Abstract
Construction methodologies for cDNA microarrays lack the ability to determine array integrity prior to hybridization, leaving the array itself a source of uncontrolled experimental variation. We solved this problem through development of a three-color cDNA array platform whereby printed probes are tagged with fluorescein and are compatible with Cy3 and Cy5 target labeling dyes when using confocal laser scanners possessing narrow bandwidths. Here we use this approach to: (i) develop a tracking system to monitor the printing of probe plates at predicted coordinates; (ii) define the quantity of immobilized probe necessary for quality hybridized array data to establish pre-hybridization array selection criteria; (iii) investigate factors that influence probe availability for hybridization; and (iv) explore the feasibility of hybridized data filtering using element fluorescein intensity. A direct and significant relationship (R2 = 0.73, P < 0.001) between pre-hybridization average fluorescein intensity and subsequent hybridized replicate consistency was observed, illustrating that data quality can be improved by selecting arrays that meet defined pre-hybridization criteria. Furthermore, we demonstrate that our three-color approach provides a means to filter spots possessing insufficient bound probe from hybridized data sets to further improve data quality. Collectively, this strategy will improve microarray data and increase its utility as a sensitive screening tool.
INTRODUCTION
Historically, studies to decipher genetic alterations have been limited to single genes or proteins. Since its introduction, the microarray platform, which can simultaneously provide expression profiling of thousands of genes, has become a mainstream component of biomedical research (1–8). However, this technology has drawn criticism due to its lack of reproducibility, which stems from normal biological variation (9), and technical problems in both target preparation, as well as array fabrication.
The generation of reliable gene expression data with cDNA microarrays requires construction of quality arrays. This task encompasses the generation of adequate amounts of concentrated probe and the printing of probes in a known ordered fashion onto prepared or purchased coated slides. These slides must possess: (i) low background fluorescence; (ii) high DNA retention capacity; and (iii) a uniform surface able to yield consistent spots of the desired size and shape. The printed array is subsequently prepared for hybridization after it is fixed/blocked in a series of steps, commonly termed post-processing, where unoccupied amine groups are converted to carboxylic moieties (10,11).
It has been demonstrated previously that insufficient amounts of support-bound probe result in an underestimation or failure to detect differential gene expression (12). In order to account for this source of potential data variability, as well as other array-fabrication based sources of data variability, we have developed a novel three-color cDNA array platform where arrays are directly visualized prior to hybridization (13,14). This is accomplished by spotting cDNA probes that are tagged during amplification with a third fluorescent dye (fluorescein) which is compatible with Cy3 and Cy5 target labeling dyes when using confocal laser scanners possessing narrow bandwidths. This three-color approach allows the assessment of slide fabrication independent of hybridization, and has provided our laboratory with a method for: (i) direct visualization of array/element morphology; (ii) quantification of probe deposition and retention on the slide surface, since the detected fluorescein signal is proportional to amount of probe present; and most importantly (iii) a means of quality control for arrays prior to hybridization for more reliable differential gene expression analysis. With this new approach we have observed that slides coated and printed together are not equivalent in terms of fluorescein intensity (i.e. DNA deposited and retained). Nor are the arrays equivalent in terms of background arising from solublized probe re-deposited elsewhere on the array during post-processing.
Previously, we observed a direct and significant relationship between pre-hybridization fluorescein signal to background measurements and post-hybridization replicate consistency, illustrating that microarray data quality can be improved through pre-hybridization slide selection based upon this quality parameter (13,14). In this report we directly evaluate the impact of differing amounts of support-bound probe on data quality, describe a novel probe tracking system for ascertaining proper plate order and orientation from culture growth, amplification and purification, through printing of probes onto the array, and finally, define a quality control threshold for support-bound probe that facilitates pre- hybridization array selection and provides a means for filtering hybridized data sets of unreliable elements.
MATERIALS AND METHODS
Library growth and tracking
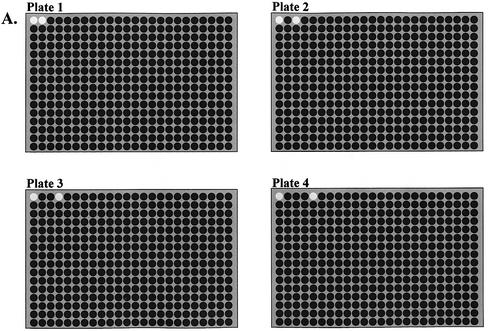
A sequence-verified human library (Research Genetics, Huntsville, AL), consisting of 41 472 clones was used as a source of probe DNA. We have opted to reformat libraries from 96- to 384-format for culture growth/archiving, PCR, purification and printing. This has reduced the number of plates of our 41 472 human clone library from 432 to a more manageable 108. The library was reformatted and subsequently manipulated using slot pin replicator tools (VP Scientific, San Diego, CA). Cultures were grown in 150 μl Terrific Broth (Sigma, St Louis, MO) supplemented with 100 mg/ml ampicillin in 384 deep-well plates (Matrix Technologies, Hudson, NH) sealed with air pore tape sheets (Qiagen, Valencia, CA) and incubated with shaking for 14–16 h. A unique asymmetric pattern of two negative controls per 384 culture plate was created by transferring the contents of the selected wells to a new 384 plate and updating the clone tracking database accordingly. The plate-specific negative control pattern was created by removing position A1 (to establish an orientation marker) and one additional plate-specific well (Fig. 1).
Figure 1.
Tracking scheme for confirmation of plate order and orientation from clone source plate to printed array. (A) Layout of asymmetric plate-specific negative controls for first four clone source plates. Position A1 of each plate is removed to serve as an orientation marker, a second negative control is used as a plate identifier. (B) 9600 element human cDNA array printed on in-house prepared poly-l-lysine-coated slide using 16 pins. Subarrays generated by each pin are labeled. Subarray (pin) 1 possesses position A1 from each source plate. The position A1 negative controls result in the absence of 24/25 elements in the first (far right) column; a laboratory plate orientation error will introduce fluorescent elements into this column. Subarray (pin) 9 (enlarged) shows a correct series of negative controls for indicated plates; other probe plates are represented in other subarrays. Improper order management of any plate at any point during array construction will disrupt this pattern. Note: observable pin clogging problem on pin 2.
Clone inserts were amplified in duplicate in 384-well format from 0.5 μl bacterial culture diluted 1:8 in sterile distilled water or from 0.5 μl purified plasmid (controls only) using 0.26 µM of each vector primer [SK865 5′-fluorescein-GTC CGT ATG TTG TGT GGA A-3′ and SK536: 5′-fluorescein-GCG AAA GGG GGA TGT GCT G-3′] (12) (Integrated DNA Technologies, Coralville, IA) in a 20 µl reaction consisting of 10 mM Tris–HCl pH 8.3, 3.0 mM MgCl2, 50 mM KCl, 0.2 mM each dNTP (Amersham, Piscataway, NJ), 1 M betaine (15,16) and 0.50 U Taq polymerase (Roche, Indianapolis, IN). Reactions were amplified with a touchdown thermal profile consisting of 94°C for 5 min; 20 cycles of 94°C for 1 min, 60°C for 1 min (–0.5°C per cycle), 72°C for 1 min; and 15 cycles of 94°C for 5 min; 20 cycles 94°C for 1 min, 55°C for 1 min, 72°C for 1 min; terminated with a 7 min hold at 72°C (17–19). PCRs analyzed for single products by 1% agarose gel electrophoresis analysis. Products from replicate plates were pooled and then purified by size exclusion filtration using the Multiscreen 384 PCR filter plates (Millipore, Bedford, MA) to remove unincorporated primer and PCR components. Forty wells of each 384-well probe plate were quantified by the PicoGreen assay (Molecular Probes, Eugene, OR) according to the manufacturers instructions. After quantification, all plates were dried down, and reconstituted at 150 ng/µl in 3% DMSO/1.5 M betaine.
Array fabrication
A single printing of 200 arrays, each possessing 9600 elements, was printed on 100 poly-l-lysine coated slides prepared in-house (2 arrays/slide) as described previously (10). Printing was conducted with a GeneMachines Omni Grid printer (San Carlos, CA) with 16 Telechem International SMP3 pins (Sunnyvale, CA) at 40% humidity and 22°C. To control pin contact force and duration, the instrument was set with the following Z motion parameters: velocity, 7 cm/s; acceleration, 100 cm/s2; deceleration, 100 cm/s2. All slides were post-processed using the previously described non-aqueous protocol (11). Slide coating, isolation of mRNA, labeling and hybridization were performed as described previously (http://cmgm.stanford.edu/pbrown/mguide/index.html). A second set of 9600 element arrays were printed on 15 different vendor-supplied coated slides (Apogent Discoveries, Waltham, MA; Asper Biotech, Redwood City, CA; Bioslide Technologies, Walnut, CA; Corning Inc., Corning, NY; Erie Scientific, Portsmouth, NH; Genetix, St James, NY; Sigma; Telechem International Inc.; Cel-Associates, Pearland, TX; Electron Microscopy Sciences, Fort Washington, PA; Polysciences Inc., Warrington, PA; Full Moon Biosystems, Sunnyvale, CA) in an attempt to identify a higher retention surface compatible with our printing and blocking protocols. Higher retention was not observed, however, the resulting low retention arrays were used to study the impact of low amounts of support-bound probe on data variability. Image files on all arrays were collected after blocking (fluorescein), and again after hybridization (Cy3 and Cy5) with a ScanArray 5000 (GSI Lumonics, Billerica, MA). Array image files were analyzed with the Matarray software (20).
RESULTS AND DISCUSSION
Tracking and array fabrication
Quality array construction requires the generation of adequate amounts of concentrated probe, and the subsequent printing probes in a known and ordered fashion onto coated glass slides. A highly optimized touchdown PCR protocol has been developed whereby 1–2 μg purified probe material is recovered from two pooled and purified 20 μl PCRs. Duplicate reactions compensate for random PCR failures, enabling overall PCR success rates, based upon gel analysis, of ∼90%. We have found that recovery of >1 μg purified probe is sufficient for printing >2000 arrays/amplification (assuming: 4 μl plate dead volume, printing at 150 ng/μl concentration, and 250 nl/pick up/100 slides using the TeleChem SMP3 pins).
The fact that the fluoresceinated array is visible prior to hybridization allows for spots that are not present on the array due to PCR failure or mechanical problems (clogged or sticking pin) to be tracked, eliminating a potential source of error between replicate slides and enables differentiation between true-negatives and false-negatives. This has lead to the development of a tracking system, which utilizes a unique pattern of negative controls for each clone source plate enabling a means to assess that all plates have had order and orientation maintained from the clone source plate through growth, PCR, pooling, purification and finally printing (Fig. 1).
Factors affecting amount of support probe
A number of critical parameters, including DNA concentration, printing buffer, slide surface, temperature, humidity and print head velocity can influence the amount of DNA deposited, retained and ultimately available for hybridization on the slide surface (11,12,21). We have found that 1.5 M betaine/3% DMSO offers the best retention under the conditions described in the Materials and Methods section (13,14).
To further investigate the parameters affecting the amount of probe deposited and retained on the slide surface, we evaluated slide placement on the arrayer deck as a potential variable. Two hundred human cDNA arrays were printed onto 100 in-house prepared poly-l-lysine slides (two consecutively printed arrays per slide, each array possessed 9600 elements). Each probe was printed 200 times from a single pin loading, in other words, 200 spots/probe were printed without returning to the probe source plate to refill the quill pin. We observed considerable variation in average array spot intensity ranging from a high of 19 107 RFU/pixel to a low of 1514 RFU/pixel (Fig. 2) that paralleled the slide printing order (R2 = 0.84). The first 100 arrays (slides 1–50) had an average fluorescein intensity per element of 11 775 ± 4354 versus 4248 ± 1237 RFU/pixel for the second 100 arrays (slides 51–100). These data indicate that arrays possessing the most support-bound DNA are those that are printed first when presumably the quill pins are at their fullest, and as the print head works across the slides on the arrayer deck, the pins become more empty, consequently, less DNA is deposited and ultimately retained on the slide surface. We investigated this phenomenon for additional printings (100 arrays/print run, two possible array formats: either a single 10 000 element or a single 20 000 element array/slide) where standardized post-blocking fluorescein images were collected for all arrays and a single lot of coated slides was used per print run. We observed an average decrease of nearly 20% in the mean fluorescein spot intensity between the first and last 20 arrays (15 415 ± 8040 versus 12 572 ± 5793 RFU/pixel).
Figure 2.
DNA retention studies using 200 human 9600 probe cDNA arrays printed over 100 poly-l-lysine slides prepared in-house. Plotted is post-blocking DNA available for hybridization (average spot fluorescein intensity RFU/pixel for each array) as a function of position on arrayer deck during printing. Linear regression line is plotted (R2 = 0.84). Slides utilized in the homotypic hybridizations illustrated in Figure 3 are indicated in red.
Impact of limiting bound probe on data quality and data filtering using the pre-hybridization fluorescein image
It is known from previous studies that limiting support-bound probe will ultimately compromise detection of differential expression (12), however, the actual amount of support-bound probe was unknown. Using tagged probes, it is possible to measure support-bound DNA based upon fluorescein intensity, and therefore define levels that may actually compromise data quality. Using the set of 9600 probe human cDNA arrays printed on vendor-supplied coated slides, as well as those printed on in-house coated slides (those plotted in Fig. 2), we set out to establish a general guideline as to how much DNA is needed per element (i.e. fluorescein intensity RFU/pixel value). This would enable the future identification of those arrays possessing insufficient bound probe, which when used as replicates would introduce experimental variability. We approached this question through the use of homotypic (self-self) hybridizations utilizing RNA extracted from cell line UACC903 and heterotypic co-hybridizations utilizing RNA extracted from UACC903 and Jurkat cells.
Homotypic experiments are useful for measuring microarray data variability since systematic noise will lead to ratio deviation from the expected value of 1. Therefore, we performed homotypic hybridizations on 9600 probe human cDNA arrays of differing average fluorescein intensities (see Fig. 2). Arrays 11/12, 31/32, 51/52, 67/68, 95/96, 129/130, 149/150, 169/170 and 189/190, (slides 6, 16, 26, 34, 48, 65, 75, 85 and 95) possessing average element fluorescein intensities after post processing of 14 692 ± 5494, 12 614 ± 4390, 13 655 ± 4274, 11 798 ± 4766, 5183 ± 1814, 5255 ± 1556, 4592 ± 1662, 4659 ± 1430 and 4676 ± 1459 RFU/pixel, respectively (see Fig. 2), were randomly selected for analysis to represent a wide range of array intensities. Arrays were then hybridized to Cy3- and Cy5-labeled UACC903 RNA. The potential variation between the multiple labeling reactions required for this experiment was normalized by pooling all the labeled targets prior to distribution over the arrays. The resulting hybridized image data was analyzed with Matarray, which employs algorithms to define quality scores for each spot on the array according to five criteria: size, signal-to-noise ratio, background level, background uniformity, and saturation (20). Based on these five scores, a composite score (qcom) is defined for each spot to give an overall assessment of its quality. Previously, we have demonstrated that the inherent variability in intensity ratio measurements correlates closely with qcom, in that high quality spots generate less variability, and therefore, removing spots with low qcom can dramatically improve the reliability of hybridization data as reflected by higher correlation coefficients between duplicate slides/spots (20). In this study, it was our objective to investigate data variability inherent to the slide itself, not variability introduced through hybridization. Therefore, all hybridized data was filtered, removing 30% of spots possessing the lowest qcom. We then calculated the standard deviation of the log ratio distribution and plotted it as a function of the rank order of printing (Fig. 3A). As the amount of support-bound probes decreases, noise increases resulting in a greater ratio distribution (Fig. 3A). The data variability due to limiting bound probe becomes more apparent when the array average fluorescein spot intensity drops to ≤5000 RFU/pixel. This observation is further supported by standard deviations of the log ratio distribution exceeding 0.35 on three of the last five arrays in the homotypic series shown in Figure 3A.
Figure 3.
Analysis of homotypic hybridizations. Labeled UACC903 cDNA hybridized 9600 element human cDNA arrays shown in Figure 2: array numbers 11/12, 31/32, 51/52, 67/68, 95/96, 129/130, 149/150, 169/170 and 189/190, (slides 6, 16, 26, 34, 48, 65, 75, 85 and 95) possessing average element fluorescein intensities after post-processing of 14 692 ± 5494, 12 614 ± 4390, 13 655 ± 4274, 11 798 ± 4766, 5183 ± 1814, 5255 ± 1556, 4592 ± 1662, 4659 ± 1430 and 4676 ± 1459 RFU/pixel, respectively. (A) Analysis of inter-slide variance: the standard deviation of the log ratio distribution plotted (y-axis) as a function of array average fluorescein spot intensity (x-axis). As the amount of bound probes decreases, noise increases resulting in a greater ratio distribution. Linear regression line is plotted. (B) Intra-slide variance: analysis of arrays 31/32, 95/96, 189/190 (slides 16, 48 and 95; top to bottom) where elements on each slide were separated into 10 groups (∼2000 spots/group) according to the rank of their fluorescein intensities. The standard deviation of the log ratio distribution for each subgroup of spots is plotted as a function of array average fluorescein intensity.
Inter-slide variation in the amount of support-bound probe is influenced by surface chemistry, slide rank order within the print run as well as other variables, however, there also exists intra-slide variation which can arise from differences in PCR quality and mechanical problems during printing (for example, see pin 2 in Fig. 1B). Since all spots, even on good arrays are not created equal, we investigated if outlying data points in homotypic hybridizations could be correlated with spot fluorescein intensity, and therefore arrays 31/32, 95/96, 189/190 (slides 16, 48 and 95) were analyzed further. Elements on each slide were separated into 10 groups (of 1920 spots/group) according to the rank of their fluorescein intensities, from low to high and the standard deviation of the log ratios was determined for each group (Fig. 3B). We observe that elements possessing higher fluorescein intensity (more bound probe) generate less variable ratio measurements. Again the amount of bound probe per element only appears to impact data quality when it drops below a spot average fluorescein intensity of ∼5000 RFU/pixel, while spots above this threshold generate equally good data. We have therefore adopted this value as a pre-hybridization quality control threshold for deeming an array suitable for use in an experiment. When printing on poly-l-lysine slides prepared in-house (1 array/slide, 100 slides/print run) we find that the majority (>80%) of arrays have an average spot intensity value after post-processing greater than 5000, as is observed for arrays 1–100 illustrated in Figure 2. The observation that data variance correlates with amount of support-bound probe supports the idea that spot intensity scores derived from the pre-hybridization fluorescein image may be useful for filtering spots that are likely to give rise to highly variable data from hybridization data.
To investigate the impact of support-bound probe on heterotypic hybridization replicate consistency and further explore the possibility of data filtering using the third dye, 80 arrays possessing a wide range of bound probe were hybridized to Cy5-labeled Jurkat and Cy3-labeled UACC903 cDNAs. Again, the potential variation between the multiple labeling reactions required for this experiment was normalized by pooling all the labeled targets prior to distribution over the arrays. All hybridized arrays possessed a post-processing fluorescein signal-to-noise value (signal/signal + noise) >0.85. Hybridized image analysis was again conducted with Matarray and spots possessing qcom scores within the lowest 30% were filtered. Correlation coefficients, using the differentially expressed genes constituting the 5% tails of the distribution, were then generated through comparison of each hybridized array to a hypothetical benchmark. Benchmarks were created for each print run [9600 probe human cDNA arrays printed on vendor supplied surfaces (n = 37 hybridized); and 9600 probe human cDNA arrays printed on in-house coated poly-l-lysine slides (n = 43 hybridized)] by first ranking arrays in terms of pre-hybridization average spot fluorescein RFU/pixel and signal-to-noise values (signal/signal + noise), as well as ranking hybridized arrays in terms of pair-wise correlation. Arrays within both the top five for pre-hybridization quality control ranking as well as within the top five for the post-hybridization pair-wise ranking were averaged to create the benchmark. Three arrays were found that met these criteria for each of the two print runs. The results of this analysis are plotted in Figure 4A and illustrates how pair-wise Pearson’s correlation coefficients increase as a function of average array support-bound probe. This relationship seems to plateau at an average array element fluorescein intensity of 5000 RFU/pixel. Once above this threshold intensity it appears possible to generate equally good data quality. We studied this relationship by fitting the data in Figure 4A with the following model:
Figure 4.
(Previous page) (A) Relationship between bound probe and replicate consistency. Red squares: arrays printed on in-house poly-l-lysine-coated slides. Black squares: arrays printed on vendor-supplied surfaces. All slides used possessed a signal-to-noise (signal/signal + noise) score >0.85. A correlation coefficient, using differentially expressed genes of the 5% distribution tails, was generated for each slide through comparison to a composite benchmark slide. A benchmark was created for each print run: 9600 probe human cDNA arrays printed on 15 vendor supplied surfaces (n = 37 hybridized); and 9600 probe human cDNA arrays printed on in-house coated poly-l-lysine slides (n = 43 hybridized). Benchmarks were constructed using hybridized arrays within the top five for fluorescein RFU and signal-to-noise and within the top five for hybridized pair-wise correlation. Pair-wise Pearson’s correlation coefficients increase as a function of average array support-bound probe (blue line, coefficient of determination for the fit is R2 = 0.73, P < 0.001). (B) Increasing replicate consistency through filtering of arrays printed on in-house poly-l-lysine-coated slides using spot fluorescein intensity (red squares illustrated in A). Average array intensity (after filtering) is plotted on the x-axis, the Pearson’s correlation coefficient of the log ratio is plotted on the y-axis. Panels 1–5 compare data when filtering spots with intensities below 5000 fluorescein RFU/pixel (red) to filtering spots with intensities below 6000 fluorescein RFU/pixel (black), 4000 fluorescein RFU/pixel (green), 3000 fluorescein RFU/pixel (blue), 2000 fluorescein RFU/pixel (cyan) and 1000 fluorescein RFU/pixel (magenta), respectively. Filtering spots possessing intensities below 5000 RFU/pixel results in higher replicate consistency.
correlation coefficient = A*log(intensity)*Δ(threshold-intensity) + C + B * Δ(intensity-threshold)
where Δ(x) is the heavy-side step function that satisfies:
The blue line in Figure 4A shows the model fit, and the coefficient of determination for the fit is R2 = 0.73 (P < 0.001), using the fitted values of 1.09, 3.99 and –3.11, for the constants A, B and C, respectively.
We investigated and found no relationship between spot fluorescein intensity and hybridized qcom (data not shown), suggesting a quality spot (possessing sufficient-bound probe, and signal/signal + noise value >0.85) is necessary, but not sufficient for high quality, reproducible hybridization data. Conversely, it is possible for a spot incapable of generating reliable data to yield an acceptable image (and therefore a high qcom score) after hybridization, therefore we explored the possibility of using fluorescein intensity-based spot filtering as a means to improve data quality. This analysis utilized the hybridized, Matarray quality-filtered, data derived from the 9600 element human cDNA printed on the in-house poly-l-lysine slides described above and illustrated in Figure 4A (red squares). We began by comparing replicate consistency when filtering from arrays spots with fluorescein intensities <6000 versus <5000 RFU/pixel (Fig. 4B, 1), and observed no difference in replicate consistency (y-axis); however, change is observed in array average spot fluorescein intensity (x-axis) since the arrays filtered at 5000 RFU/pixel stringency possess more lower intensity spots, thereby lowering the average. These data indicate that spots with intensities between 5000–6000 RFU/pixel generate data equal in consistency to spots possessing intensities >6000 RFU/pixel. However, when comparing the replicate consistency observed when filtering spots with intensities <5000 RFU/pixel to filtering intensities of <4000 RFU/pixel, <3000 RFU/pixel, <2000 RFU/pixel and <1000 spots, a decrease in correlation coefficients is observed among those arrays with more moderate array average fluorescein spot intensities, as the stringency is relaxed and more lower intensity spots are included in the analysis (Fig. 4B, 2–5).
In Figure 4B this effect is not observed on arrays with high amounts of support-bound probe (intensities of >7000 RFU/pixel) since these arrays have relatively few spots of low intensity therefore, as a percentage, very few spots are filtered. The variation observed in these arrays possessing high amounts of support-bound probe is likely due to differences in hybridization, washing and image collection, illustrating the potential value of an automated hybridization instrument (all arrays in this study were manually hybridized under a glass coverslip). However, these slides with high amounts of support-bound probe do have some missing spots, and since the arrays in this analysis were printed together they, by and large, share the same missing spots. If this analysis possessed arrays from multiple printings and multiple probe preparations, correlation coefficients on arrays with high amounts of bound probe would likely be more dependent on, and benefit from, third dye filtering.
The variability of microarray data can arise from both biological and technical sources. In this report, we have shown that the array itself can be a source of considerable variability. Our three-color approach allows pre-hybridization quality assessment as well as post-hybridization data filtering. Pre-hybridization quality control-based selection can greatly reduce data variability since slides with: (i) high background due to probe solublized and redistributed over the slide surface during post-processing or (ii) low amounts of support-bound probe or (iii) high variation in spot morphology/deposition across the array can be avoided. Based on the observations described here and those in our previous report (14), we have established putative slide acceptance criteria: array mean element intensity >5000 RFU/pixel, coefficient of variation (CV) intensity <10%, mean signal-to-noise score (signal/signal + noise) >0.85, and CV of spot size <20%. On average, >80% slides of a print run will meet these criteria yielding arrays that are able to detect gene expression changes as low as 1.5-fold. On pre-selected slides, however, there are still spots that do not meet these criteria; therefore, data filtering using the third-dye can be beneficial. We believe that our novel visualization approach has broad application, improving microarray data reproducibility not only for laboratories using cDNA arrays but, potentially, those spotting oligonucleotide probes as well.
REFERENCES
- 1.Dhanasekaran S.M., Barrette,T.R., Ghosh,D., Shah,R., Varambally,S., Kurachi,K., Pienta,K.J., Rubin,M.A. and Chinnaiyan,A.M. (2001) Delineation of prognostic biomarkers in prostate cancer. Nature, 412, 822–826. [DOI] [PubMed] [Google Scholar]
- 2.Garber M.E., Troyanskaya,O.G., Schluens,K., Petersen,S., Thaesler,Z., Pacyna-Gengelbach,M., van de Rijn,M., Rosen,G.D., Perou,C.M., Whyte,R.I., Altman,R.B., Brown,P.O., Botstein,D. and Petersen,I. (2001) Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl Acad. Sci. USA, 98, 13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedenfalk I., Duggan,D., Chen,Y., Radmacher,M., Bittner,M., Simon,R., Meltzer,P., Gusterson,B., Esteller,M., Kallioniemi,O.P., Wilfond,B., Borg,A. and Trent,J. (2001) Gene-expression profiles in hereditary breast cancer. N. Engl. J. Med., 344, 539–548. [DOI] [PubMed] [Google Scholar]
- 4.Trent J.M., Stanbridge,E.J., McBride,H.L., Meese,E.U., Casey,G., Araujo,D.E., Witkowski,C.M. and Nagle,R.B. (1990) Tumorigenicity in human melanoma cell lines controlled by introduction of human chromosome 6. Science, 247, 568–571. [DOI] [PubMed] [Google Scholar]
- 5.Southern E.M., Maskos,U. and Elder,J.K. (1992) Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics, 13, 1008–1017. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T., Perou,C.M., Tibshirani,R., Aas,T., Geisler,S., Johnsen,H., Hastie,T., Eisen,M.B., van de Rijn,M., Jeffrey,S.S. et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclass with clinical implications. Proc. Natl Acad. Sci. USA, 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 8.Hegde P., Qi,R., Gaspard,R., Abernathy,K., Dharap,S., Earle-Hughes,J., Gay,C., Nwokekeh,N.U., Chen,T., Saeed,A.I., Sharov,V., Lee,N.H., Yeatman,T.J. and Quackenbush,J. (2001) Identification of tumor markers in models of human colorectal cancer using a 19,200-element complementary DNA microarray. Cancer Res., 61, 7792–7797. [PubMed] [Google Scholar]
- 9.Pritchard C.C., Hsu,L., Delrow,J. and Nelson,P.S. (2001) Project normal: defining normal variance in mouse gene expression. Proc. Natl Acad. Sci. USA, 98, 13266–13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen M. and Brown,P. (1999) DNA arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 11.Diehl F., Grahlmann,S., Beier,M. and Hoheisel,J. (2001) Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res., 29, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue H., Eastman,P.S., Wang,B.B., Minor,J., Doctolero,M.H., Nuttall,R.L., Stack,R., Becker,J.W., Montgomery,J.R., Vainer,M. and Johnston,R. (2001) An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res., 29, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hessner M.J., Dowling,K., Kokanovic,O., Meyer,L., Nye,S.H., Wang,X., Waukau,J. and Ghosh,S. (2001) Use of fluoroscein-labeled probes as a quality control tool for cDNA microarrays. Am. J. Hum. Genet., 69 (Suppl.), 468. [Google Scholar]
- 14.Hessner M.J., Wang,X., Hulse,K., Meyer,L., Wu,Y., Nye,S.H., Guo,S.-W. and Ghosh,S. (2002) Three color cDNA microarrays: quantitative assessment through the use of fluoroscein-labeled probes. Nucleic Acids Res., 31, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henke W., Herdel,K., Jung,K., Schnorr,D. and Loening,S. (1997) Betaine improves the PCR amplification of GC-rich sequences. Nucleic Acids Res., 25, 3957–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rees W., Yager,T., Korte,J. and Von Hippel,P. (1993) Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry, 32, 137–144. [DOI] [PubMed] [Google Scholar]
- 17.Don R.H., Cox,P.T., Wainwright,B.J., Baker,K. and Mattick,J.S. (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res., 19, 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecker K.H. and Roux,K.H. (1996) High and low annealing temperatures increase both specificity and yield in touchdown and stepdown PCR. Biotechniques, 20, 478–485. [DOI] [PubMed] [Google Scholar]
- 19.Roux K.H. and Hecker,K.H. (1997) One-step optimization using touchdown and stepdown PCR. Methods Mol. Biol., 67, 39–45. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Ghosh,S. and Guo,S.-W. (2001) Quantitative quality control in microarray image processing and data acquisition. Nucleic Acids Res., 29, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegde P., Qi,R., Abernathy,K., Gay,C., Dharap,S., Gaspard,R., Hughes,J.E., Snesrud,E., Lee,N. and Quackenbush,J. (2000) A concise guide to cDNA microarray analysis. Biotechniques, 29, 548–550, 552,–554, 556. [DOI] [PubMed] [Google Scholar]