing method that allows large numbers of single DNA strands to be sequenced, we show that modifications of C, as well as to a lesser extent of G, residues cause such misincorporations. Experiments where oligonucleotides containing modified bases are used as templates in amplification reactions suggest that both of these types of misincorporations can be caused by deamination of the template bases. New DNA sequencing methods in conjunction with knowledge of misincorporation processes have now, in principle, opened the way for the determination of complete genomes from organisms that became extinct during and after the last glaciation.
Keywords: pyrosequencing, deamination, DNA damage, wolves, mammoth
DNA sequences determined from the remains of ancient organisms make unique contributions to our understanding of biological and genetic change over time, because they allow a direct view of past genetic variation and thus obviate the need to rely solely on inference from DNA sequences in extant organisms to reconstruct molecular evolutionary change. Although limited to cases where well preserved specimens are found by archaeologists or are preserved in museum collections, the determination of DNA sequences from long-dead organisms has now become routine (1, 2). For example, ancient DNA sequences have clarified the phylogenetic relationships of many extinct species (3–6), contributed to our understanding of population changes caused by the last glacial maximum (7–9), and allowed the viral variants responsible for past pandemics to be identified (10).
However, the analysis of ancient DNA is challenging, and technical issues set limits for the age and types of DNA sequences that can be retrieved. Cloning in plasmid vectors (11, 12) was the first approach used to study ancient DNA, but this was soon replaced by the PCR (13, 14). The PCR has the advantage that it often starts from more than one template molecule and thus represents a statistical sample of the targeted DNA sequence (13). It also has the advantage that specific sequences can be retrieved repeatedly from the same specimen to confirm results. As a consequence, almost all ancient DNA sequences published to date are determined from DNA amplified by PCR. However, several artifacts can cause differences between ancient DNA sequences determined by PCR and the DNA sequences that were present in the organisms when alive. Nucleotide misincorporations by DNA polymerases during amplification, which may be induced by chemical modifications in the ancient DNA, are one source of artifacts (15–23), as is “jumping PCR,” i.e., template switching induced by DNA damage that cause recombination between different template molecules during PCR (24).
Contamination by modern DNA, which may be introduced during handling of the specimen or subsequent laboratory procedures, represents another source of artifacts in studies of ancient DNA (15, 25). Although this problem can largely be avoided by adhering to published and widely accepted guidelines for ancient DNA work (1, 2, 26), ancient human remains represent a troublesome exception to this. Because human DNA is pervasively present in the environment at archaeological excavations, in museums, and in laboratory facilities, contaminating human DNA can be amplified from almost every DNA extract prepared from ancient specimens, provided that the PCR performed is sensitive enough (1, 25). As a consequence, when ancient human remains are studied, it becomes impossible to distinguish contaminants from endogenous DNA sequences. By contrast, this is not a problem when non-human organisms are studied, because primers targeting DNA sequences from these organisms often will not amplify human DNA sequences, and when this nevertheless occurs, human DNA sequences can generally be identified as contaminants from their similarity to the human genome sequence.
Except for studies involving ancient human remains, nucleotide misincorporations therefore account for the majority of potential errors in ancient DNA sequences. This is particularly the case when amplifications start from very few template molecules. If a nucleotide misincorporation occurs when a single DNA strand is replicated during the first cycle of PCR, all molecules produced during subsequent cycles will carry this error. As a consequence, the final PCR product will be homogeneous, although it carries an erroneous sequence (19). This problem can be reduced to a level that does not undermine most biological conclusions, if the amplification of any particular DNA sequence is repeated to ensure that each nucleotide position is determined in two or more independent amplifications (19).
Recently, large-scale genomic approaches have been applied to the study of ancient DNA. In such approaches, total DNA from an ancient specimen is isolated, and subsequently a large number of random fragments are sequenced either after cloning in a plasmid vector (27) or directly after PCR from single molecules (28). In such cases, each DNA sequence is derived from a single ancient molecule. Because any nucleotide misincorporation that occurs during initial replication in bacteria or during PCR will be present as an unambiguous, but incorrect, base in the sequences produced, these approaches are especially prone to errors due to nucleotide misincorporations. Furthermore, because each individual DNA sequence is only determined once, such errors can only be described statistically unless ancient genomes are sequenced to such a high redundancy that each sequence position is determined from multiple sequence reads allowing nucleotide misincorporations to be identified and distinguished from allelic variants. For two reasons, this will, in most cases, remain impossible in the near future. First, the average size of the ancient DNA sequences present in ancient specimens is almost always <100 nt (14, 27). Second, only a few percent of the DNA sequences determined from the total DNA extracted from ancient remains are derived from the organism in question, whereas the rest are from organisms that have colonized the specimen either shortly after its death or during its subsequent deposition (27), although some permafrost specimens contain endogenous DNA in higher relative abundance (28). Thus, until sequencing costs are drastically reduced, high coverage is not likely to be achieved when ancient genomes are studied. Ancient DNA sequences determined from high-throughput approaches are therefore potentially much more affected by errors due to nucleotide misincorporations than DNA sequences determined by targeted approaches using PCR amplifications that are repeated several times.
Knowledge about nucleotide misincorporation patterns are thus of great relevance for the high-throughput approaches that are beginning to be used to study ancient genomes, because such knowledge will allow the extent of various types of misincorporations to be estimated statistically for such data. Misincorporation patterns are also of interest, because radically different misincorporation patterns have been reported in amplification products from ancient remains. Hofreiter et al. (19) observed almost exclusively C/G-to-T/A transitions in mitochondrial sequences derived from Pleistocene cave bears. In contrast, Gilbert et al. (21, 22) observed ≈62% such substitutions but in addition ≈31% T/A-to-C/G transitions and ≈7% transversions when they analyzed mitochondrial control region sequences from ancient human remains, and Hansen et al. (20) and Binladen et al. (23) similarly observed substantial amounts of T/A-to-C/G transitions and transversions in addition to C/G-to-T/A transitions when analyzing mitochondrial and nuclear sequences derived from ancient human and animal remains, respectively. To shed light on the patterns of nucleotide misincorporations that occur when ancient DNA samples are enzymatically amplified, we have investigated DNA sequences determined from late Pleistocene wolves by sequencing of cloned PCR products as well as from a Siberian mammoth by high-throughput pyrosequencing on the 454 platform (29). We also use synthetic DNA templates that carry specific molecular lesions to test whether these cause errors compatible with the patterns of misincorporations observed in the ancient DNA sequences.
Results
Substitution Patterns in DNA Amplifications from Ancient Wolves.
We extracted DNA from 62 bone and teeth samples from wolves that vary in age between 50 and 50,000 years B.P. and originate from 14 different geographic localities (Fig. 5, which is published as supporting information on the PNAS web site). From each extract, we attempted to amplify a 100-bp-long fragment of the mitochondrial control region (including primers) spanning the bases 15612–15668 in the dog mitochondrial genome (between the primers) (GenBank accession no. U96639). Although 27 samples failed to yield any canid-like amplification products, the target DNA sequence could be repeatedly amplified for 29 of the 62 wolf specimens. The remaining six samples yielded PCR products that could not be replicated in later amplification attempts and were therefore excluded from further analysis. Such a failure rate is typical of most studies of ancient remains (4, 8, 30).
From each of the 29 remaining specimens, we cloned 2 or more PCR products per specimen and sequenced 5–16 clones from each amplification product. If all clones sequenced from one PCR product carried substitutions that distinguished them from all clones of a second independent PCR product, the amplification was repeated and the nucleotide state observed twice was considered to be the endogenous one, whereas the other nucleotide state was deemed to represent a misincorporation (19). We then compared clones from each individual PCR product and identified additional misincorporations by looking for substitutions in each of the cloned sequences, compared with the consensus sequence of the amplification. If more than one clone carried a particular substitution, they were deemed to come from the same misincorporation event. Thus, substitutions observed in all clones of a certain PCR product but not in at least two other PCR products, as well as substitutions present in one or several clones from a single PCR product, were counted to yield the total number of nucleotide misincorporations observed from a specimen.
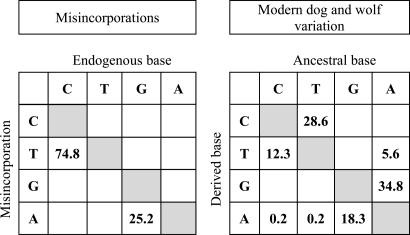
In all, we analyzed 1,058 clone sequences from 108 PCR products and identified a total of 115 nucleotide misincorporations as defined above. Five of these were consistent differences between all clones from distinct amplification products and thus represent misincorporations that are likely to have occurred during the first cycle of PCR when exclusively ancient DNA template molecules are present, whereas the rest were seen among the clones from single amplification products. They can thus represent errors made either when ancient templates were replicated or errors introduced when newly synthesized molecules were replicated. However, the high frequency with which misincorporations were observed suggests that a large fraction of them are caused by the ancient nature of the template DNA. Strikingly, all 115 misincorporations represented C/G-to-T/A transitions (Fig. 1).
Fig. 1.
Substitution pattern observed among the misincorporations seen in the PCR products from 29 ancient wolves (Left) and among modern dog and wolf mtDNA sequences (Right) from Savolainen et al. (33) (dogs) and Vila et al. (31, 32) (wolves). Numbers represent percentages, and the “ancestral” sequence for the dogs and wolves is approximated with the consensus sequence for all sequences studied.
To investigate whether contamination of DNA from different modern or ancient dogs could have confounded the results, we examined differences between the DNA sequences inferred to be endogenous to each of the 29 ancient wolves and the consensus of all these sequences. Among 80 nucleotide differences seen, C/G-to-T/A transitions comprised 15%, whereas the remaining 85% were T/A-to-C/G transitions. When 198 modern wolf and dog mtDNA control region sequences (31–33) are analyzed in the same way, C/G-to-T/A transitions make up 31% of all substitutions seen, whereas T/A-to-C/G transitions make up 63% and transversions make up the remaining 6% (Fig. 1). Thus, the pattern seen among the substitutions deemed to represent nucleotide misincorporations in the ancient DNA sequences differs from the variation among modern wolf and dog sequences (X2 = 237.3, df = 4, P = 3.6 × 10−50), as well as from the variation among ancient wolves (X2 = 153.6, df = 4, P = 3.5 × 10−32). Contamination with modern wolf and/or dog DNA can therefore not explain the extreme substitution pattern observed among amplification products from the ancient wolf samples, where exclusively C/G-to-T/A transitions are observed. Rather, a large fraction or all of the latter substitutions are likely to derive from chemical modifications present in the ancient DNA.
Large-Scale Pyrosequencing.
To further investigate the nucleotide misincorporations in amplifications of ancient samples, we analyzed large-scale parallel pyrosequencing data generated by a 454 instrument. In this procedure, total DNA extracted from a 43,000-year-old mammoth bone (34) from the Bol'shaya Kolopatkaya river (lat 70°N, long 151°E), Russia, was ligated to biotinylated linkers, and single DNA strands were attached to Sepharose beads, amplified by PCR, and subjected to pyrosequencing on the 454 platform (29). This approach has two main advantages over the clone-based approach described above. First, each sequence read derives from one single-stranded DNA molecule, and the sequence read allows the actual template strand to be inferred. Therefore, when the ancient DNA sequence is compared with an extant species, it is possible to know what base was read off the ancient DNA molecule when it was replicated. For example, whereas C-to-T and G-to-A differences need to be pooled in the analysis of the PCR products from the ancient wolves above, because it is not known whether the initial strand that was replicated in the PCR carried a C or a G (Fig. 1), the 454-based pyrosequencing makes it possible to observe each of these sequence differences independently. Second, because each experiment generates large amounts of DNA sequence, it is possible to detect even rare types of misincorporations that may be missed when smaller numbers of clones of PCR products are analyzed. However, unlike analyses of defined regions amplified by PCR primers, with this method, one cannot choose beforehand which regions to sequence. Therefore, after sequencing, it is first necessary to determine which sequences derive from the target species rather than other organisms that have colonized the organism after its death, and to generate reliable alignments of the target species to one or more other genomes. Once this is done, the extent of DNA sequence divergence and misincorporations can be estimated statistically.
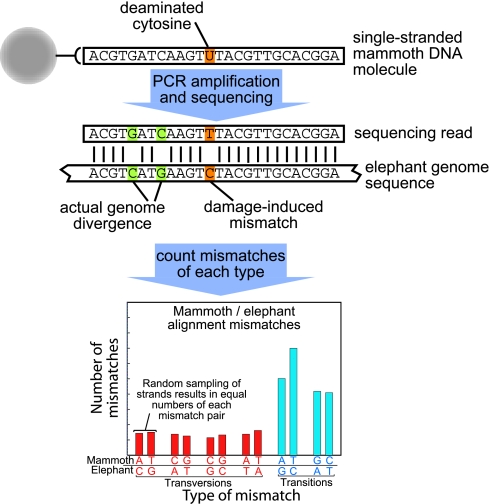
In designing a statistical analysis to infer misincorporation rates, we exploit the double-stranded nature of DNA and investigate two aspects of the data. First, for each type of mismatch observed when the mammoth reads are compared with the closest extant genome sequence available, the African elephant, there is a strand-equivalent mismatch that would have been observed had the other strand been sequenced instead. For example, each read containing a mammoth T aligned to an elephant C would have been observed as a read containing a mammoth A aligned to an elephant G, if the complementary strand had been sequenced. Thus, unless misincorporations occur, the number of any particular nucleotide differences between mammoth and elephant should equal the number of reciprocal strand-equivalent differences, where the mammoth sequence carries the complementary nucleotide (Fig. 2), provided that the frequency with which DNA strands are captured in the sequencing reaction is independent of their base composition. Because the numbers of As and Ts are nearly identical among the 146,733 mammoth nucleotide positions analyzed, as are the number of Cs and Gs, this condition is met, and this aspect of the data can be used to detect nucleotide misincorporations.
Fig. 2.
Schematic illustration showing how 454 sequencing captures individual single-stranded mammoth molecules. When these are compared with elephant sequences, any reciprocal base-equivalent differences between the genomes will be present in statistically equal numbers across many reads. This is illustrated by the two G–C and C–G differences shown in green. Any modification in the ancient DNA, for example a deaminated C that yields U (orange), will yield an excess of differences relative to the reciprocal difference, as seen for the T–C vs. A–G differences below.
The second aspect of the data we use to detect misincorporations relies on the fact that nucleotide differences where, for example, the mammoth carries a T and the elephant a G versus where the mammoth carries a G and the elephant a T, should be equal in number if the rate and patterns of nucleotide substitutions along the evolutionary lineages leading to the mammoth and the elephant were similar. Thus, any difference in number between pairs of such reciprocal differences indicates either that the pattern of substitutions changed in one species (a fairly unlikely event when closely related species are studied) or that nucleotide misincorporations contribute to one of the two types of differences.
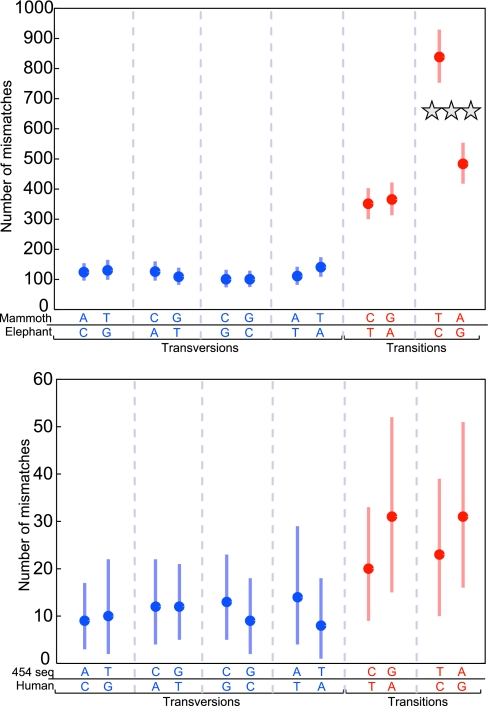
In our analysis of the DNA sequences from the late Pleistocene mammoth, we first identified sequences that are most similar to the African elephant genome or the human genome (see Materials and Methods). These were deemed to be ancient mammoth and human sequences contaminating the specimen, respectively. A total of 2,983 nucleotide differences were observed within the 1,800 alignments to the extant African elephant genome (Fig. 3Upper) and 192 differences within the 59 alignments to the human genome (Fig. 3 Lower). Among the elephant alignments, a total of 2,040 transitions and 943 transversions were seen, yielding a transition-to-transversion ratio of 2.16, typical of closely related mammalian genomes (35). Interestingly, when each pair of strand-equivalent nucleotide differences between the mammoth and the elephant was compared, none differed significantly, except T/C and A/G differences where the mammoth sequences had 73.3% more C-to-T than G-to-A differences. This finding indicates that many of the nucleotide residues in the ancient DNA that differ to the elephant genome by being read as a T where the elephant carries a C have been modified to appear as T during the amplification process. This excess of C-to-T mismatches was not observed within the 59 alignments to the human genome (Fig. 3 Lower), indicating that it does not affect the more recent human DNA that contaminates the specimen.
Fig. 3.
Number of base mismatches of each type, grouped into strand-equivalent pairs for the mammoth and elephant comparisons (Upper) and the human comparisons (Lower). The circle indicates the observed number of mismatches of each type. The lines above and below are the 99% confidence intervals obtained for 10,000 bootstrap replicates of the alignments. Note that positions where the mammoth carries a T and the elephant carries a C are significantly more numerous than positions where the mammoth carries an A and the elephant carries a G. Note also that the positions where the mammoth carries an A and the elephant carries a G are significantly more numerous than positions where the mammoth carries a G and the elephant carries an A. Among the 59 human sequences analyzed, there are no significantly elevated mismatches.
When pairs of reciprocal differences are compared, none of them differ significantly in number within a pair except where the mammoth carries an A and the elephant a G, which occurs ≈26% more frequently than positions where the mammoth carries a G and the elephant an A. That rates and patterns of substitutions are unlikely to have changed among these closely related species (an assumption that is supported by the observation that among the transversions, no indications of any differences on the elephant and mammoth lineages are seen) suggests that some modification affects G residues in the ancient DNA and causes some of them to be read as A's in these experiments.
Misincorporations Induced by Deamination.
It has been previously shown that deamination of cytosine residues causes C-to-T transitions in DNA sequences determined from ancient DNA (19). We therefore assume that this explains the excess of C/G-to-T/A differences seen in the wolf sequences, as well as the excess of C-to-T differences between the elephant and mammoth described above. However, both guanine and adenine residues may also become deaminated, and it has been suggested that deamination of adenine residues may cause misincorporations in which A appears as G during PCR from ancient extracts (21, 22).
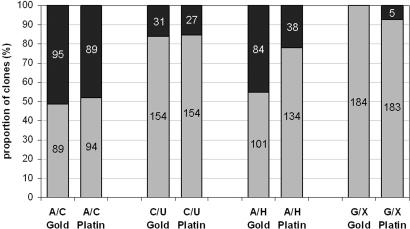
To determine the extent to which deamination of cytosine, adenine, and guanine residues causes DNA polymerases used in PCR of ancient DNA to misincorporate nucleotides, we used three pairs of oligonucleotides to serve as templates in amplifications. The two oligonucleotides in each pair are identical except that one carries either a cytosine residue (C) or its deaminated form, a uracil residue (U), at one position; another carries an adenine residue (A) or its deaminated form, a hypoxanthine residue (H); and the third pair carries a guanine residue (G) or its deaminated form, a xanthine residue (X) (Fig. 6, which is published as supporting information on the PNAS web site). These oligonucleotides were used as templates in PCRs with two DNA polymerase reagents, AmpliTaq Gold (Applied Biosystems, Foster City, CA) and Platinum Taq High Fidelity (Invitrogen, Carlsbad, CA), which are both commonly used in ancient DNA research. From each amplification product, 21 to 47 clones were sequenced. When the oligonucleotides carrying C, A, and G, respectively, were used individually, the correct nucleotide, G, T, and C, respectively, were incorporated on the other strand in all clones analyzed (data not shown). In contrast, when the oligonucleotide carrying the U was used, both polymerase reagents inserted A residues opposite the template sites carrying a U residue in all clones sequenced (data not shown). Similarly, when the oligonucleotide carrying H was used, both polymerase reagents inserted C residues in all clones sequenced (data not shown), whereas when the oligonucleotide carrying X was used, AmpliTaq Gold inserted C in all clones sequenced while Platinum Taq High Fidelity inserted T residues in 18 of 47 clones sequenced (38%) and C in the rest. Thus, U and H cause misincorporations whenever they serve as templates for the polymerase reagents tested. By contrast, X causes no misincorporations when it serves as a template for AmpliTaq Gold and causes misincorporations only in a minority of cases when Platinum Taq High Fidelity is used.
We then tested whether the polymerase reagents preferentially amplify a nucleotide that naturally occurs in DNA or its deaminated form when both are present in a PCR. As a control, we first mixed the two nucleotides carrying the unmodified bases (A and C) in equal amounts. In this case, both DNA polymerase reagents amplify the two templates with equal efficiency and without any detectable misincorporations, as illustrated in Fig. 4. When C and its deaminated form U are available in equal amounts, both DNA polymerase reagents yield ≈15% (SD = 0.7% and 2.1%, respectively) misincorporations where U is read as T. Therefore, both reagents amplify the unmodified base C residues with ≈70% higher efficiency than U residues.
Fig. 4.
Summary of the oligonucleotide competition amplifications. Each bar represents the ratio of nucleotides found among clones from amplification products generated from a mixture of two oligonucleotides, given by the abbreviations below the bars. Misincorporated nucleotides are indicated by the black upper part of each bar, and numbers give the numbers of clones sequenced. The polymerase reagents are indicated as “Gold” for AmpliTaq Gold and “Platin” for Platinum Taq High Fidelity.
When A and its deaminated form H are both available, AmpliTaq Gold yields misincorporations where C residues are incorporated in 45% (SD = 0.7%) of the clones sequenced, whereas Platinum Taq High Fidelity does so in 22% (SD = 1.4%) of such clones. Thus, whereas AmpliTaq Gold shows no strong preference for amplifying a template containing an A residue over an H residue, Platinum Taq High Fidelity is ≈55% more efficient in amplifying the unmodified base.
Finally, when G and X are both available, AmpliTaq Gold yields no misincorporated residues among the 184 clones sequenced, whereas Platinum Taq High Fidelity yields 2.7% (SD = 2.1%) of misincorporated T residues. Although the extent to which the polymerase reagents prefer the unmodified templates cannot be determined in this case, it is clear that AmpliTaq Gold incorporates only the “correct” base C when it encounters X, whereas Platinum Taq High Fidelity yields misincorporations of T residues.
In summary, although the DNA polymerase reagents used in the amplifications from the Pleistocene wolf samples (AmpliTaq Gold) and in the pyrosequencing experiments (Platinum Taq High Fidelity) behave similarly in causing misincorporations as a result of deaminated C residues (U), they behave differently in that AmpliTaq Gold yields approximately twice as many misincorporations where a deaminated A residue (H) is read as a G residue as does Platinum Taq High Fidelity. Consequently, if deaminated A residues would have occurred at any appreciable frequency in the wolf DNA samples analyzed, we would have detected them as nucleotide misincorporations. When the two polymerase reagents encounter deaminated G residues (X), only Platinum Taq High Fidelity, which is used in the pyrosequencing experiments, yields misincorporations where G residues are misread as A residues.
Discussion
When DNA sequences were amplified from 29 wolf remains from 8 European and 6 Asian localities that vary in age from 50 to ≈50,000 years, all nucleotide misincorporations detected were changes from C/G to T/A base pairs. Misincorporations were found in samples from all localities and all ages (Fig. 5), including the single sample that is 50 years old, whereas only two samples yielded no misincorporations in the amplifications analyzed. Thus, C/G-to-T/A changes are obviously the predominant nucleotide misincorporation that affects amplifications from ancient DNA sequences retrieved from a variety of different localities and times.
A novel massively parallel pyrosequencing method (29) that allows the sequencing of large numbers of single-template DNA strands allowed us to determine whether C/G-to-T/A misincorporations in ancient DNA sequences are caused by miscoding lesions involving predominantly cytosine or guanine residues. To do this, we generated data from a late Pleistocene mammoth from Siberia. The results (Fig. 3 Upper) show that cytosine rather than guanine residues are the predominant cause of these transitions. If no lesions affect guanine residues, then as much as ≈40% of all nucleotide differences where the mammoth carries a T and the elephant a C are caused by misincorporations. If guanine residues are also affected by some lesion (see below), this proportion is even higher.
For several reasons, the lesion affecting cytosine residues is very likely to be deamination. First, the deaminated form of cytosine, uracil, has been shown to occur in ancient DNA extracts (14, 17), and when ancient DNA is treated with uracil DNA glycosylase that removes uracil from DNA, C/G-to-T/A misincorporations are not observed (19). Furthermore, the results presented in Fig. 4 show that uracil residues yield the expected misincorporations in amplifications with the polymerase reagent used in the amplifications from the wolf samples, as well as the reagent used in 454-based pyrosequencing from the mammoth sample. No other strand-equivalent nucleotide difference suggests modification of any additional base (Fig. 3 Upper). However, it is noteworthy that the amounts of A–G differences between the mammoth and the elephant are larger than G–A differences. Assuming that the rate of these nucleotide substitutions along the mammoth and elephant evolutionary lineages are similar, this result suggests that some lesion in the mammoth affects G residues and causes them to be read as A residues during PCR. This phenomenon is seen also in large-scale pyrosequencing reads from cave bears and Neandertals (R.E.G., unpublished data). Thus, two different lesions in ancient DNA are likely to be responsible for the high rate of C/G-to-T/A transitions seen in the high-throughput data from the mammoth. One of these is deamination of cytosine residues that causes C to be read as T, and the other is a hitherto unknown lesion affecting guanine residues. This modification may be deamination, because deamination causes G residues to be read as A residues by the polymerase reagent used in the pyrosequencing experiments. By contrast, it causes no misincorporations when the polymerase reagent used in the PCR from the wolf samples is used. This is in agreement with previous work where xanthine has been shown to cause predominantly chain termination by Taq polymerase (36) but leads to misincorporation of T residues by DNA polymerase I (37).
Interestingly, there is no evidence that any lesion-causing transversions affect these nucleotides, because all strand-equivalent transversional differences are of equal magnitude, as are all different types of transversions (Fig. 3 Upper). This may be unexpected, in view of the fact that G residues are prone to oxidation to 8-hydroxyguanine in vivo, which yield G-to-T transversions. Future work is necessary to clarify whether this is not frequent in ancient samples or whether 8-hydroxyguanine may become further modified to lesions that block DNA replication in ancient DNA. In the case of T and A residues in the mammoth DNA, there is no reason to assume that they have suffered large rates of miscoding lesions unless they are both affected by different lesions that cause transitions at rates that are statistically indistinguishable (Fig. 3 Upper). This observation is in agreement with biochemical studies, which have shown that deamination of adenine residues is ≈40-fold slower than deamination of cytosine residues (38).
Thus, there is no evidence from these experiments for any lesion that affects A residues and causes them to be read as G residues, as has been suggested previously (20). This observation is further supported by the experiments, where deaminated adenine residues, i.e., hypoxanthine, are used as templates in PCR, which demonstrate that the DNA polymerases used would have yielded misincorporations if hypoxanthine residues were present in the ancient DNA (Fig. 4). It is unclear why others have observed large amounts of T/A-to-C/G transitions among the substitutions interpreted as misincorporations when amplifying ancient DNA (20–23). It may be that some unknown lesion other than deamination affects A or T residues and contributes to the observations by these authors. In fact, an indication that this may be the case comes from an experiment where we used Platinum Taq High Fidelity (the polymerase reagent used by these authors) to amplify DNA sequences from three wolf samples. It was found to yield almost 60% more misincorporations than AmpliTaq Gold, and almost the entire increase in misincorporations was due to T/A-to-C/G misincorporations (Fig. 7, which is published as supporting information on the PNAS web site). It may thus be that under certain conditions, Platinum Taq High Fidelity yields misincorporations that it does not yield under other conditions, e.g., the parallel pyrosequencing experiment (Fig. 3 Upper). However, because human mitochondrial control region sequences were analyzed in the studies that found T/A-to-C/G misincorporations to predominate over C/G-to-T/A misincorporations (20–22), one possibility is that some of the T/A-to-C/G substitutions observed may be due to contamination by contemporary human DNA (25, 26, 39).
The results presented in Fig. 3 Upper show that direct large-scale pyrosequencing of the DNA extracted from ancient remains and the comparison of the DNA sequences to complete genome sequences of closely related organisms provide a powerful means not only to determine ancient DNA sequences (28), but also to determine the patterns of nucleotide misincorporation caused by lesions in the ancient DNA. Knowledge about such patterns is necessary to better interpret the results of such approaches as long as the coverage of shotgun approaches is not sufficient to ascertain severalfold coverage of each nucleotide position. It should be noted, though, that misincorporation patterns can be determined from such large-scale approaches only when a genome from a closely related extant species is available, because if the extant genome to which the ancient DNA sequences are compared is too distantly related, phylogenetic differences between the genomes, as well as putative differences in patterns and rates of substitutions in the organisms, make the detection of base-equivalent and reciprocal differences difficult. For example, when cave bear DNA sequences are compared with the dog genome, these types of analyses are difficult, whereas for the mammoth and Neandertal genomes, the elephant and human genomes, respectively, offer suitable comparisons (R.E.G., unpublished data). In the near future, we expect that novel large-scale and cost-effective sequencing methods in conjunction with high-quality genome sequences from many extant species will make analyses of the genomes of several extinct species possible.
Materials and Methods
Ancient DNA Extraction, Amplification, and Sequencing.
Approximately 100 mg of material was removed from each of 62 wolf (Canis lupus) bone and teeth specimens from 14 different localities in Europe and Asia (Table 1, which is published as supporting information on the PNAS web site). Each sample was ground by using mortar and pestle and incubated in 2.5 ml of 0.5 M EDTA (pH 8.0)/0.5% N-laurylsarcosine/1% poly(vinylpyrolidon)/0.25 mg/ml proteinase K/50 mM DTT/2.5 mM N-phenacylthiazolium bromide for 16 h at 37°C under rotation. After centrifugation for 5 min at 4,000 rpm in a Megafuge 1.0R (Heraeus, Duesseldorf, Germany), the supernatant was recovered and added to a mixture of 5 M guanidinium thiocyanate, 0.04 mM Tris, 0.025 M sodium chloride, and 50 μl of silica suspension, prepared as in refs. 40 and 41. After incubation under rotation for 1 h at 37°C, followed by centrifugation for 2 min at 4,000 rpm (Megafuge 1.0R), the silica pellet was washed once with 5 M GuSCN/0.04 mM Tris/0.025 M sodium chloride and once by using New Wash solution (Qbiogene, Irvine, CA). After drying the pellet for 5 min at 56°C, DNA was eluted at 56°C in 10 mM Tris/1 mM EDTA for 8 min.
Five microliters of each extract was added to 15 μl of PCR mix consisting of 2.5 units of AmpliTaq Gold, 1.25× AmpliTaq Gold PCR buffer, 2.5 mM MgCl2, 1 mM dNTPs, and 0.25 μM primers HVR-wolf-F (5′-ATA TTA TAT CCT TAC ATA GGA CAT-3′) and HVR-wolf-R (5′-ATT AAG CCC TTA TTG GAC T-3′). Amplifications were performed in an MJ thermocycler (Bio-Rad, Hercules, CA) with a 3-min activation step at 94°C, followed by 60 cycles of 93°C for 30 s, 50°C for 60 s, and 72°C for 30 s, and finished with a final extension at 72°C for 15 min. Amplification products were visualized in agarose gels and cloned by using the TOPO TA cloning kit (Invitrogen). When primer artifacts were present or the desired products were present in low abundance, PCR products were isolated from the agarose gel by using the QIAquick gel extraction kit (Qiagen, Valencia, CA) and either directly cloned or reamplified as above, except that the initial activation step of the PCR was prolonged to 7 min and 40 amplification cycles were used. To avoid contamination, the extraction and PCR preparation (except the addition of template to reamplification reactions) were carried out in a laboratory exclusively used for ancient DNA analyses where protective clothing, face shields, bleach treatment of surfaces and instruments, UV radiation, air filtration, positive pressure, and other measures are used to avoid contamination of the experiments with extraneous DNA. To monitor laboratory contamination, at least one mock control for every four samples was carried along during the extraction procedure, as well as a minimum of two water controls per PCR amplification. At least five clones from each amplification product were sequenced in an ABI PRISM 3730 capillary sequencer using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
Pyrosequencing on the 454 Platform.
Sequence reads from 454 were analyzed by database similarity searches as follows. Each sequence in the 454 database was first compared with all of the other sequences in the database to identify repeat sequences. Occasionally, the 454 emulsion PCR or signal reading will generate repeat sequences. These repeats do not derive from discrete molecules and thus are not independent sequence reads. Such repeats were identified by single-linkage clustering of all sequences containing 95% identity over 95% of each sequence length. From each cluster, the single best-aligning sequence was used for analysis.
Each sequence read was used as a query to search the elephant genome (www.broad.mit.edu/ftp/pub/assemblies/mammals/elephant), the mammoth mitochondrial genome (42), the dog genome (43), the mouse genome (44), the human genome (45), the env division of GenBank sequences, and the nt GenBank database from the National Center for Biotechnology Information. BLAST (blastall 2.2.12) searches were performed with the following options: –K 100 –b 10 –v 100 – I T –e 0.001 –F F –a 2. For each query sequence, the single best BLAST hit was analyzed. Hits from within a database were compared by e-value. Hits across different databases were compared by taking the total number of aligned residues. Sequence reads whose best hits were against the elephant genome (putative mammoth sequences) or against the human genome (putative human contaminants) were further analyzed. Only alignments longer than 30 nt were analyzed, because shorter alignments are not sufficient to correctly identify the source species (our unpublished observation). Within these alignments, the numbers of mismatches of each type were counted. To generate 99% confidence intervals for the number of each type of mismatch, the alignments were randomly sampled, with replacement 10,000 times, and the numbers of each type of mismatch were counted in each sample.
Oligonucleotide Amplifications.
The oligonucleotides containing the deaminated bases uracil and hypoxanthine, as well as cytosine and adenine, were ordered from Operon Biotechnologies (Cologne, Germany), and the oligonucleotides containing xanthine and guanine were synthesized by G.E.W. and J.T. Approximately 150 pg of oligonucleotide template (either one or two oligonucleotides in equal amounts) were used in amplifications of 20 μl consisting of 1 unit of AmpliTaq Gold or 1 unit of Platinum Taq High Fidelity, respectively, 1× AmpliTaq Gold PCR buffer or 1× High Fidelity PCR buffer, respectively, 2.5 mM MgCl2 or 2.5 mM MgSO4, respectively, 1 mM dNTPs, and 0.25 μM primers Oligo-F (5′-CCA CAG TAT TAT GTC CGT-3′) and Oligo-R (5′-GCT TTA ACT TCC GTA GTG-3′). Amplifications were performed in an MJ thermocycler with a 3-min activation step at 94°C, followed by 30 cycles of 93°C for 25 s, 50°C for 60 s, and either 72°C for 45 s in the case of AmpliTaq Gold or 68°C for 45 s when Platinum Taq High Fidelity was used. For the amplification of guanine- and xanthine-containing oligonucleotides, the primers Xan-1 (5′-GCG CGC CCA TCT AT-3′) and Xan-2 (5′-AGT TGT CAG AAG CAA ATG TAA-3′) were used in amplification reactions with similar chemical composition to those of the prior oligonucleotide amplifications. The reactions were performed in 40 cycles initiated by a 9-min activation step for AmpliTaq Gold and a 3-min activation step for Platinum Taq High Fidelity, respectively, followed by five cycles of 93°C for 25 s, 50°C for 45 s, and either 72°C for 45 s in the case of AmpliTaq Gold or 68°C for 45 s in the case of Platinum Taq High Fidelity. Further five cycles of 93°C for 25 s, 37°C for 45 s, and either 72°C for 45 s in case of AmpliTaq Gold or 68°C for 45 s in case of Platinum Taq High Fidelity were carried out and subsequently followed by 30 cycles of 93°C for 25 s, 50°C for 45 s, and either 72°C for 45 s in case of AmpliTaq Gold or 68°C for 45 s in case of Platinum Taq High Fidelity, with a final extension for 15 min at either 72°C for AmpliTaq Gold or 68°C for Platinum Taq High Fidelity. Amplification products were cloned and sequenced as described above.
Supplementary Material
Acknowledgments
We thank L. Kordos (Geological Museum of Hungary, Budapest, Hungary), V. Dimitrijevic (University of Belgrade, Belgrade, Serbia), T. Engel (Naturhistorisches Museum Mainz/Landessammlung für Naturkunde Rheinland-Pfalz, Mainz, Germany), M. Germonpré (Institut Royale des Sciences Naturelles de Belgique, Brussels, Belgium), N. Schmidt-Kittler (University of Mainz, Mainz, Germany), D. Nagel (University of Vienna, Vienna, Austria), and E. Willerslev (Copenhagen, Denmark) for paleontological samples; Barbara Hoeber, Barbara Hoeffner, and Antje Weihmann for sequencing; Matthias Meyer, Nadin Rohland, Johannes Krause, Adrian Briggs, Leif Andersson, and Tomas Lindahl for helpful discussions and comments; and the Max Planck Society for financial support.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequences reported in this paper have been deposited in the GenBank (accession nos. DQ852634–DQ852662) and EMBL (accession nos. CAAM01000001–CAAM01073172) databases.
See accompanying Profile on page 13575.
References
- 1.Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 2.Willerslev E, Cooper A. Proc Biol Sci; 2005. pp. 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorenson MD, Cooper A, Paxinos EE, Quinn TW, James HF, Olson SL, Fleischer RC. Proc Biol Sci; 1999. pp. 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loreille O, Orlando L, Patou-Mathis M, Philippe M, Taberlet P, Hanni C. Curr Biol. 2001;11:200–203. doi: 10.1016/s0960-9822(01)00046-x. [DOI] [PubMed] [Google Scholar]
- 5.Orlando L, Leonard JA, Thenot A, Laudet V, Guerin C, Hanni C. Mol Phylogenet Evol. 2003;28:485–499. doi: 10.1016/s1055-7903(03)00023-x. [DOI] [PubMed] [Google Scholar]
- 6.Karanth KP, Delefosse T, Rakotosamimanana B, Parsons TJ, Yoder AD. Proc Natl Acad Sci USA. 2005;102:5090–5095. doi: 10.1073/pnas.0408354102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard JA, Wayne RK, Cooper A. Proc Natl Acad Sci USA. 2000;97:1651–1654. doi: 10.1073/pnas.040453097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, Sher AV, Pybus OG, Gilbert MTP, Barnes I, Binladen J, et al. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- 9.Hofreiter M, Serre D, Rohland N, Rabeder G, Nagel D, Conard N, Munzel S, Pääbo S. Proc Natl Acad Sci USA. 2004;101:12963–12968. doi: 10.1073/pnas.0403618101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, et al. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. Nature. 1984;312:282–284. doi: 10.1038/312282a0. [DOI] [PubMed] [Google Scholar]
- 12.Pääbo S. Nature. 1985;314:644–645. doi: 10.1038/314644a0. [DOI] [PubMed] [Google Scholar]
- 13.Pääbo S, Wilson AC. Nature. 1988;334:387–388. doi: 10.1038/334387b0. [DOI] [PubMed] [Google Scholar]
- 14.Pääbo S. Proc Natl Acad Sci USA. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pääbo S. In: PCR Protocols and Applications: A Laboratory Manual. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. San Diego: Academic; 1990. pp. 159–166. [Google Scholar]
- 16.Handt O, Höss M, Krings M, Pääbo S. Experientia. 1994;50:524–529. doi: 10.1007/BF01921720. [DOI] [PubMed] [Google Scholar]
- 17.Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Pääbo S. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 19.Hofreiter M, Jaenicke V, Serre D, Haeseler Av A, Pääbo S. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen A, Willerslev E, Wiuf C, Mourier T, Arctander P. Mol Biol Evol. 2001;18:262–265. doi: 10.1093/oxfordjournals.molbev.a003800. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert MT, Hansen AJ, Willerslev E, Rudbeck L, Barnes I, Lynnerup N, Cooper A. Am J Hum Genet. 2003;72:48–61. doi: 10.1086/345379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert MTP, Willerslev E, Hansen AJ, Barnes I, Rudbeck L, Lynnerup N, Cooper A. Am J Hum Genet. 2003;72:32–47. doi: 10.1086/345378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binladen J, Wiuf C, Gilbert MT, Bunce M, Barnett R, Larson G, Greenwood AD, Haile J, Ho SY, Hansen AJ, et al. Genetics. 2006;172:733–741. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pääbo S, Irwin DM, Wilson AC. J Biol Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 25.Malmstrom H, Stora J, Dalen L, Holmlund G, Gotherstrom A. Mol Biol Evol. 2005;22:2040–2047. doi: 10.1093/molbev/msi195. [DOI] [PubMed] [Google Scholar]
- 26.Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S. Nat Rev Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- 27.Noonan JP, Hofreiter M, Smith D, Priest JR, Rohland N, Rabeder G, Krause J, Detter JC, Pääbo S, Rubin EM. Science. 2005;309:597–599. doi: 10.1126/science.1113485. [DOI] [PubMed] [Google Scholar]
- 28.Poinar HN, Schwarz C, Qi J, Shapiro B, MacPhee RDE, Buigues B, Tikhonov A, Huson DH, Tomsho LP, Auch A, et al. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 29.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohland N, Pollack JL, Nagel D, Beauval C, Airvaux J, Pääbo S, Hofreiter M. Mol Biol Evol. 2005;22:2435–2443. doi: 10.1093/molbev/msi244. [DOI] [PubMed] [Google Scholar]
- 31.Vila C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 32.Vila C, Amorim I, Leonard J, Posada D, Castroviejo J, Petrucci-Fonseca F, Crandall K, Ellegren H, Wayne R. Mol Ecol. 1999;8:2089–2103. doi: 10.1046/j.1365-294x.1999.00825.x. [DOI] [PubMed] [Google Scholar]
- 33.Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 34.Römpler H, Rohland N, Lalueza-Fox C, Willerslev E, Kuznetsova T, Rabeder G, Bertranpetit J, Schöneberg T, Hofreiter M. Science. 2006;313:62. doi: 10.1126/science.1128994. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z, Yoder AD. J Mol Evol. 1999;48:274–283. doi: 10.1007/pl00006470. [DOI] [PubMed] [Google Scholar]
- 36.Sismour AM, Lutz S, Park JH, Lutz MJ, Boyer PL, Hughes SH, Benner SA. Nucleic Acids Res. 2004;32:728–735. doi: 10.1093/nar/gkh241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuenschell GE, O'Connor TR, Termini J. Biochemistry. 2003;42:3608–3616. doi: 10.1021/bi0205597. [DOI] [PubMed] [Google Scholar]
- 38.Karran P, Lindahl T. Biochemistry. 1980;19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 39.Serre D, Langaney A, Chech M, Teschler-Nicola M, Paunovic M, Mennecier P, Hofreiter M, Possnert G, Pääbo S. PLoS Biol. 2004;2:313–317. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Höss M, Pääbo S. Nucleic Acids Res. 1993;21:3913–3914. doi: 10.1093/nar/21.16.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, Barnes I, Lister AM, Ebersberger I, Pääbo S, Hofreiter M. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- 43.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, III, Zody MC, et al. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 44.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 45.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.