Abstract
Background
Hox genes encode transcription factors that are involved in pattern formation in the skeleton, and recent evidence suggests that they also play a role in the regulation of endochondral ossification. To analyze the role of Hoxc-8 in this process in more detail, we applied in vitro culture systems, using high density cultures of primary chondrocytes from neonatal mouse ribs.
Results
Cultured cells were characterized on the basis of morphology (light microscopy) and production of cartilage-specific extracellular matrix (sulfated proteoglycans and type II Collagen). Hypertrophy was demonstrated by increase in cell size, alkaline phosphatase activity and type X Collagen immunohistochemistry. Proliferation was assessed by BrdU uptake and flow cytometry. Unexpectedly, chondrocytes from Hoxc-8 transgenic mice, which exhibit delayed cartilage maturation in vivo [1], were able to proliferate and differentiate normally in our culture systems. This was the case even though freshly isolated Hoxc-8 transgenic chondrocytes exhibited significant molecular differences as measured by real-time quantitative PCR.
Conclusions
The results demonstrate that primary rib chondrocytes behave similar to published reports for chondrocytes from other sources, validating in vitro approaches for studies of Hox genes in the regulation of endochondral ossification. Our analysis of cartilage-producing cells from Hoxc-8 transgenic mice provides evidence that the cellular phenotype induced by Hoxc-8 overexpression in vivo is reversible in vitro.
Keywords: skeletal development, Hox genes, Hoxc-8, chondrocyte, primary cells, endochondral ossification, transgenic mice, gene expression
Background
Endochondral ossification is the process by which mesenchymal cells condense at specific sites and differentiate into chondrocytes, forming the cartilage anlagen that are the model for the future bone. The cells in the center of the anlagen, which are initially immature, undergo an ordered differentiation program [2](also called chondrocyte maturation [3]): the chondrocytes proliferate, become pre-hypertrophic, and then undergo hypertrophy and matrix calcification. The calcified cartilage is then invaded by blood vessels that bring osteoblasts and osteoclasts, and bone is formed. Each step of cartilage maturation occurs in a precise and tightly regulated manner [4]. Disruptions of this process cause abnormalities in cartilage and bone formation [5,6]. Endochondral ossification occurs in embryonic skeletal formation, in skeletal growth and fracture healing.
Homeobox genes of the Hox class are required for proper patterning of skeletal elements [7]. The functional role of Hox genes in skeletal growth and development has been clearly demonstrated, but how they control the differentiation of specific tissues is not well understood. Hox genes encode transcription factors that regulate the expression of yet unidentified target genes [8]. In order to identify such target genes and to better understand the role of Hox genes in cartilage differentiation and maturation, we established in vitro culture systems for primary mouse rib chondrocytes.
Previously, we generated transgenic mice that overexpress the homeobox transcription factor Hoxc-8 in the thoracic region, where Hoxc-8 is normally expressed ([1] and unpublished results). The transgenic mice exhibit profound cartilage defects, predominantly in ribs and vertebral column, and severity of defects depends on transgene dosage. The abnormal cartilage is characterized by an accumulation of proliferating chondrocytes and reduced cartilage maturation. The structural rigidity of rib cartilage is greatly compromised, fatally interfering with pulmonary function, and vertebral cartilage is so weak that the skeleton often disassembles during skeletal preparation [1]. These results suggest that Hoxc-8 continues to regulate skeletal development well beyond pattern formation in a tissue-specific manner, presumably by controlling the progression of cells along the chondrocyte differentiation pathway. We found a similar phenotype upon overexpression of Hoxd-4 in our transgenic system (Kappen, manuscript in preparation), whereas overexpression of the LIM-homeodomain transcription factor Isl-1 did not cause abnormalities in cartilage but other developmental defects [1,9]. The observation that cartilage is affected by misregulation of Hoxc-8 and Hoxd-4, but not by a divergent homeobox gene, indicates that the capacity to regulate cartilage differentiation is specific to homeobox genes of the Hox subclass. It also suggests that Hox genes could be involved in human chondrodysplasias and other cartilage disorders. We envisioned that well-defined in vitro culture systems would allow us to further characterize the cellular and molecular basis of abnormal chondrocyte differentiation in Hox transgenic mice. Detailed knowledge of regulatory mechanisms in endochondral ossification will be essential for strategies to manipulate chondrocyte proliferation, differentiation and maturation in skeletal growth and development, osteochondrodysplasias and fracture healing.
The in vitro chondrocyte culture systems we utilized here consisted of high-density cultures of primary rib chondrocytes from neonatal mice. The micromass culture system [10] provides the three-dimensional environment needed for chondrogenesis, cartilage maturation and hypertrophy. The system also allows the investigation of the continuous program of differentiation, maturation and hypertrophy in the same culture. The cellular phenotype in culture was characterized by morphology and extra-cellular matrix (ECM) production. Chondrocyte maturation was assessed on the basis of cell proliferation, cellular hypertrophy, alkaline phosphase activity and expression of Collagen type II and type X. Apoptosis was investigated by TUNEL (terminal deoxynucleotidyl transferase-mediated deoxyuridine triposphate nick end labeling). High density bulk cultures were used to assess the capacity of chondrocytes from Hoxc-8 transgenic mice for cell proliferation and differentiation. Proliferation of chondrocytes in vivo was assayed by BrdU incorporation, and gene expression was analyzed by real-time quantitative PCR.
Results
We previously reported [1] that Hoxc-8 transgenic mice exhibit profound defects in cartilage, particularly in the rib cage and vertebral column. The cartilage is structurally insufficient and weak, contains fewer hypertrophic chondrocytes, displays much reduced staining for sulfated proteoglycans, and consists predominantly of immature chondrocytes, with a high fraction of proliferating cells. These results point to a role of Hoxc-8 in regulation of cartilage maturation and chondrocyte differentiation. A similar phenotype is found in Hoxd-4 transgenic mice ([1] and Kappen, manuscript in preparation) indicating that Hox transcription factors regulate chondrocyte development. To examine this function in more detail, we employed a culture system that facilitates chondrocyte maturation and cartilage formation, and closely recapitulates the in vivo situation [11]. The micromass system has been used extensively for studies of cartilage formation from limb mesenchyme [10-13] and also for culture of chondrocytes in high density [14]. Here, we have adapted the system to neonatal mouse rib chondrocytes, to study the role of Hoxc-8 in chondrocyte maturation.
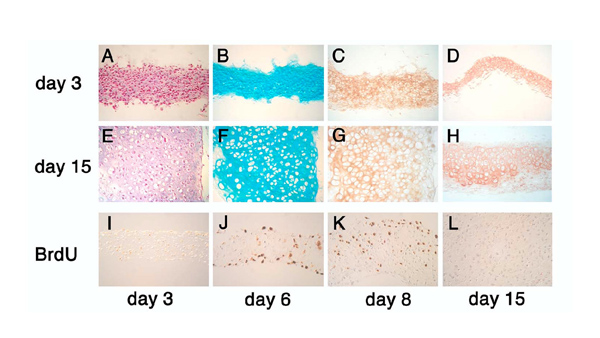
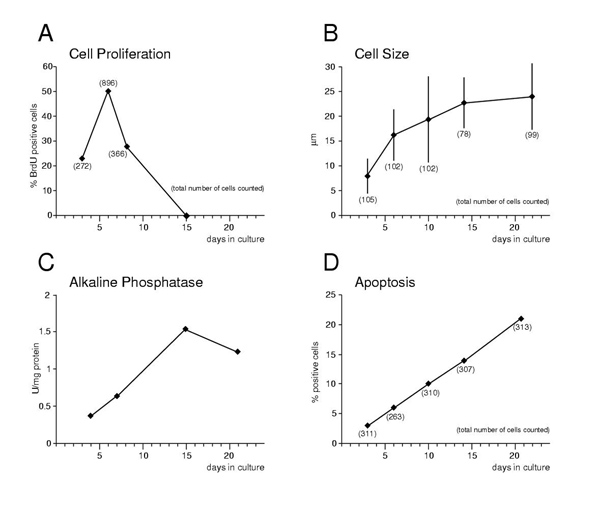
As established previously, chondrocytes in micromass culture are able to progress along their differentiation pathway to hypertrophy [14]. In cultures of neonatal rib chondrocytes, we find essentially the same results. As shown in Figure 1, histology (Panels A and E) and immunohistochemistry for Collagens II and X (Figure 1, Panels C, D and G, H) revealed appropriate cellular morphology, as well as increase in cell size towards hypertrophy (Figure 1, Panels E, G, H), and extracellular matrix production (Figure 1, Panel F). BrdU incorporation identified proliferating cells early in the culture (Figure 1, Panels J, K), while virtually no dividing cells were detected by day 15 (Figure 1, Panel L). The proportion of proliferating cells was lower at the beginning of the culture (Figure 2, Panel A), reflecting our observation that cells may need about 1–2 days to adapt to the culture conditions. Maximal proliferation was found on days 3 and 4, consistent with the presence of predominantly proliferating cells in the culture (compare Figure 2, Panel A and Figure 1, Panel I). Manifesting their progression to hypertrophy, the size of cells increased (Figure 2, Panel B), with the majority of cells progressed to hypertrophy by day 15 (see also Figure 1). Concommittant with cartilage ECM deposition, there was an increase in Alkaline Phosphotase activity (Figure 2, Panel C). As cells matured towards hypertrophy, there was also increased cell death in our micromass cultures (Figure 2, Panel D). Apoptosis, as assessed by TUNEL, was present from day 3 on, and the percentage of positive cells increased with time in culture. Similar results were reported by other authors [15-17]. These results demonstrate that the cultured rib chondrocytes behaved in a similar manner as chondrocytes in vivo. Taken together, primary rib chondrocytes from mouse neonates grown in micromass culture display cellular differentiation, proliferation, maturation and hypertrophy, recapitulating the chondrocyte maturation program in vivo. These results establish the utility of this in vitro system to investigate the role of Hox genes and other transcription factors in endochondral ossification.
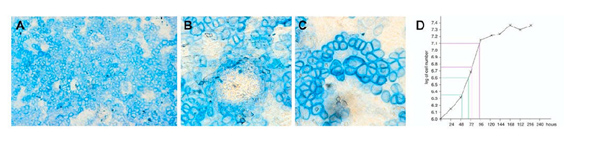
Figure 1.
Micromass cultures of primary rib chondrocytes. Morphology of micromass cultures was examined at day 3 (A-D) and day 15 (E-H) by light microscopy of paraffin-embedded sections stained with hematoxylin and eosin (A and E), Alcian blue (B and F), Collagen II (C, G), and Collagen X (D, H). Cell proliferation was assessed by BrdU uptake as detected by immunohistochemistry on days 3 (I), 6 (J), 8 (K) and day 15 (L), respectively.
Figure 2.
Quantitative parameters of primary rib chondrocytes in micromass culture. Panel A: Cell Proliferation was assessed by BrdU incorporation as detected by immunohistochemistry on sections of micromass cultures harvested after 24 hours of BrdU exposure. At day 15, no positive cells were detected (see also Figure 1, Panel L). The major phase of growth occurs between days 3 and 4, 72 to 96 hours after plating. Panel B: Cell size of chondrocytes was measured by photomicrography on sections, and mean cell size increases over time. Panel C: Alkaline phosphotase activity in protein extracts from micromass cultures was measured as the release of p-nitrophenol from p-nitrophenylphosphate by absorbance at 405 nm. One unit of enzyme activity will produce one μ mole of p-nitrophenol per minute. Panel D: Apoptosis was assessed by TUNEL staining on sections from micromass cultures, and was found to increase over time.
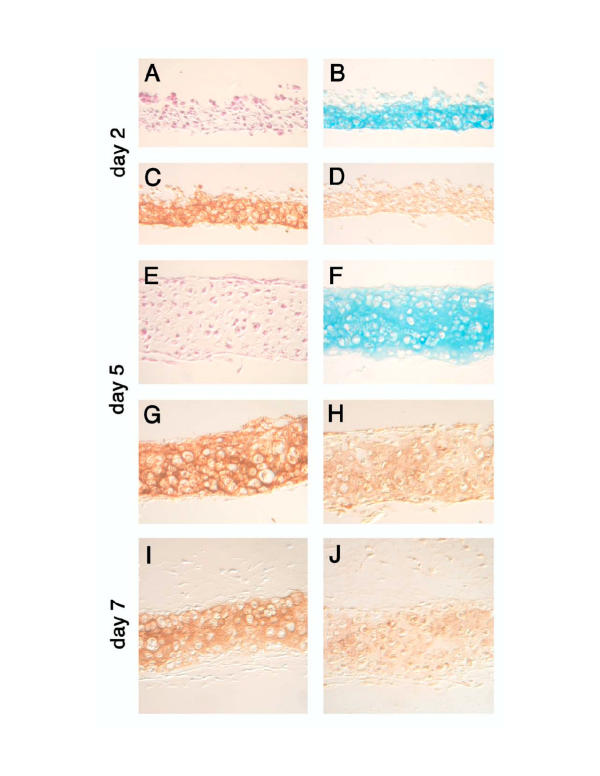
Rib chondrocytes from neonatal Hoxc-8 transgenic mice were cultured in the micromass system, and the cultures were examined for the same parameters described above. Figure 3 shows that Hoxc-8 transgenic chondrocytes morphologically appeared normal and were able to produce Collagens II and X, and sulfated proteoglycans (Panels C, G, I, D, H, J and B, F respectively). There was no discernable difference in the ability of Hoxc-8 transgenic chondrocytes to grow or mature in this system (compare to Figure 1, Panels A-H). These data indicate that chondrocytes from Hoxc-8 transgenic mice have the capacity to differentiate and mature normally under the culture conditions described here. This finding was unexpected in light of the severe cartilage defects in Hoxc-8 transgenic mice [1]. However, the possibility remained that, despite apparently normal maturation, proliferation of chondrocytes could be affected by the Hoxc-8 overexpression. Such an effect would be consistent with the accumulation of immature, proliferating chondrocytes in Hoxc-8 transgenetic mice [1]. We therefore characterized cell proliferation in Hoxc-8 transgenic chondrocytes.
Figure 3.
Micromass cultures of Hoxc-8 transgenic cells. Primary rib chondrocytes were prepared from newborn mice and cultured in micromass as described. Histological staining with (Hematoxylin & Eosin Panels A, E), and Alcian Blue, (Panels B, F) and immunohistochemical detection of Collagen II (Panels C, G, I) and Collagen X (Panels D, H, J) on sections prepared on day 2 (Panels A-D), day 5 (Panels E-H), and day 7 (Panels I, J) do not indicate differences to control chondrocyte cultures (see Figure 1) even at early timepoints after plating.
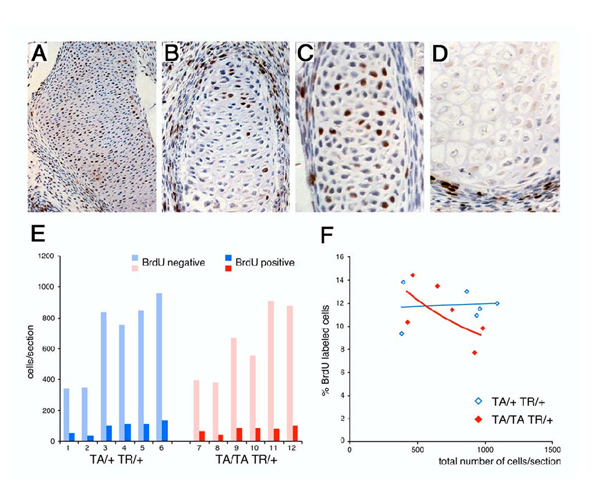
In vivo labeling with BrdU was performed to investigate the proliferative status of chondrocytes in Hoxc-8 transgenic mice. Previously, we [1] found that upon overexpression of Hoxc-8, the vertebral cartilages accumulate immature chondrocytes, and we showed that these cells are proliferating cells by virtue of staining for Proliferating Cell Nuclear Antigen (PCNA). The accumulation of proliferating chondrocytes could be explained in four different scenarios: (i) There is a block in differentiation that prevents cells from entering hypertrophy. The proportion of hypertrophic cells was indeed reduced in Hoxc-8 transgenic mice, but progression to hypertrophy was not completely inhibited. (ii) There is increased recruitment of precursor cells into the chondrocyte cell lineage. This proposition is difficult to investigate, and no evidence is available at this stage. (iii) The rate of cell division is increased. Then, the cell cycle duration of proliferating chondrocytes should be shorter. (iv) The cell cycle time of proliferating chondrocytes is lengthened, resulting in a greater number of cells in the steady-state pool of proliferating cells. To distinguish these latter two possibilities, we analyzed BrdU incorporation into chondrocytes in Hoxc-8 transgenic mice in vivo. Multiple sections were examined from each embryo and showed incorporation of BrdU four hours after labeling. Figure 4 shows representative results for the immunohistochemical detection of BrdU incorporation in cartilage structures (Panels A, D). To directly compare cell proliferation between embryos of different genotype, sections of vertebral centers were assessed for BrdU incorporation. Figure 4, Panel E shows results from embryos isolated at day 15.75 post coitum. The differences in cell number/section of vertebral center indicate that individual embryos were more progressed in overall growth than others, and thus had larger vertebral centers with more cells overall. Maturation was consistent within a given litter (compare samples 1, 2, to 7, 8 and 5, 6 to 11, 12, respectively). The fraction of cells positive for BrdU incorporation was between 7.8 and 14.4%, with a trend towards lower rates of incorporation in developmentally older embryos with higher Hoxc-8 transgene expression levels (TA/TA TR/+; Figure 4, Panel F). While additional measurements at later stages of development would be needed to confirm whether this is a continuous trend for cartilage development in late pregnancy in Hoxc-8 overexpressing animals, these results were initially unexpected: The phenotype of Hoxc-8 transgenic animals at 17.5 days (approximately 2 days later than assayed here) is characterized by accumulation of proliferating cells. Intuitively, the expectation would be that greater rates of BrdU incorporation should be found. However, accumulation of proliferating cells can also result from a slow-down of the cell cycle, in form of a lower rate of BrdU incorporation. It is intriguing to note that reduced BrdU incorporation is particularly detected in developmentally older animals where the vertebral centers are made up of a larger number of cells, reflecting a longer duration since commitment of the precursors to chondrocyte fate. It should also be noted that both genotypes assayed here develop cartilage defects, which may explain the relatively moderate differences. A prediction from these data is that on subsequent days, or upon longer labeling intervals, even lower rates of BrdU incorporation should be detected. Quantification of such findings, and in vivo measurements of cell cycle duration, however, require pulse-chase experiments [18,19], which involve large numbers of animals and are very time-consuming.
Figure 4.
BrdU incorporation in Hoxc-8 transgenic animals. Incorporation of BrdU in Hoxc-8 transgenic animals was detected by Horseradish Peroxidase (HRP) immunohistochemistry on sections from embryonic day 15.75 animals. Sections were cut at 5 μm thickness and counterstained with Hematoxylin. Quantitation of BrdU incorporation was obtained by counting cells in the vertebral centers on multiple sections from the same individual (see Methods). Panel A: Cartilage of the humerus (50 × magnification). Panel B: Cartilage of the third rib (200 ×). Panel C: Cartilage of the sixth rib (400 ×). Panel D: Hypertrophic cells of the scapula (200 ×). BrdU incorporation was only detected in proliferating and prehypertrophic, but not in hypertrophic cells. Panel E: BrdU incorporation (dark colored bars) in vertebral center chondrocytes (light colored bars: cells without BrdU incorporation) of embryos at embryonic day 15.5 to 16.5. Data were obtained from animals with moderate (blue; genotype TA/+ TR/+) and high (red; genotype TA/TA TR/+) Hoxc-8 expression. Panel F: Fraction of cells labeled by BrdU incorporation as a function of size of vertebral centers in animals with moderate (blue, open symbol) and high (red, closed symbol) Hoxc-8 transgene expression levels (genotypes correspond to colors as before).
Instead, we sought to investigate the proliferative capacity of chondrocytes from Hoxc-8 transgenic mice by using in vitro assays. We performed cell cycle assays on primary chondrocytes isolated from rib cages of transgenic and control neonates. Chondrocytes were placed into high density bulk cultures [20], where they initially continued to proliferate, and later differentiated, with cartilage nodules present by 14 days after plating. Figure 5 shows representative cultures (Figure 5, Panels A-C), and the growth curve for rib chondrocytes in high density bulk cultures (Figure 5, Panel D). From this, we were able to estimate a mean doubling time of 17–19 hours. This conforms well to previous published measurements for chick limb chondrocytes [21] and rat chondrocytes in vitro [22]. The linear phase of growth was observed between days 3 and 4, and therefore the 72 hour time point was chosen for analysis of cell cycle kinetics of Hoxc-8 transgenic chondrocytes in vitro. First, we established the Fluorescence Activated Cell Sorting (FACS) assay using mouse embryonic fibroblasts, and then we assayed rib chondrocytes from Hoxc-8 transgenic and control mice in high density bulk cultures. From these cultures, cells were removed at various timepoints after BrdU incorporation and subjected to Flow cytometry. Figure 6 (Panels A-E") shows a typical result for fibroblasts. The same readout was used for cultures of primary rib chondrocytes (Figure 6, Panels F-O). As cells go through the cell cycle, DNA content increases, and this is reflected in a concomitant increase in BrdU incorporation. Upon division, the signal for DNA content is reduced by half, while overall BrdU incorporation increases with every subsequent cell cycle. These assays allowed us to determine the fraction of cells in each stage of the cell cycle (Figure 7, Panel A), and established FACS analysis as a valuable tool for the analysis of chondrocyte proliferation.
Figure 5.
Primary mouse rib chondrocytes in high density bulk cultures. Panel A: Representative culture at 14 days stained with Alcian Blue (100 × magnification); Panel B: Cartilage nodule (200 ×); Panel C: Hypertrophic cells (200 ×); Panel D: Growth curve for primary rib chondrocytes in high-density bulk culture. The linear phase of growth occurs between 48 and 96 hours of plating. From the slope of the curve a mean doubling time of 17 hours (distance between the two green or the two purple lines, respectively) during the linear phase can be calculated.
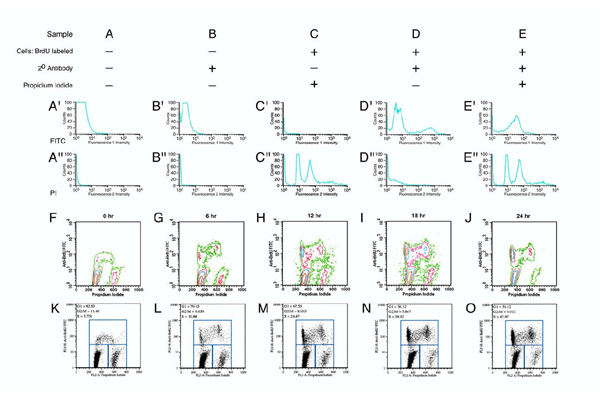
Figure 6.
Cell proliferation assays by Fluorescence Activated Cell Sorting (FACS). Cell proliferation assays were done with BrdU incorporation, which was detected by staining of isolated nuclei with a FITC-fluorochrome-coupled monoclonal antibody. DNA content in nuclei was determined on the basis of fluorescence intensity for Propidium Iodide. The approach was first validated for primary embryonic fibroblast cultures (Panels A-E") and then applied to primary chondrocytes (Panels F-O). Cells were incubated (Panels A, B) in standard medium (DMEM, 10% FCS, high glucose), or in medium with BrdU (Panels C, D, E) for 18 hours, and removed from the dishes by trypsinization. Preparation of nuclei and staining for BrdU and Propidium Iodide were done as described in the methods. The fluorescent signals in samples A and B (Panels A', A", B', B") were very low, as expected for cells without BrdU incorporation and without PI labeling. Sample C showed no green fluorescence (Panel C') due to absence of the BrdU-specific antibody during staining. Yet Propidium Iodide (Panel C") labels two populations of nuclei, those with 1n DNA content (peak at medium red fluorescence intensity) and those with 2n DNA content (high red fluorescence intensity) that are in S-Phase. Sample D was labeled only for BrdU incorporation and shows the majority of nuclei unlabeled (Panel D; low green fluorescence intensity) while a smaller fraction incorporated BrdU (Panel D', high green fluorescence intensity). No signal for DNA content was obtained (Panel D") consistent with absence of PI. Sample E was double-labeled and showed signal for both BrdU (Panel E') and PI (Panel E"). Panels F-O: Cell cycle analysis of primary rib chondrocytes incubated after 72 hours of culture with BrdU for various length of time. Panels F-J: Contour plots display fluorescence intensity for BrdU incorporation (Y-axis) and DNA content (X-axis), with contours representing increasingly higher number of cells with a given fluorescence intensity (orange, highest cell number; green lowest cell number). DNA content and BrdU incorporation increase with DNA synthesis, and DNA content is reduced by half at cell division. Cell that recently underwent DNA synthesis are intensely labeled for BrdU (FITC, Y-axis), and may have 2n DNA content (high Propidium Iodide, X-axis) before, and 1n DNA content (low Propidium Iodide) after cell division. The proportion of cells in the BrdUhi PIlow group will increase with multiple cell divisions. Panels K-O: Dot plot representation of the same data with gating to quantitate fractions of cells in each group: cells with high intensity for BrdU-staining are in S-phase, cells with high Propidium Iodide but low BrdU-staining intensity are in G2/M-phase, and cells low for both Propidium Iodide and BrdU-staining are in G1 phase of the cell cycle.
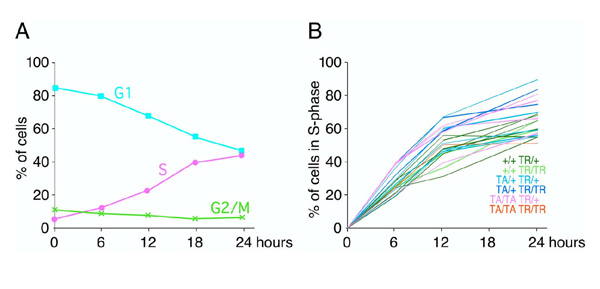
Figure 7.
Proliferation studies for Hoxc-8 transgenic cells. From the data in Figure 6, the proportion of cells in each phase of the cell cycle can be estimated, and a representative graph is shown in Panel A (turquoise: G-phase; green: G2/M-phase; pink: S-phase). With increasing time of incubation, BrdU incorporation increases, labeling cells that actively proliferate. After 24 hours, up to 40% of cells have entered the DNA synthesis phase of the cell cycle. Panel B shows the proportion of chondrocytes in S phase in correlation to Hoxc-8 transgene expression level. The Hoxc-8 transgene is not expressed (REF) in TR only cells (dark and light green), moderately expressed in cells from animals with TA/+ TR/+ (light blue) and TA/+ TR/TR (dark blue) genotype (REF, REF) and more strongly expressed in cells from TA/TA TR/+ (pink) and TA/TA TR/TR (red) animals (REF, REF). Similar fractions of cells are in S-phase in all samples by 24 hours, and the rate of entry into S-phase (slope of curves) is very similar, indicating that cells of all genotypes exhibit proliferative capacity irrespective of expression of the Hoxc-8 transgene.
Interestingly, when primary chondrocytes from Hoxc-8 transgenic mice were assayed, they showed the same cell cycle kinetics as chondrocytes from control animals. Essentially similar proportions of cells in S phase were found in all cultures (Figure 7, Panel B). This was regardless of levels of expression of the Hoxc-8 transgene (Figure 7, Panel B) which increases with increasing transgene dosage [1,23]. These data indicate that, after appropriate time in culture, transgenic chondrocytes were able to proliferate normally, at least over the 24 hours duration of the assay.
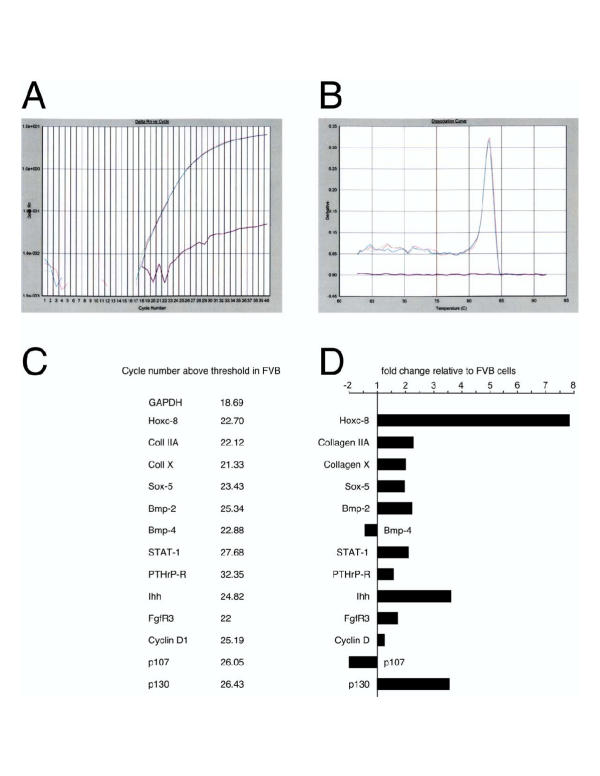
Given our inability to distinguish Hoxc-8 transgenic cells from controls by parameters for either cell differentiation or proliferation, we sought to determine which molecular differences exist in our transgenic mice associated with the documented deficiencies in cartilage maturation [1]. To this end, we analyzed gene expression in freshly isolated primary rib chondrocytes from Hoxc-8 transgenic animals [1]. Real-time quantitative PCR was performed. As shown in Figure 8, Hoxc-8 expression is elevated in Hoxc-8 transgenic mice, as expected [23]. Differences also exist between controls and transgenics in expression levels for several genes involved in cartilage development. For example, Collagen II expression (splice form A) is increased, as is Ihh mRNA, and Bmp-4 and p107 expression are decreased. Taken together, these results establish alterations in gene expression upon overexpression of the Hoxc-8 transgene in the developing skeleton.
Figure 8.
Gene expression in rib chondrocytes from Hoxc-8 and Hoxd-4 transgenic mice. Real-time quantitative PCR was performed as described. Panel A shows typical curves for real-time detection of amplification. Panel B shows a melting curve analysis for the PCR products, which is indicative of the quality of the reaction. For each reaction, the fidelity of amplification was assessed by inspection of the amplification and melting curves. Panel C lists the cycle number above the threshold level (Ct) for each primer pair at which product was detected in a representative experiment in FVB controls. Low Ct values reflect higher expression levels, high Ct values correspond to lower expression levels. Panel D shows gene expression measurements in Hoxc-8 transgenic samples. GAPDH cDNA levels in each sample were used to standardize measurements, and gene expression levels are normalized to the level of each gene found in primary chondrocytes from normal FVB control mice. The results are plotted as "fold change relative to FVB" with decreased expression in negative (left on X-axis) iand increased expression in positive values (right on X-axis). The FVB control sample would always be 1, and therefore was not plotted. RNA extracts were prepared from postnatal day 0 mouse rib chondrocytes from normal FVB newborns or animals transgenic for Hoxc-8, and equal amounts of cDNA were pooled from 4–5 individual animals for each sample.
The biological corollaries of changes in gene expression profiles are noteworthy in that they may provide evidence on the molecular targets of Hox genes in cartilage, as well as on the consequences of Hox gene overexpression. In Hoxc-8 transgenic chondrocytes, we found increased expression of Collagen II splice form A mRNA [24]. This is consistent with our earlier finding of at least 2-fold increased Collagen II mRNA expression by in situ hybridization and increased numbers of immature chondrocytes in Hoxc-8 transgenic mice [1]. Sox-5, which is normally expressed in proliferating chondrocytes [25,26], is elevated in Hoxc-8 transgenic cells, again consistent with accumulation of immature chondrocytes. Unexpected was the elevated expression of Collagen X mRNA, a marker for hypertrophic cells. As fewer hypertrophic cells are present in Hoxc-8 transgenics, and cartilage maturation is delayed in Hoxc-8transgenic animals [1], one would expect to see decreased Collagen X mRNA levels [27]. Yet, the data clearly indicate that mRNA expression is elevated in transgenic rib chondrocytes. This could be either from increased Collagen X mRNA transcription within prehypertrophic and hypertrophic cells or from ectopic activation of Collagen X transcription in proliferating chondrocytes. Whether either of these possibilities would also increase Collagen X expression at the protein level remains to be investigated. Interestingly, Collagen X expression has been shown to be induced by Bmp-2 [28], which is overexpressed in our transgenic mice. Bmp-2 signaling has a positive effect on chondrocyte proliferation [29], and can induce Indian hedgehog (Ihh) expression in responsive cells [30]. We find Ihh mRNA elevated in Hoxc-8 transgenic chondrocytes (see Figure 8). Ihh can induce Bmp expression in proliferating cells, further increasing positive effects on chondrocyte proliferation [31]. At the same time, Ihh decreases the rate of progression to hypertrophy [32,33], again consistent with an accumulation of proliferating cells in our transgenic animals. Whether the elevated Ihh mRNA levels originate from increased expression in cells that make Ihh normally (prehypertrophic and hypertrophic chondrocytes) or from ectopic activation by the Hox transcription factors in proliferating cells, remains to be investigated. Greater numbers of prehypertrophic cells are unlikely to account for elevated expression of prehypertrophic cell markers: histological sections from Hoxc-8 transgenic animals show no increase in prehypertrophic cells [1], and PTHrP receptor mRNA levels are unchanged in cells from transgenic animals (see Figure 8). These data also suggest that PTHrP signaling [34-36] is only moderately affected by overexpression of Hoxc-8. We found the expression of p107 mRNA and Bmp-4 mRNA to be decreased, which, if reflected in reduced protein levels, would be consistent with the phenotype of delayed cartilage maturation in Hox transgenic animals: As Bmp-4 was shown to have a positive effect on hypertrophy and inhibitory effect on proliferation [37,38], decreased Bmp-4 expression would be expected to promote chondrocyte proliferation. p107 has been shown to be required for exit from the cell cycle [39,40], and reduced p107 expression is associated with deregulated proliferation. Also, chondrocytes without p107 expression fail to respond to inhibitory signals from FGFs [41]. The relationship to elevated Fgf receptor 3 mRNA levels in Hoxc-8 transgenic cells, however, remains to be clarified. The same caveat applies to elevated expression in Hoxc-8 transgenic cells of p130, which is also known to be linked to cell cycle exit and FGF response [39]. While the profile of gene expression changes in Hoxc-8 transgenic mice is suggestive of interactions between the various gene products, further studies will be required to mechanistically link the gene expression differences to the maturational delay in the Hoxc-8 transgenic cartilage.
In summary, the gene expression profiles of primary chondrocytes from Hoxc-8 transgenic animals display notable differences in the expression of regulators of chondrocyte maturation in a pattern largely consistent with delayed chondrocyte maturation in vivo. These results now allow us to investigate whether any of these molecules is directly regulated by Hox transcription factors in chondrocytes. It is also remarkable, as apparent from our cell culture results, that cells with obvious molecular differences downstream of Hoxc-8 expression can be modulated to resemble a normal chondrocyte phenotype after a few days in culture.
Discussion
Our studies of chondrocytes from Hoxc-8 transgenic mice reveal several interesting results: in vivo, chondrocytes remain relatively immature, with few progressing to hypertrophy in Hoxc-8 transgenic mice. Combined with the fact that some, albeit fewer, hypertrophic cells are present, the accumulation of proliferating chondrocytes suggests that Hoxc-8 regulates the rate of progression of chondrocytes to maturity [1]. In line with this interpretation, the in vivo BrdU incorporation assays suggested that cell cycle progression of chondrocytes may be decreased in transgenic animals with higher Hoxc-8 transgene expression levels. If fewer immature cells enter the cell cycle in a given time interval, this is consistent with, and would explain the increased steady-state numbers of chondrocytes upon Hoxc-8 overexpression. This could occur through two different mechanisms: (i) Hoxc-8 actively recruits cells into the chondrocyte lineage and maintains them in the proliferating state; or (ii) Hoxc-8 inhibits a specific step in chondrocyte maturation. In order to distinguish between these possibilities, we sought to employ cell culture assays with well-established parameters for chondrocyte differentiation and maturation. Interestingly, chondrocytes from Hoxc-8 tansgenic animals behaved undistinguishable from control cells under all conditions tested. No differences were found in capacity to differentiate in micromass (see Figure 3), or in high density cultures (not shown), and proliferative indices are comparable (see Figure 7), irrespective of transgene dosage. The latter could be explained by the fact that the FACS analyses were done at the time of greatest proliferation, three days after culture initiation, which may allow transgenic cells to recover to a normal phenotype. However, we did not detect any differences in the first few days of culture by microscopy or histology. Cellular differences must exist at the time the cells are taken from the animal (which would be expected to die as a Hoxc-8 transgenic), and indeed molecular differences exist (see Figure 8). Yet, the cell culture conditions either quickly reverse cellular phenotype or support normal chondrocyte maturation. Alternatively, it is conceivable that the Hoxc-8 transgene could be silenced as cells are isolated from the animal, returning cells to their normal phenotype. There is little evidence to support this possibility as Hoxc-8 is expressed normally in our chondrocyte cultures (data not shown), and the promoter used to drive the transgene contains a cartilage-specific element (Cormier and Kappen, unpublished results) that is active in chondrocytes. Yet, even if transgene expression was reduced in culture, the important implication of our results is that the downstream biological effects of at least 6 days of Hoxc-8 overexpression in vivo are reversible. From these results, we postulate that the cartilage defect in Hoxc-8 transgenic mice is mediated by absence of factors/signals that are present in the culture medium. Serum-free conditions and supplementation with candidate molecules would be one strategy to investigate this further. An interesting, albeit preliminary, finding from our gene expression studies is that chondrocytes from Hoxc-8 transgenic animals display gene expression profiles that are indicative for immature chondrocytes, but not totally identical. This suggests that both Hox transcription factors may, in addition to the delay in chondrocyte maturation, induce specific changes in gene expression. By extending the collection of candidate genes, or screening on microarrays, it may thus be possible to identify specific targets for Hox transcription factors in chondrocytes.
Conclusions
Our results support the conclusion that factors in serum-containing medium provide the environment in which Hoxc-8 transgenic chondrocytes are capable of reverting to a normal cellular phenotype. Thus, Hoxc-8 induced changes in chondrocytes are reversible under certain conditions. This has important implications for the identification of the transcriptional targets of Hox transcription factors. In our system, the biological effects of Hox genes in chondrocytes can be modulated by external factors, and this finding points to the importance of gene-environment interactions as modulators of Hox gene function.
Methods
Preparation of mouse rib chondrocytes
The preparation of primary rib chondrocytes from neonatal mice is a modification of the method used by Lefebvre et al. [20]. Briefly, newborn FVB mice were sacrificed, and the ventral regions of rib cages were dissected in sterile conditions to release soft tissues. Rib cages were transferred to phosphate buffered saline (PBS) and washed extensively. Cells were dissociated in 50 ml tubes by enzymatic digestion with 0.25% Collagenase (Worthington Biochemical, Collagenase Type 2 – CLS 2) and 0.25% Trypsin/EDTA (Invitrogen/Gibco BRL) for 1 hour at 37°C, or until the soft tissues were floating in the solution. The solution was aspirated, leaving the rib cages on the bottom of the tube. The ribcages were washed with PBS, and fresh 0.25% Collagenase in PBS was added for 1 hour at 37°C, or until the rib cages were dissociated. The enzymatic reaction was stopped with an equal volume of tissue culture medium containing 10% FCS. The cells were filtered through a 70 μm cell strainer (Becton Dickinson) and pelleted by centrifugation in 50 ml conical tubes (maximum volume 25 ml) at 1200 rpm for 5 minutes. The pellet was washed with PBS, cells were counted, and adjusted for desired cell density.
Primary chondrocyte high density bulk cultures
For analysis of cell cycle parameters, we kept primary chondrocytes in high density bulk cultures, as this allows for recovery of cells for flow cytometry. These cultures undergo chondrocyte maturation with differentiation to hypertrophy and formation of cartilage modules. Briefly, cells were plated on 35 mm dishes coated with 0.1% gelatin at a final concentration of 3 × 105/dish, in DMEM, high glucose, 10% FCS and 25 μg/ml L-ascorbic acid. Removal of cells was done by trypsinization. For Alcian Blue staining, cells were fixed in 70% Ethanol for 30 minutes and then incubated for one hour with the Alcian Blue staining solution, which consisted of 15 mg Alcian Blue 8GX (Sigma) in 30 ml of 95% Ethanol and 20 ml Glacial acetic acid. Cells were rinsed again with 70% Ethanol, and dehydrated through an Ethanol series before coverslipping on the dish.
Primary high density micromass cultures
For micromass cultures [12], cell density was adjusted to 25 × 106 cells/ml. 103 l of cell suspension was plated in the center of a culture well (24-well plate, Midwest Scientific). The cultures were incubated at 37°C in a humidified incubator with 5% CO2 for 1 1/2 to 2 hours to allow for cell adhesion and attachment. Medium was added very slowly and consisted of a 1:1 mixture of DMEM and F12 (Life Technologies/Gibco BRL) supplemented with 10% fetal calf serum (Hyclone), 5000 U/ml Penicillin, 53 g/ml Streptomycin (Life Technologies/Gibco BRL), 25 μg/ml L-ascorbic acid (Life Technologies/Gibco BRL) and 10 mM β-Glycerophosphate (Sigma). Medium was replaced with fresh medium every other day [11,13,42].
Assessment of morphological and histological parameters of micromass cultured cells
During the first 2 or 3 days, micromass cultures were observed directly by phase contrast light microscopy. After that, cultures were harvested, washed in PBS, fixed in 4% Paraformaldehyde for 45 minutes, dehydrated in graded series of Ethanol and paraffin-embedded for subsequent analysis. We used histochemical staining with Hematoxylin and Eosin to ascertain cell morphology; and Alcian Blue at pH 1 [43] to detect cartilage matrix sulfated glucosaminoglycans as described before [1]. The production of Collagen type II was assessed by immunohistochemistry using a monoclonal antibody against mouse type II Collagen (Chemicon), and secondary antibody and colorimetric reaction kit (Histostain Plus, Zymed). Cell proliferation was assessed by BrdU incorporation. Cells were incubated with 3 mM of BrdU (Sigma), harvested after 24 hrs and paraffin-embedded. Cells with BrdU incorporation were detected by immunohistochemistry with a monoclonal antibody against BrdU (BrdU staining kit, Zymed). Cell size was determined by measuring cell diameter microscopically, and mean cell size was calculated. Alkaline phosphatase activity was measured as follows: Cells were rinsed in cold TBS (Tris-buffered Saline), scrapped from the plate, homogenized in 0.05% Triton X-100 in Tris-HCl (pH 7.5) on ice, and the sample was centrifuged at 1200 rpm for 5 minutes at 4°C. Protein concentration in the supernatant was assessed by Micro BCA assay (Pierce Chemicals). Alkaline phosphatase activity in the supernatant was determined spectrophotometrically at 405 nm wavelength as the amount of p-nitrophenol released from the substrate p-nitrophenyl phosphate (ALP10, Sigma). Type X Collagen deposition in extracellular matrix was analyzed by immunohistchemistry using a monoclonal antibody against mouse type X Collagen (Research Diagnostics, Inc.) and the same secondary reagents as described above. Apoptosis was assessed by terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate (dUTP) nick end labeling (TUNEL) (In situ Cell Death Detection Kit, POD, Boehringer-Manheim).
Transgenic mice
The generation of Hoxc-8 transgenic mice and the characterization of their phenotype have been published [1]. Briefly, Hoxc-8 expressing mice are generated in a binary transgenic mouse system [44] by crossing a transactivator strain (harboring a transgene in which expression of the transcriptional transactivator VP16 is under control of the Hoxc-8 promoter; [45]) to a transresponder strain (that carries as a transgene the Hoxc-8 cDNA linked to the ICP4 immediate early gene promoter, which is silent in mice in the absence of VP16; [45,46]). Upon combination of both transgenes, the Hoxc-8 transresponder transgene is activated, and Hoxc-8 becomes overexpressed [23]. This system also allows the generation of transgenic mice with increased gene dosage by superimposition of transgene loci to homozygosity [23,44]. For the studies reported here, we used the T239 transactivator and the IE-c8-254 transresponder strains. All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee at UNMC.
Hoxd-4 transgenic mice were generated using the same transactivator strain crossed to the IE-d4-70 transresponder strain [1]. Maintenance and genotyping of transgenic mice was done as described before [1,23]. Unless otherwise noted, cells and samples were obtained from animals hemizygous for both transgenes.
All transgenes were maintained on a homogenous FVB inbred genetic background, and normal FVB mice were used as controls unless mentioned otherwise.
In vivo BrdU incorporation
To assess chondrocyte proliferation in Hoxc-8 transgenic mice, we performed in vivo BrdU labeling experiments. BrdU labeling reagent (Amersham RPN202) in PBS was injected into pregnant females at 0.01 ml/gr body weight. Four hours later, dams were sacrificed, embryos were isolated and fixed in 4% paraformaldehyde overnight at 4°C. They were then dehydrated through an alcohol series and embedded in paraffin for histological sectioning. Sections of 10 μm thickness were rehydrated and incubated for 10 minutes with one part 30% H2O2: 9 parts MeOH and processed in HIER (Biotek Solutions), denaturing and blocking solutions following the manufacturer's protocol. Staining of sections for BrdU incorporation was done using the Zymed BrdU staining kit with Horseradish Peroxidase, and development with DAB for 5 minutes. Counterstaining was done with Hematoxylin for one minute. Sections of vertebral structures were matched between pairs of embryos from the same litter according to anatomical landmarks of other tissues on the same section. Cells in skeletal structures were counted from at least 5 (samples 1,2,7,8) and more than 10 (all other samples) consecutive slides for each animal and cell counting was done independently by two technologists without knowledge of sample genotype. Data were collected from three independent experiments and were averaged to cells/section to normalize for potential growth differences between litters. Exponential trendlines were added by regression analysis as implemented in Microsoft Excel.
Assessment of cell proliferation in vitro by BrdU incorporation and flow cytometry
Primary rib chondrocytes were prepared as described above and 3 × 105 cells/well were placed into culture in 6-well plates. Medium was changed daily. Proliferation was assessed by cell counting, and maturation was ascertained visually as cartilage nodule formation, and histochemically by Alcian Blue staining. Cell cycle kinetics were measured by BrdU incorporation and flow cytometry for DNA content at various time points after initiation of the cultures. Cells that had not been incubated with any reagents or cells that were exposed only to anti-BrdU antibody and Propidium Iodide served as controls. For BrdU incorporation studies, BrdU labeling reagent (Amersham) was diluted 1:1000 in medium to a final concentration of 10 μM BrdU and 1 μM FdU, and sterilized through a 0.22 μm filter. 2 ml of staining solution were added to each well of a 6-well plate, followed by incubation for 0, 6, 12, 18, or 24 hours as indicated. To remove the cells, wells were washed with PBS, and 2 ml Trypsin-EDTA (Gibco/BRL) was added for at least five minutes at 37°C. The reaction was inactivated by an equal volume of serum-containing medium, and after vigorous pipetting, cells were collected into Falcon 2052 (12 × 75 mm, Becton-Dickinson) tubes. Cells were pelleted at 400 g for five minutes without brakes, the supernatant was decanted and the pellet was loosened by vortexing at setting 4. Cells were washed with 1% BSA/PBS and repelleted. During cell resuspension, 1 ml ice-cold 70% Ethanol was added, in which cells were either stored at -20°C for up to three days, or processed after 20 minute fixation by repelleting. Resuspension was done in 1 ml 2 N HCl/0.5% Triton X-100, and samples were incubated for 30 minutes at room temperature with vortexing every five minutes, followed by repelleting. After removal of the supernatant, nuclei were resuspended in 1 ml 0.1 M Na2B4O7 (pH 8.5) for two minutes at room temperature, and washed with 1 ml 0.5% Tween-20/1% BSA/PBS. Then, 20 μl FITC-coupled anti-BrdU antibody (Becton-Dickinson) was added, followed by 30 minute incubation at room temperature in the dark. After another wash with 1 ml 0.5% Tween-20/1% BSA/PBS, nuclei were resuspended in 0.25 ml PBS containing 5 μg/ml Propidium Iodide, filtered through 1.2 ml Costar cluster tubes (Costar #4410), or 6 ml tubes with 35 μm strainer cap (Falcon 2235, Becton-Dickinson) in case of volumes greater than 0.5 ml. Flow cytometry was done on a FACScan (Becton-Dickinson) and results were analyzed using FlowJo software (TreeStar).
Gene expression studies
To measure gene expression quantitatively, RNA was extracted from primary rib chondrocytes as follows: cells were harvested as described above, rinsed in PBS and kept in Trizol at -80°C. Extracts were thawed, homogenized in a PowerGen 125 homogenizer for 1 minute, centrifuged at 2000 rpm for 30 seconds. The supernatant was transferred to fresh tubes, 20% of volume of chloroform was added, and the sample was vortexed for 20 seconds, followed by centrifugation at 12000 rpm for 5 minutes at 4°C. The top layer was removed to fresh tubes for precipitation with 0.5 ml Isopropyl Alcohol, and centrifuged at 12000 rpm for 10 minutes at 4°C. The supernatant was removed and the pellet was washed three times with 75% EtOH. After final removal of supernatant, the pellet was air dried for 1–2 minutes and dissolved in nanopure H2O by gentle pipetting and, where necessary, by incubation at 37°C for 10 minutes. RNA concentration was measured by spectrophotometry. Reverse transcription of RNA was performed using the Superscript II first strand synthesis system for RT-PCR (Gibco BRL), following the supplier's instructions. Purification of cDNA was done using the Qiagen PCR Purification Kit, and cDNA concentration was measured by spectrophotometry. Primers for amplification were designed using Primer Express software (ABI) with the following parameters: TM requirements: min. TM 58°C, max. TM 60°C, optimal TM 59°C; GC content requirements: min. % GC 20, max.% GC 80; length requirements: min. length 9, max length 40, optimal length 20; amplicon requirements: min. TM 0°C, max. TM 85°C, min. length 50, max. length 150. The following primer pairs (Genbank accession numbers and coordinates on the mRNA sequence given in brackets) were used to amplify the specific cDNAs for
Bcl-2 (NM_009741); forward primer: 5'-CGGAGACGAGTTCAACGAAAC-3'(681–701); reverse primer: 5'-TGTAAGATAACCATTTGAGGGTGG-3'(770–747).
Bone Morphogenetic Protein-2 (Bmp-2; NM_007553); forward primer: 5'-CCTCAAGTCCAGCTGCAAGAG-3'(1223–1243); reverse primer: 5'-GGTGCCACGATCCAGTCATT-3'(1300–1281).
Bone Morphogenetic Protein-4 (Bmp-4; D14814); forward primer: 5'-GCACTGCCGCAGCTTCTC-3'(5630–5647); reverse primer: 5'-CACTGACAGAAAACAAGGCATATAATAA-3'(5727–5700).
Collagen II splice form A (NM_001844); forward primer: 5'-AATGGGCAGAGGTATAAAGATAAGGA-3'(24–49); reverse primer: 5'-CATTCCCAGTGTCACACACACA-3'(99–78).
Collagen X (X67348); forward primer: 5'-CAAACGGCCTCTACTCCTCTGA-3'(1937–1958); reverse primer: 5'-CGATGGAATTGGGTGGAAAG-3'(2065–2046).
Dlx 5 (NM_01156); forward primer: 5'-ACGCGCGGAGTTGGC-3'(437–451); reverse primer: 5'-CTTGATCTTGGATCTTTTGTTCTGAA-3'(518–493)
Fibroblast growth factor receptor 3 (Fgfr3; NM_008010); forward primer: 5'-GAGTCTACACCCACCAGAGTGATGT-3'(2200–2224); reverse primer: 5'-AGCCCCCCAGCGTAAAGAT-3'(2271–2253).
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Xm_194302) forward primer: 5'-CCAGAACATCATCCCTGCATC-3'(674–694); reverse primer: 5'-GGTAGGAACACGGAAGGCC-3'(794–776)
Hoxc-8 (NM_010466); forward primer: 5'-CAACACTAACAGTAGCGAAGGACAAG-3' (595–620); reverse primer: 5'-CAAGGTCTGATACCGGCTGTAAGT-3' (727–704).
Indian hedgehog (Ihh; U85610); forward primer: 5'-CCCCAACTACAATCCCGACA-3'(543–562); reverse primer: 5'-TCATGAGGCGGTCGGC-3'(607–592).
p107 (U27177); forward primer: 5'-TGGATTATTGAAGTTCTCGATTTGC-3'(1624–1648); reverse primer: 5'-ATGCTGTTCAGATGTTTCACCATG-3'(1736–1713)
Parathyroid Hormone/Parathyroid hormone related peptide Receptor (PTH/PTHrP-R; L28108); forward primer: 5'-GAAAGAATAAAGCAAAAGCGAGACA-3'(232–256); reverse primer: 5'-AGGGAGCTCTGACATCGGG-3'(299–281).
Sox-5 (NM 011444); forward primer: 5'-ATGGTGTGGGCGAAAGATGA-3'(1802–1821); reverse primer: 5'-GGCGGGCCTGCTCCT-3'(1946–1932).
All real-time PCR reactions were performed in triplicate on an ABI Prism 7000 in SYBR Green Master Mix (Applied Biosystems), with denaturation at 94°C for 15 seconds, annealing at 55°C for one minute and extension at 60°C for one minute in a total of 40 cycles. 100 nM of each primer were used and 10 ng of template cDNA. Measurements were done in triplicate for each sample, and for each sample, the cycle number (Ct) was determined at which signal above threshold was detected. The values were averaged and standardized to measurements for GAPDH cDNA in the same sample by subtraction: CtGENE - CtGAPDH = ΔCt. This value for each gene in transgenic samples was then normalized for the value of the same gene in the control sample by a second subtraction: ΔCttransgenic - ΔCtcontrol = ΔΔCt. The value for "fold expression" relative to the control is obtained by the formula f = 2ΔΔCt. The resulting data were expressed as "fold change" relative to the gene expression level in the control which was prepared from an FVB mouse.
Acknowledgments
Acknowledgements
SAC performed the in vivo BrdU labeling studies and FACS analyses, MAM did the analyses involving micromass cultures, and CK conceived and supervised the studies, wrote the manuscript and secured funding from the Arthritis Foundation (to C.K.), the Aircast Foundation (to C.K.), NIH-F32AR008545 (to S.C.), NIH-NIAMS RO3-AR44945 (to C.K.), and NIH-NIDCR R21-DE014523-01 (to C.K.). We thank Diane Costanzo, Lauren Greenlee, Jean Kloss, Kristin Smith, Catherine Talmadge, Teresa Tinder and Hope West for technical assistance, M. Anita Jennings for histology, Jill Martin for cell sorting, and Saralyn Fisher for secretarial services. We also are grateful to Drs. Veronica Lefebvre, Benoit de Crombrugghe, H.K. (Tina) Dinh and J. Michael Salbaum for technical advice and discussions.
Contributor Information
Stephania A Cormier, Email: cormier.stephania@mayo.edu.
Maria Alice Mello, Email: mellom@ep.niams.nih.gov.
Claudia Kappen, Email: ckappen@unmc.edu.
References
- Yueh YG, Gardner DP, Kappen C. Evidence for regulation of cartilage differentiation by the homeobox gene Hoxc-8. Proc Natl Acad Sci USA. 1998;95:9956–9961. doi: 10.1073/pnas.95.17.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancedda R, Descalzi-Cancedda F, Castagnola P. Chondrocyte differentiation. Int Rev Cytol. 1995;159:265–358. doi: 10.1016/s0074-7696(08)62109-9. [DOI] [PubMed] [Google Scholar]
- Hickok NJ, Haas AR, Tuan RS. Regulation of chondrocyte differentiation and maturation. Microsc Res Techn. 1998;43:174–190. doi: 10.1002/(SICI)1097-0029(19981015)43:2<174::AID-JEMT9>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13:721–727. doi: 10.1016/S0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Olsen BR. Heritable diseases of the skeleton. Part I: Molecular insights into skeletal development-transcription factors and signaling pathways. FASEB J. 1997;11:125–132. doi: 10.1096/fasebj.11.2.9039954. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Olsen BR. Heritable diseases of the skeleton. Part II: Molecular insights into skeletal development-matrix components and their homeostasis. FASEB J. 1997;11:227–233. [PubMed] [Google Scholar]
- Capecchi MR. Function of Homeobox Genes in Skeletal Development. Ann NY Acad Sci. 1996;785:34–37. doi: 10.1111/j.1749-6632.1996.tb56241.x. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Scott MP. Downstream of the homeotic genes. New Biologist. 1992;4:5–15. [PubMed] [Google Scholar]
- Muller YL, Yueh YG, Yaworsky PJ, Salbaum JM, Kappen C. Caudal dysgenesis in Isl-1 in transgenic mice. FASEB J. 2003. [DOI] [PMC free article] [PubMed]
- Cottrill CP, Archer CW, Wolpert L. Cell sorting and chondrogenic aggregate formation in micromass culture. Dev Biol. 1987;122:503–515. doi: 10.1016/0012-1606(87)90314-9. [DOI] [PubMed] [Google Scholar]
- Mello MA, Tuan RS. High density micromass cultures of embryonic limb bud mesenchymal cells: an in vitro model of endochondral skeletal development. In vitro Cell Dev Biol. 1999;35:262–269. doi: 10.1007/s11626-999-0070-0. [DOI] [PubMed] [Google Scholar]
- Ahrens PB, Solursh M, Reiter RS. Stage-related capacity for limb chondrogenesis in cell culture. Dev Biol. 1977;60:69–82. doi: 10.1016/0012-1606(77)90110-5. [DOI] [PubMed] [Google Scholar]
- DeLise AM, Stringa E, Woodward WA, Mello MA, Tuan RS. Embryonic limb mesenchyme micromass culture as an in vitro model for chondrogenesis and cartilage maturation. Methods Mol Biol. 2000;137:359–375. doi: 10.1385/1-59259-066-7:359. [DOI] [PubMed] [Google Scholar]
- Kameda T, Koike C, Saitoh K, Kuroiwa A, Iba H. Analysis of cartilage maturation using micromass cultures of primary chondrocytes. Dev Growth Differ. 2000;42:229–236. doi: 10.1046/j.1440-169x.2000.00508.x. [DOI] [PubMed] [Google Scholar]
- Farnum CE, Wilsman NJ. Histochemical evidence of DNA fragmentation characteristic of apoptosis in hypertrophic chondrocytes. Trans Orthop Res Soc. 1995;20:77. [Google Scholar]
- Hatori M, Klatte KJ, Teixeira CC, Shapiro IM. End labeling studies of fragmented DNA in the avian growth plate: evidence of apoptosis in terminally differentiated chondrocytes. J Bone Miner Res. 1995;10:1960–1968. doi: 10.1002/jbmr.5650101216. [DOI] [PubMed] [Google Scholar]
- Zenmyo M, Komiya S, Kawabata R, Sasaguri Y, Inoue A, Morimatsu M. Morphological and biochemical evidence for apoptosis in the terminal hypertrophic chondrocytes of the growth plate. J Pathol. 1996;180:430–433. doi: 10.1002/(SICI)1096-9896(199612)180:4<430::AID-PATH691>3.3.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Farnum CE, Wilsman NJ. Determination of proliferative characteristics of growth plate chondrocytes by labeling with bromodeoxyuridine. Calc Tissue Int. 1993;52:110–119. doi: 10.1007/BF00308319. [DOI] [PubMed] [Google Scholar]
- Wilsman NJ, Farnum CE, Green EM, Lieferman EM, Clayton MK. Cell cycle analysis of proliferative zone chondrocytes in growth plates elongating at different rates. J Orthop Res. 1996;14:562–572. doi: 10.1002/jor.1100140410. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, De Crombrugghe B. Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 1994;14:329–335. doi: 10.1016/0945-053X(94)90199-6. [DOI] [PubMed] [Google Scholar]
- Giaretti W, Moro G, Quarto R, Bruno S, Di Vinci A, Geido E, Cancedda R. Flow cytometric evaluation of cell cycle characteristics during in vitro differentiation of chick embryo chondrocytes. Cytometry. 1988;9:281–290. doi: 10.1002/cyto.990090403. [DOI] [PubMed] [Google Scholar]
- Favaretto AL, Lins CE, Felipe MS, Da Cruz WB. [Characterization of a chondrocyte primary culture from rib cartilage of the rat] Rev Bras Biol. 1989;49:731–736. [PubMed] [Google Scholar]
- Rundle CH, Macias MP, Gardner DP, Yueh YG, Kappen C. Transactivation of Hox Gene Expression in a VP16-Dependent Binary Transgenic Mouse System. Biochim Biophys Acta. 1998;1398:164–178. doi: 10.1016/S0167-4781(98)00046-3. [DOI] [PubMed] [Google Scholar]
- Sandell LJ, Morris N, Robbins JR, Goldring MB. Alternatively spliced type II procollagen mRNAs define distinct populations of cells during vertebral development: differential expression of the amino-propeptide. J Cell Biol. 1991;114:1307–1319. doi: 10.1083/jcb.114.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9:S69–75. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/S1534-5807(01)00003-X. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Pang MK, Zhou S, Cowan SK, Kong RY, Pfordte T, Olsen BR, Sillence DO, Tam PP, Cheah KS. Abnormal compartmentalization of cartilage matrix components in mice lacking collagen X: implications for function. J Cell Biol. 1997;136:459–471. doi: 10.1083/jcb.136.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk SW, Luvalle P, Leask T, Leboy PS. A BMP responsive transcriptional region in the chicken type X collagen gene. J Bone Miner Res. 1998;13:1521–1529. doi: 10.1359/jbmr.1998.13.10.1521. [DOI] [PubMed] [Google Scholar]
- Enomoto-Iwamoto M, Iwamoto M, Mukudai Y, Kawakami Y, Nohno T, Higuchi Y, Takemoto S, Ohuchi H, Noji S, Kurisu K. Bone morphogenetic protein signaling is required for maintenance of differentiated phenotype, control of proliferation, and hypertrophy in chondrocytes. J Cell Biol. 1998;140:409–418. doi: 10.1083/jcb.140.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp A. Interaction of growth factors regulating chondrocyte differentiation in the developing embryo. Osteoarthritis Cartilage. 2001;9:S109–17. [PubMed] [Google Scholar]
- Minina E, Wenzel HM, Kreschel C, Karp S, Gaffield W, McMahon AP, Vortkamp A. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development. 2001;128:4523–4534. doi: 10.1242/dev.128.22.4523. [DOI] [PubMed] [Google Scholar]
- Karp SJ, Schipani E, St-Jacques B, Hunzelman J, Kronenberg H, McMahon AP. Indian hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development. 2000;127:543–548. doi: 10.1242/dev.127.3.543. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüppner H. Role of parathyroid hormone-related peptide and Indian hedgehog in skeletal development. Pediatr Nephrol. 2000;14:606–611. doi: 10.1007/s004670000343. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM, Lanske B, Kovacs CS, Chung UI, Lee K, Segre GV, Schipani E, Jüppner H. Functional analysis of the PTH/PTHrP network of ligands and receptors. Rec Progr Horm Res. 1998;53:283–301. [PubMed] [Google Scholar]
- Chung UI, Lanske B, Lee K, Li E, Kronenberg H. The parathyroid hormone/parathyroid hormone-related peptide receptor coordinates endochondral bone development by directly controlling chondrocyte differentiation. Proc Natl Acadf Sci USA. 1998;95:13030–13035. doi: 10.1073/pnas.95.22.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Boorla S, Frendo JL, Hogan BL, Karsenty G. Skeletal abnormalities in doubly heterozygous Bmp4 and Bmp7 mice. Dev Genet. 1998;22:340–348. doi: 10.1002/(SICI)1520-6408(1998)22:4<340::AID-DVG4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Rossi F, MacLean HE, Yuan W, Francis RO, Semenova E, Lin CS, Kronenberg HM, Cobrinik D. p107 and p130 Coordinately regulate proliferation, Cbfa1 expression, and hypertrophic differentiation during endochondral bone development. Dev Biol. 2002;247:271–285. doi: 10.1006/dbio.2002.0691. [DOI] [PubMed] [Google Scholar]
- Beier F, Leask TA HS, Chow C, Taylor AC, Lee RJ, Pestell RG, Ballock RT, LuValle P. Cell cycle control genes in chondrocyte proliferation and differentiation. Matrix Biol. 1999;18:109–120. doi: 10.1016/S0945-053X(99)00009-8. [DOI] [PubMed] [Google Scholar]
- Laplantine E, Rossi F, Sahni M, Basilico C, Cobrinik D. FGF signaling targets the pRb-related p107 and p130 proteins to induce chondrocyte growth arrest. J Cell Biol. 2002;158:741–50. doi: 10.1083/jcb.200205025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskey AL, Stiner D, Doty SB, Binderman I, Leboy P. Studies of mineralization in tissue culture: optimal conditions for cartilage calcification. Bone Miner. 1992;16:11–36. doi: 10.1016/0169-6009(92)90819-Y. [DOI] [PubMed] [Google Scholar]
- Lev R, Spicer S. Specific Staining of sulfate groups with alcian blue at low pH. J Histochem Cytochem. 1964;12:309. doi: 10.1177/12.4.309. [DOI] [PubMed] [Google Scholar]
- Kappen C. The VP16-dependent binary system for inducible gene expression in transgenic mice. In: John Wiley & Sons, editor. Genetic manipulation of receptor expression and function. Accili D; 1999. pp. 69–92. [Google Scholar]
- Gardner DP, Byrne GW, Ruddle FH, Kappen C. Spatial and temporal regulation of a LacZ reporter transgene in a binary transgenic mouse system. Transg Res. 1996;5:37–48. doi: 10.1007/BF01979920. [DOI] [PubMed] [Google Scholar]
- Yaworsky PJ, Kappen C. Heterogeneity of neural progenitor cells revealed by enhancers in the nestin gene. Dev Biol. 1999;205:309–321. doi: 10.1006/dbio.1998.9035. [DOI] [PMC free article] [PubMed] [Google Scholar]