Abstract
60S and 40S ribosomal subunits are assembled in the nucleolus and exported from the nucleus to the cytoplasm independently of each other. We show that in vertebrate cells, transport of both subunits requires the export receptor CRM1 and Ran·GTP. Export of 60S subunits is coupled with that of the nucleo- cytoplasmic shuttling protein NMD3. Human NMD3 (hNMD3) contains a CRM-1-dependent leucine-rich nuclear export signal (NES) and a complex, dispersed nuclear localization signal (NLS), the basic region of which is also required for nucleolar accumulation. When present in Xenopus oocytes, both wild-type and export-defective mutant hNMD3 proteins bind to newly made nuclear 60S pre-export particles at a late step of subunit maturation. The export-defective hNMD3, but not the wild-type protein, inhibits export of 60S subunits from oocyte nuclei. These results indicate that the NES mutant protein competes with endogenous wild-type frog NMD3 for binding to nascent 60S subunits, thereby preventing their export. We propose that NMD3 acts as an adaptor for CRM1–Ran·GTP-mediated 60S subunit export, by a mechanism that is conserved from vertebrates to yeast.
Keywords: NMD3/nuclear transport/nucleolus/ribosome/60S subunit
Introduction
Biogenesis of eukaryotic ribosomes occurs primarily within the nucleolus and requires numerous trans-acting factors that modify and process the primary rRNA transcript and promote ribosome assembly (Hadjiolov, 1985; Warner, 1990; Maxwell and Fournier, 1995; Olson et al., 2000; Leary and Huang, 2001; Fatica and Tollervey, 2002). In vertebrates, the mature 40S and 60S subunits contain, respectively, 18S rRNA and 28S, 5.8S and 5S rRNAs plus a total of ∼80 ribosomal proteins (r-proteins) (Wool et al., 1995). The major steps of rRNA maturation have been defined (Savino and Gerbi, 1990; Sollner-Webb et al., 1996), but little is known about how the resulting subunits are exported to the cytoplasm (Bataille et al., 1990).
Active nuclear export of RNA and protein is mediated by cargo-specific export receptors that pass through the nuclear pore complexes (NPCs) embedded in the nuclear envelope (Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Macara, 2001). Most RNAs and RNPs interact only indirectly with their export receptors via bi-functional adaptor proteins that contain a cargo-binding domain and a nuclear export signal (NES), which is recognized by the export receptor (Dahlberg and Lund, 1998; Izaurralde and Adam, 1998; Nakielny and Dreyfuss, 1999). Members of the importin-β family of export receptors, such as CRM1 (Fornerod et al., 1997; Stade et al., 1997), bind their cargo (plus adaptor) only in the presence of a small G-protein complexed with GTP, Ran·GTP. After the export complex enters the cytoplasm, Ran·GTP is hydrolyzed to Ran·GDP, causing release of the cargo; the adaptors, receptors and Ran are then recycled back into the nucleus, to support continued export (Lund and Dahlberg, 2001).
Recently, Nmd3p, a protein conserved between Eukarya and Archaea (Belk et al., 1999; Ho and Johnson, 1999), was shown to be essential for export of 60S subunits in Saccharomyces cerevisiae (Ho et al., 2000; Gadal et al., 2001). Although the gene’s name suggests a role in nonsense-mediated decay (NMD) of mRNA, Nmd3p is unlikely to play a direct role in this pathway. Nmd3p is located predominantly in the cytoplasm, associated with free 60S subunits (Ho and Johnson, 1999), but also shuttles between the nucleus and cytoplasm. The NES and the nuclear localization signal (NLS) required for shuttling are located in a C-terminal domain of Nmd3p, which is absent from archaeal NMD3 homologs. NES-deficient mutants of Nmd3p are dominant-negative inhibitors of cell growth, blocking a late step in 60S subunit biogenesis (Belk et al., 1999; Ho et al., 2000). The nuclear export of Nmd3p is dependent on Crm1p (Xpo1p) function, and inactivation of this export receptor leads to accumulation of 60S subunits within the nucleus (Ho et al., 2000; Stage-Zimmermann et al., 2000; Gadal et al., 2001). These observations have led to the model that Nmd3p binds to newly made 60S subunits in the nucleus, thereby facilitating their export to the cytoplasm via the CRM1 pathway.
Here we have asked if the NMD3 protein of higher eukaryotes acts as an export adaptor for 60S subunits in the manner proposed for Nmd3p in yeast. We identified the regions of human NMD3 (hNMD3) that are important for its intracellular localization in transfected Hela cells, and determined the effects of wild-type and transport-defective hNMD3 proteins on ribosome export from Xenopus oocyte nuclei. Our results show that nuclear export of the nascent 60S subunit is coupled to export of NMD3 in vertebrate cells, and indicate that the association of NMD3 with the pre-export 60S particles allows for recognition by the export receptor CRM1.
Results
Nucleo-cytoplasmic shuttling of GFP–hNMD3
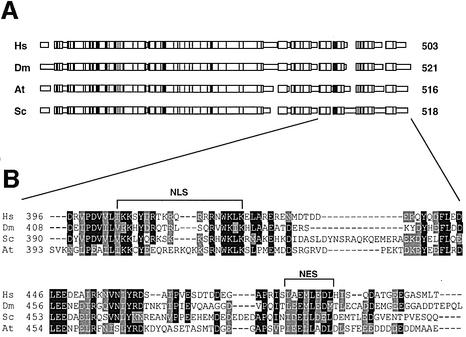
The amino acid sequences of hNMD3 and yeast Nmd3p share 47% identity (Belk et al., 1999; Ho and Johnson, 1999; Figure 1). Significantly, several regions in the C-terminal domain of hNMD3 have strong sequence and position homology to the proposed nuclear transport signals of yeast Nmd3p (yNmd3p). To determine if these conserved motifs affect the localization of NMD3, we generated chimeric genes encoding green fluorescent protein (GFP) fused to various forms of hNMD3 with wild-type or mutant C-terminal domains.
Fig. 1. NMD3 is conserved between Eukarya and Archaea. (A) Schematic diagram showing the conserved nature of the eukaryotic NMD3 proteins. This alignment was made using MACAW. Blocks of amino acid similarities are indicated by wide boxes, and positions of identity among all four sequences are indicated by vertical bars. (B) Sequence alignment of the C-terminal domains of Homo sapiens (Hs), Drosophila melanogaster (Dm), Saccharomyces cerevisiae (Sc) and Arabidopsis thaliana (At) NMD3 proteins. Conserved (black) and similar (gray) amino acid residues are indicated. NLS and NES indicate the regions resembling a basic nuclear localization signal and a leucine-rich nuclear export signal, respectively. Accession numbers are: Hs (ADD27716), Dm (AE003423.1), At (AAD24816) and Sc (S48909).
The ∼83 kDa GFP–hNMD3 chimeric protein was found in both the cytoplasm and nucleus of transiently transfected HeLa cells (Figure 2A). Because this protein is too large to enter the nucleus by diffusion, it must contain an NLS (see also below, Figure 6). Within the nucleus, GFP–hNMD3 was present in both the nucleoplasm and the nucleoli. Upon short treatment of the transfected cells with leptomycin B (LMB), an inhibitor of CRM1-mediated nuclear export (Wolff et al., 1997; Kudo et al., 1998), the nucleoplasmic and nucleolar accumulation of the protein was strongly increased (Figure 2B), indicating that GFP–hNMD3 shuttles between the nucleus and cytoplasm and that its export is mediated by CRM1.
Fig. 2. hNMD3 is a nucleo-cytoplasmic shuttling protein. Wild-type and NES-deficient mutant GFP–hNMD3 proteins were expressed in transiently transfected HeLa cells, and their intracellular distributions were determined by direct fluorescence microscopy after 16 h of expression (A, C and E) and after further treatment of the transfected cells with leptomycin B (3 ng/ml) (+LMB) for 1.5 h (B, D and F). Bottom: comparison of the leucine-rich regions of human and yeast NMD3 proteins with the known NESs of PKI and Rev proteins. Western blot analyses using antibodies to GFP showed that more than ∼95% of the GFP was in the chimeric protein (data not shown), so fluorescence reflects the distribution of the intact fusion protein.
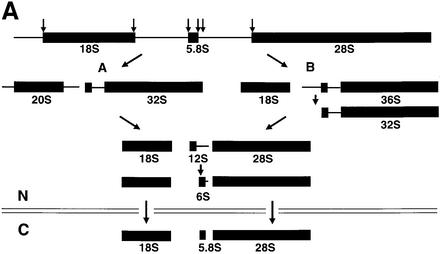
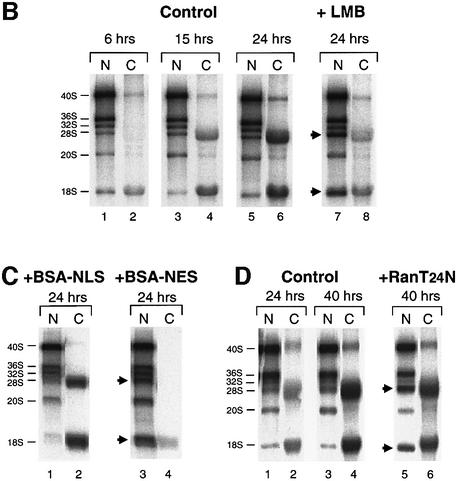
Fig. 6. Export of ribosomal subunits in Xenopus oocytes requires CRM1 and Ran·GTP. (A) Outline of the major pathways of rRNA processing in Xenopus (Savino and Gerbi, 1990), including the conversion of 12S rRNA to 6S rRNA, a precursor of 5.8S rRNA, that is matured after export to the cytoplasm (our unpublished results). (B and C) Requirement for the export receptor CRM1. (B) Oocytes were labeled with [32P]GTP at 0 h, and treatment with LMB (400 ng/ml) was initiated at 6 h. The intracellular distributions of newly made rRNAs in control and LMB-treated oocytes were monitored at the indicated times by analysis of 0.5 oocyte equivalents of total nuclear (N) and cytoplasmic (C) RNAs in a 1.2% agarose gel. (C) PKI NES peptides conjugated to BSA (NES–BSA) were injected into nuclei 1 h prior to labeling with [32P]GTP for 24 h, and labeled rRNAs of control and treated oocytes were analyzed as in (A). (D) Requirement for Ran·GTP. Oocytes were labeled with [32P]GTP at 0 h, and RanT24N was injected into the cytoplasm at 24 h; rRNAs of control and treated oocytes were analyzed after 24 and 40 h of labeling, as indicated. The gel mobilities of precursor and mature rRNAs are indicated, and arrowheads show the nuclear accumulation of mature rRNAs upon inhibition of ribosomal subunit export.
Sequences promoting nuclear export
The sequence of amino acids 480–489 of hNMD3 resembles a leucine-rich NES (Figure 2; Fischer et al., 1995; Wen et al., 1995). Substitution of the leucine residues at positions 480, 484 and 487 with alanine (NESmut) or deletion of amino acids 480–489 (ΔNES) produced mutant proteins that accumulated in both the nucleoplasm and nucleoli, even in the absence of LMB (Figure 2C–F). Mutant proteins with single (L487A) or double (L480A, L487A) alanine substitutions also displayed decreased NES activity, with the extent of nuclear accumulation correlating with the number of leucines mutated (not shown). Thus, amino acids in the 480–489 region constitute part of the NES needed for efficient export of NMD3.
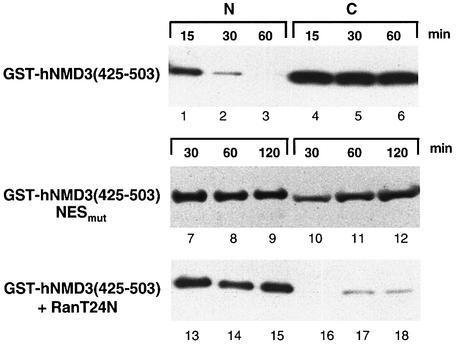
To test directly if this region of hNMD3 acts as an NES, a different chimeric reporter protein was used, consisting of the C-terminal 79 amino acids of hNMD3 fused to GST [GST–hNMD3(425–503)]; we note that GST alone does not diffuse out of Xenopus laevis oocyte nuclei (Askjaer et al., 1999). Upon nuclear injection, the GST fusion protein was exported rapidly to the cytoplasm when the presumptive NES was intact (Figure 3, lanes 1–6) but not when the it was mutated (lanes 7–12) or deleted (not shown). Also, the chimeric protein did not appear in the cytoplasm when the nuclear pool of Ran·GTP was depleted (lanes 13–18) (by injection of RanT24N, an inhibitor of the nuclear Ran GTP/GDP exchange factor RCC1; Izaurralde et al., 1997), showing that the protein was exported in a Ran·GTP-dependent manner. These results are consistent with the proposal that the leucine-rich 480–489 region of hNMD3 functions as an NES for CRM1–Ran·GTP-mediated export of hNMD3.
Fig. 3. The NES region of hNMD3 directs Ran·GTP-dependent nuclear export of a heterologous protein in Xenopus oocytes. GST fusion proteins containing the wild-type or NESmut version of the C-terminal 79 amino acids (residues 425–503) of hNMD3 were injected into nuclei of control oocytes (top and middle panels) or oocytes pre-injected with RanT24N (bottom panel) to inhibit Ran·GTP-dependent export, and export was monitored with time. One oocyte equivalent of nuclear (N) and cytoplasmic (C) extracts was analyzed by western blotting with anti-GST antibodies. Note the shorter time course for the top (15, 30 and 60 min) versus the middle and bottom (30, 60 and 120 min) panels. GST alone does not exit the nucleus during this time course (Askjaer et al., 1999; unpublished data).
Sequences affecting nuclear localization
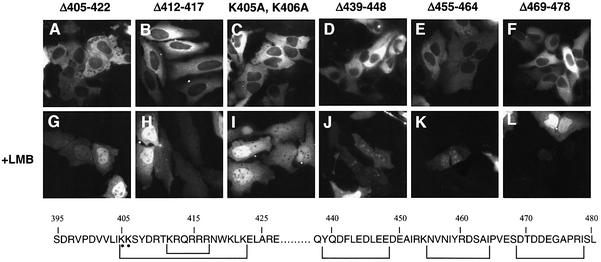
The region of hNMD3 between amino acids 405 and 422 is highly basic and resembles canonical NLSs found in other proteins (Boulikas, 1993). GFP–hNMD3 mutant proteins that lacked this entire region (Δ405–422; Figure 4A) or a portion of it (e.g. Δ412–417; Figure 4B and Supplementary table S-I available at The EMBO Journal Online), or that had alanines instead of lysines at positions 405 and 406 (K405A,K406A) (Figure 4C) did not accumulate in the nucleus. Thus, the entire basic amino acid region appears to be necessary for the steady-state nuclear localization seen with the wild-type chimeric protein.
Fig. 4. The NLS of hNMD3 is complex. GFP–hNMD3 fusion proteins containing the indicated mutations were expressed in HeLa cells as in Figure 2, and protein localizations were monitored without (A–F) or with further treatment with LMB (G–L). The relevant amino acid sequences of the C-terminal domain of hNMD3 are shown below.
Surprisingly, several non-basic regions located adjacent to this basic domain (i.e. amino acids 396–404, 439–447, 455–464 and 469–478) were also required for nuclear accumulation. Mutant proteins lacking any one of these additional regions failed to accumulate in the nucleus (Figure 4D–F; Supplementary table S-I), showing that amino acids located throughout the C-terminal domain govern the intracellular distribution of hNMD3. In spite of their cytoplasmic localization, the mutant proteins must still shuttle between the nucleus and cytoplasm, as they accumulated in the nucleus when export was inhibited by LMB (Figure 4G–L) or mutation of the NES (Supplementay table S-I). We conclude that the NLS is complex, with several elements each contributing to the efficiency of import and hence to the nucleo-cytoplasmic distribution of the protein.
Sequences affecting nucleolar localization
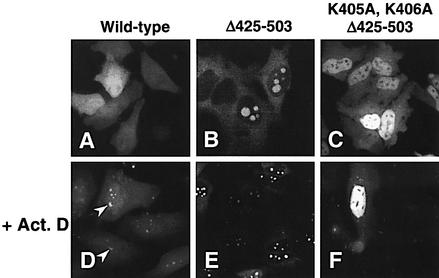
The intranuclear localization of the mutant proteins in the presence of LMB (Figure 4G–L) or upon mutation of the NES (Supplementary table S-I) revealed that amino acids located in the 396–422 region of GFP–hNMD3 acted as a nucleolar localization signal (NoLS). Proteins lacking basic amino acids in the region 405–422 accumulated primarily in the nucleoplasm rather than in the nucleolus (Figure 4G–I). In contrast, a mutant protein containing this basic region but lacking amino acids 425–503 (i.e. both the non-basic NLS sequences and the NES) accumulated prominently in nucleoli but not in the nucleoplasm (in ∼25% of the transfected cells) or solely in the cytoplasm (Figure 5B); the low percentage of cells manifesting nuclear localization of this protein may have resulted from the absence of the non-basic components of the NLS. Surprisingly, mutation of only two basic amino acids of the NLS (K405A,K406A, Δ425–503) caused exclusion from the nucleolus and led to nucleoplasmic localization in all cells (Figure 5C; see also Figure 4I). Thus, accumulation of hNMD3 in nucleoli requires some of the basic amino acids in the 405–422 region but not amino acids in the 425–503 region (see also Figure 4J–L).
Fig. 5. hNMD3 contains sequences required for nucleolar localization. Wild-type and mutant GFP–hNMD3 proteins were expressed as in Figure 2, and protein localizations were monitored without (A–C) or with treatment with actinomycin D (0.04 ng/ml) for 1.5 h (+ActD) (D–F). Note that treatment with ActD causes the accumulation of GFP–hNMD3 proteins in nucleolar cap structures (arrowheads in D), provided the proteins contain sequences required for nucleolar entry.
Accumulation in the nucleolus of both the wild-type and the NES-deficient mutant (Δ425–503) protein increased when the synthesis of rRNAs was blocked by treatment with actinomycin D (ActD) (Figure 5D and E). Thus, exit of hNMD3 from nucleoli might depend on continued production of ribosomes. ActD itself is unlikely to be directly responsible for the accumulation of GFP–hNMD3 in the nucleolus, since the mutant protein hNMD3Δ425–503,K405,K406 (see above) was excluded from nucleoli even in the presence of ActD (Figure 5F). The ActD treatment also resulted in localization of some of the GFP–hNMD3 to nucleolar cap structures (arrowheads in Figure 5D) similar to those described for other nucleolar shuttling proteins (Andersen et al., 2002).
CRM1 and Ran·GTP function in the export of ribosomal subunits
In yeast, export of Nmd3p and 60S ribosomal subunits is coupled (Ho et al., 2000; Gadal et al., 2001). We therefore asked if export of 60S subunits and NMD3 is coupled in metazoans. To do that, we developed a manipulatable ribosome export assay using Xenopus oocytes, which contain highly amplified, very actively transcribed rRNA genes (Savino and Gerbi, 1990). Since oocyte nuclei and cytoplasm can be separated cleanly (Lund and Paine, 1990), the intracellular distributions of newly made endogenous 28S and 18S rRNAs can be used as indicators of the export of 60S and 40S subunits, respectively. In these cells, steady-state labeling of the different forms of nuclear rRNA precursors (Figure 6A) is achieved only after several hours of incubation with [α-32P]GTP, and the nuclear maturation and export of 18S rRNA is much faster than that of 28S rRNA; this results in the preferential appearance of labeled 18S rRNA in the cytoplasm at early times (6 and 15 h) after addition of label (Figure 6B, lanes 1–4; see Dunbar and Baserga, 1998).
Mature newly made rRNAs accumulated in the nuclei of oocytes treated with LMB (Figure 6A, compare lanes 5 and 6 with lanes 7 and 8), indicating that CRM1 function (see Fornerod et al., 1997) is required for export of both 60S and 40S subunits. Likewise, export of both 28S and 18S rRNAs was inhibited by nuclear injection of competitor bovine serum albumin (BSA)–NES peptide conjugates (Figure 6C) but not by control BSA–NLS peptide conjugates. Export was also inhibited by depletion of Ran·GTP (by injection of RanT24N; Figure 6D). Thus, the CRM1–Ran·GTP system is required for export of both 60S and 40S subunits in Xenopus oocytes.
A role for NMD3 in the export of 60S ribosomal subunits from oocyte nuclei
The requirement for CRM1 function in ribosome export is consistent with a model, developed for yeast, that 60S-associated Nmd3p provides an NES that is recognized by Crm1p during export of 60S subunits (Ho et al., 2000; Gadal et al., 2001). To test directly if NMD3 functions this way in metazoans, we monitored the effects of exogenous wild-type and mutant NMD3 proteins on ribosome metabolism in oocytes.
Wild-type and several of the mutant hNMD3 proteins characterized above, but lacking GFP, were made by injection of mRNAs into oocyte cytoplasms, and protein synthesis was monitored by labeling with [35S]methionine plus cysteine (Figure 7). In all cases, the 35S-labeled hNMD3 proteins accumulated in the cytoplasm (dots at lanes 2, 4, 6, 10, 12 and 14), and their nuclear levels resembled the extents of intranuclear accumulation of the corresponding GFP chimeras observed in HeLa cells. Proteins that were present in the nucleoplasm of HeLa cells, such as wild-type hNMD3 (wt) and NES-deficient mutant proteins (Figure 2A–C) with or without an NLS (K405A,K406A,NESmut; Supplementary table S-I), accumulated prominently in oocyte nuclei (Figure 7, lanes 1, 3, 9 and 11), whereas hNMD3Δ425–503, which was largely absent from the nucleoplasm of HeLa cells (Figure 5B), was present in only low amounts in oocyte nuclei (Figure 7, lane 5). However, the small amounts of hNMD3Δ425–503 that did accumulate within nuclei appeared to be nucleolar both in HeLa cells (Figure 5B) and in oocytes (our unpublished results). The mutant protein lacking both the NLS and NoLS regions (Δ405–503) could not be detected in oocyte nuclei (lanes 13 and 14).
Fig. 7. Intracellular distributions of hNMD3 proteins in Xenopus oocytes. Oocytes were injected with m7G-capped mRNAs encoding the indicated wild-type and mutant hNMD3 proteins and labeled with [35S]methionine for 20–24 h; nuclear (N) and cytoplasmic (C) extracts (1 and 0.5 oocyte equivalents, respectively) were analyzed by SDS–PAGE in 8% (lanes 1–8) or 10% (lanes 9–16) gels and by autoradiography. Dots indicate the newly made exogenous hNMD3 proteins. hNMD3(Δ425–503) accumulates to a much lower level in the nucleus than hNMD3(NESmut) (see text), but nonetheless is a very effective inhibitor of 60S subunit export (compare with Figure 8A, lanes 9–12).
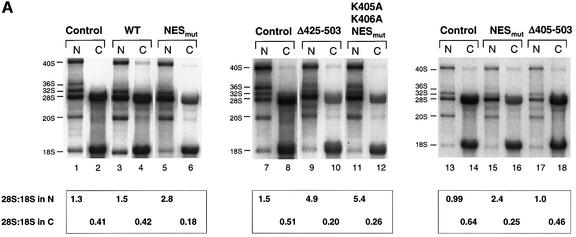
The presence of either wild-type or mutant hNMD3 proteins in the nucleus led to changes in the pattern of processing intermediates of 28S rRNA (as monitored after the labeling of nuclear rRNA precursors had reached steady state; Figure 8A). In contrast, the metabolism of 18S rRNA was not significantly affected by the exogenous hNMD3 proteins, confirming that 18S and 28S rRNAs are processed and exported independently of one another. This independence allowed us to use 18S rRNA to normalize the extent of 28S rRNA export (see legend to Figure 8).
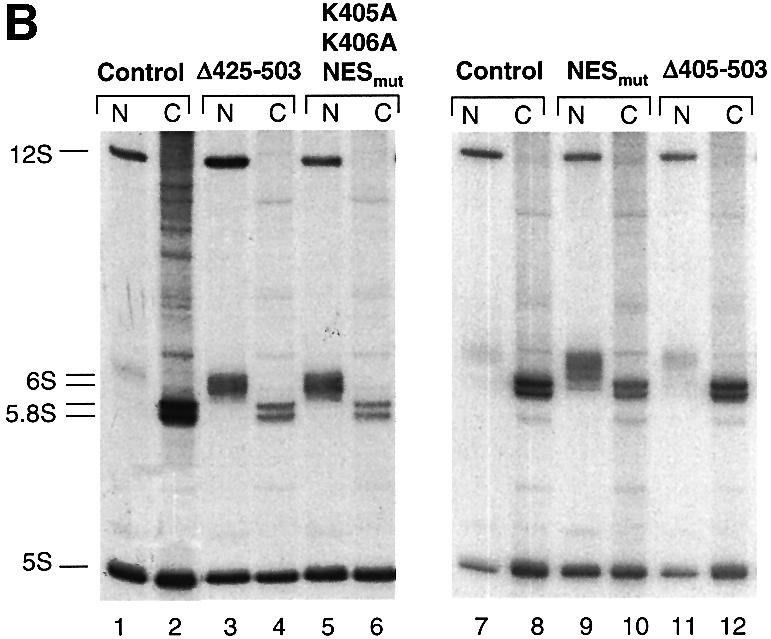
Fig. 8. NES-deficient hNMD3 proteins are dominant-negative inhibitors of 60S subunit export. (A) Analyses of large rRNAs. Lanes 1–12: oocytes were pre-labeled with [32P]GTP for ∼6 h prior to injection of m7G-capped mRNAs encoding the indicated hNMD3 proteins, and the intracellular distributions of newly made rRNAs were analyzed after 40–48 h of labeling, as in Figure 6. Lanes 13–18: oocytes were injected with hNMD3 mRNAs 2 h prior to labeling with [32P]GTP, and rRNAs were analyzed after 48 h of labeling. The molar ratios of newly made 28S to18S rRNAs within the nucleus (28S:18S in N) or cytoplasm (28S:18S in C) were determined by quantification of the 32P-labeled rRNAs by phosphorImager analyses; the cytoplasmic ratio of 28S:18S rRNAs is <1.0 in control oocytes due to the slower rate of maturation of 28S rRNA than 18S rRNA (compare with Figure 6B). (B) Analyses of small rRNAs. Lanes 1–12: total RNAs (corresponding to lanes 7–18 in A) were fractionated on 8% denaturing polyacrylamide gels for analyses of 12S and 6S rRNAs (the nuclear precursors of 5.8S rRNA; compare with Figure 6A) and the mature 5S and 5.8S rRNAs. The extra bands seen in the cytoplasm of control oocytes (lane 2) are non-specific degradation products.
Expression of wild-type hNMD3 left export of 28S rRNA largely unchanged but led to increased levels of the mature 28S rRNA in the nucleus at the apparent expense of the 36S and 32S pre-rRNAs, (Figure 8A, compare lanes 1 and 2 with lanes 3 and 4). Because excess hNMD3 in the nucleus did not accelerate or retard export of 28S rRNA, NMD3 apparently is neither limiting nor inhibiting for 60S subunit export in Xenopus oocytes. The increased rate of processing of precursor 28S rRNA is likely to be an indirect effect of excess nuclear hNMD3 since association of hNMD3 with 60S subunits occurs at a late stage of the maturation process (see below).
Unlike wild-type hNMD3, the NES-deficient mutant proteins (NESmut and Δ425–503) strongly inhibited export of 60S subunits, as shown by the increase in the nuclear accumulation of 28S rRNA (relative to that of 18S rRNA) and the concomitant reduction in the cytoplasmic levels of 28S rRNA (Figure 8A, lanes 7 and 8, and 9 and 10). In contrast, a mutant protein lacking both NLS and NES function (Δ405–503) affected neither 60S subunit maturation nor export (Figure 8A, lanes 13 and 14, and 17 and 18). Thus, NES-deficient hNMD3 proteins act as inhibitors of 60S subunit export, provided they gain access to the nucleus. Interestingly, mutant protein that is not expected to enter nucleoli, NESmut,K405A,K406A (Supplementay table S-I), also inhibited 60S subunit export (lanes 11 and 12), showing that entry into the nucleolus may not be necessary for inhibition.
We propose that, like comparable mutants of yeast Nmd3p, NES-deficient mutants of human NMD3 protein inhibit export of 60S subunits by competing with endogenous wild-type NMD3 for a binding site on nuclear 60S subunits. As a consequence, the nascent 60S subunits are deprived of an NES-containing export adaptor.
Association of hNMD3 with nascent 60S ribosomal subunits
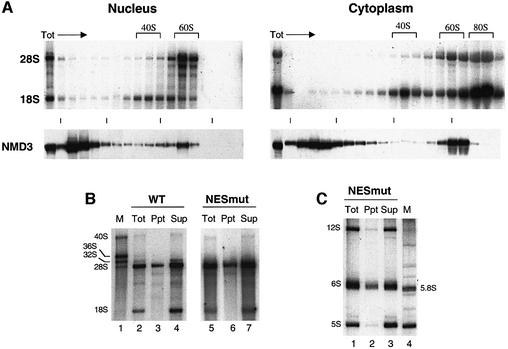
To test directly if hNMD3 binds to 60S subunits in oocytes, we monitored ribosome association of wild-type and NES-deficient forms of hNMD3 that were fused to maltose-binding protein (MBP–hNMD3). The injected MBP fusion proteins (made in bacteria) had the same nucleo-cytoplasmic distributions and the same effects on 60S subunit export as the comparable untagged hNMD3 proteins produced in oocytes (data not shown). As shown by sucrose gradient centrifugation, MBP–hNMD3 associated with 60S subunits in both nucleoplasmic and cytoplasmic extracts, but not with 40S subunits nor 80S ribosomes (Figure 9A); also, this pattern of association was not affected by the presence or absence of a functional NES on the hNMD3 (data not shown). As expected, anti-MBP antibodies specifically co-precipitated 28S rRNA but not 18S rRNA from both nucleoplasmic (Figure 9B) and cytoplasmic (not shown) extracts. The lack of pre-28S rRNAs from the immunoprecipitated ribosomes (Figure 9B) demonstrated that hNMD3 bound only to subunits that were in the late stages of maturation.
Fig. 9. hNMD3 associates with nascent 60S ribosomal subunit in both the nucleus and the cytoplasm. (A) Nucleoplasmic and cytoplasmic extracts of oocytes injected with MBP–hNMD3-NESmut fusion protein and labeled with [32P]GTP were fractionated on 10–40% sucrose gradients, and the distributions of newly made rRNAs (top panels) and MBP–hNMD3 (bottom panels) were determined by agarose gel electrophoresis and western blotting with anti-MBP antibodies, respectively. In the experiment shown, the nucleoplasmic extract was prepared from oocytes treated with VSV M protein (an inhibitor of nuclear export; Her et al., 1997) to increase the levels of 40S and 60S export complexes in the nucleoplasm, but both wild-type and NESmut hNMD3 proteins are also associated with nascent 60S subunits in nucleoplasmic extracts of untreated control oocytes [unpublished data; compare with (B)]. (B and C) Nuclear extracts of oocytes injected with wild-type (WT) or NESmut MBP–hNMD3 proteins were immunoprecipitated with anti-MBP antibodies, and the large (B) and small (C) rRNAs of the bound (Ppt) and unbound (Sup) fractions and the total (Tot) extract were analyzed by gel electrophoresis as in Figure 8. M: marker rRNAs as indicated. Note that the nucleoplasmic extracts are devoid of nucleoli, which contain the majority of the 32S, 36S and 40S precursor rRNA, and which are abundant in total nuclear RNAs (Figure 6B).
We were unable to detect a presumptive pre-export complex containing nuclear 60S subunits, NMD3, CRM1 and Ran·GTP (data not shown). Perhaps such complexes rapidly dissociated during sample preparation upon exposure to cytoplasmic RanGAP. Similar observations have been made for yeast (Nissan et al., 2002; A.Johnson, unpublished).
The nuclear 60S subunits that were precipitated by anti-MBP antibodies were highly enriched in 6S rRNA, a normally short-lived, immediate precursor of 5.8S rRNA (Figure 9C). The association of this precursor with the nuclear 60S subunits is consistent with our finding that in oocytes, the processing of 6S to 5.8S rRNA occurs only upon delivery of the 60S subunit to the cytoplasm and does not occur when subunit export is inhibited by treatment with LMB or vesicular stomatitis virus (VSV) matrix (M) protein (E.Lund, C.R.Trotta and J.E.Dahlberg, in preparation; compare with Figure 6A). The presence of NES-deficient hNMD3 proteins that could enter the nucleus resulted in a sharp increase in the nuclear levels of 6S rRNA, and a concomitant decrease in the levels of cytoplasmic 5.8S rRNA (Figure 8B, lanes 3–6 and lanes 9 and 10), confirming that the mutant protein had inhibited export of 60S subunits (compare with Figure 8A). In contrast, the nuclear levels of 12S rRNA [a long-lived, earlier precursor of 5.8S rRNA (Peculis and Steitz, 1993; compare with Figure 6A] showed no consistent change (Figure 8B), and very little of the 12S rRNA was co-immunoprecipitated by the anti-MBP antibodies (Figure 9C); the absence of co-precipitated labeled 5S rRNA is due to the recruitment of stored, unlabeled 5S rRNAs into newly made 60S subunits in oocytes (Dixon and Ford, 1982; Allison et al., 1991). We conclude that NES-deficient hNMD3 proteins interfere with the biogenesis of 60S subunits at a very late stage in their production and that the nuclear target of hNMD3 is the mature, pre-export 60S ribosomal subunits.
Discussion
Using a combination of in vivo approaches, we have demonstrated that NMD3 functions in 60S ribosome export in a manner that is conserved from human to yeast. Our data are consistent with a model in which hNMD3, like its yeast ortholog yNmd3p, is actively imported into the nucleus where it binds to newly assembled 60S ribosomal subunits and acts as an adaptor for 60S subunit export via the CRM1 pathway.
Complex localization signals in hNMD3
We showed that hNMD3 is a nucleo-cytoplasmic shuttling protein with a C-terminal domain that contains complex signals for both nuclear import and export. Amino acid sequences responsible for the intracellular trafficking were studied using chimeric GFP–hNMD3 proteins expressed in transiently transfected HeLa cells. Comparable results were obtained when the levels of expression were high or low, indicating that overexpression of the exogenous protein did not significantly perturb the normal distribution of NMD3.
Mutation or deletion of the leucine-rich NES (amino acids 480–489) led to nuclear plus nucleolar accumulation of the mutant GFP–hNMD3 reporter proteins. Similar changes in the intracellular localization of wild-type GFP–hNMD3 occurred in the presence of LMB (Figure 2), indicating that CRM1 is the export receptor for hNMD3. The C-terminal domain of yNmd3p contains a comparable NES (Ho et al., 2000; Gadal et al., 2001) that requires amino acids I493, L497 and L500 (comparable with L480, L484 and L487 of hNMD3; compare with Figure 1; J.Hedges and A.Johnson, unpublished data).
The C-terminal region of hNMD3 (amino acids 425–503) containing the NES was able to direct very efficient, Ran·GTP-dependent export of a chimeric GST–reporter protein from Xenopus oocyte nuclei (Figure 3). However, deletion (or mutation) of the sequences corresponding to amino acids 480–489 from the GST chimera did not abolish its export, suggesting that other amino acids in this region also contribute to the activity of the NES. It is unclear if the remaining export activity is due to a weak interaction with CRM1 or to interaction of the chimeric protein with another export receptor.
The conserved region of basic amino acids (Figure 1) that resembles a bipartite NLS (residues 405–422) (Robbins et al., 1991) provides NLS activity, as shown by the predominantly cytoplasmic localization of both human and yeast mutant NMD3 proteins lacking some or all of these amino acids (Figure 4; J.Hedges and A.Johnson, unpublished data). Surprisingly, several mutations outside the putative NLS that were dispersed throughout the C-terminal domain of GFP–hNMD3 also caused cytoplasmic accumulation (Figure 4). All of the NLS-defective mutant proteins exhibited a low level of NLS activity when export was blocked by LMB or mutation of the NES (Figure 4 and Supplementary table S-I), but it is unclear if this residual activity arose entirely from hNMD3 or was due in part to cryptic NLS activity of the GFP moiety of the fusion proteins (I.G.Macara, personal communication).
The non-basic sequences could contribute to nuclear accumulation either by upregulating NLS activity or by downregulating NES activity (Kaffman and O’Shea, 1999). It is unlikely that these sequences affect NES function, since their deletion (e.g. Δ439–448) from the reporter protein GST–hNMD3(426–503) did not appear to increase (or decrease) the rate of export from oocyte nuclei (unpublished data). NLS activity could be modulated through changes in the way the basic NLS region is presented to the nuclear import machinery or through covalent modification. However, NLS activity of GFP–hNMD3 is unlikely to be modulated by tyrosine phosphorylation as nuclear accumulation was not affected when three conserved tyrosine residues (Y408,Y439, Y459) were mutated to phenylalanines (unpublished data); also, no evidence has been obtained indicating that yNmd3p is phosphorylated (J.Hedges and A.Johnson, unpublished data).
Amino acid sequences responsible for accumulation of hNMD3 in nucleoli overlap the basic amino acid region of the NLS, including lysines K405 and K406, but they do not extend C-terminally beyond position 425 (Figures 4G–L, and 5B and C). This NoLS could promote nucleolar retention of hNMD3 or it could act as a nucleolar entry signal (or both). Inhibition of rRNA synthesis by ActD, which prevents retention of many proteins that interact with nucleolar RNA (e.g. Dundr et al., 1995; Zirwes et al., 2000), increased, rather than decreased the accumulation of GFP–hNMD3 in nucleoli and nucleolar cap structures in an NoLS-dependent manner (Figure 5D–F). This result raises the possibility that exit of hNMD3 from nucleoli requires ongoing synthesis and assembly of ribosomal subunits, and that the NoLS functions directly in entry of the protein into nucleoli, rather than in retention. It is unclear if NoLSs that have been identified recently in the protein subunit of telomerase (hTERT) (Yang et al., 2002; Wong et al., 2002) and protein components of the RNase MRP protein complex (van Eenennaam et al., 2001; Jarrous, 2002) function in a similar manner. Exit of hNMD3 also may involve the leucine-rich NES and CRM1 function (Figures 2 and 5), but a direct role of CRM1 in its intra-nuclear trafficking has not been demonstrated.
Thus, the steady-state intracellular distribution of hNMD3 is influenced by interactions of its NES, NLS and NoLS, encoded in the C-terminal domain, with cellular factors. It remains unclear how the activities of these complex signals normally are regulated and balanced, within the cell.
Role of hNMD3 in 60S ribosome biogenesis
We have shown that the same basic mechanisms for export of 60S ribosomal subunits are conserved between humans, frogs and yeast. 60S subunit export requires functional CRM1 and Ran·GTP (Figure 6), and NMD3 plays a key role in this process in all of these organisms (Figure 8). This is consistent with our observation that hNMD3 can substitute for Nmd3p in yeast cells (A.W.Johnson, unpublished).
Both wild-type and NES-deficient hNMD3 bind to the newly made 60S subunits in the nucleus (Figure 9), but only the NES-deficient hNMD3 inhibits export of 60S subunits (Figure 8). The continued export in the presence of exogenous wild-type hNMD3 shows that an excess of NMD3 in the nucleus is not inhibitory. We propose that the mutant protein blocks 60S subunit export by forming pre-export particles that lack an NES. In yeast, a similar dominant-negative phenotype of NES-deficient yNmd3p is suppressed by fusion of an exogenous NES [from protein kinase I (PKI)] to the N-terminus of the protein, showing that export of 60S subunits correlates with the acquisition of an NES through binding of Nmd3p (Ho et al., 2000). We conclude that in both metazoans and yeast, NMD3 acts as an export adaptor whose NES is recognized by the export receptor CRM1.
Increasing the level of wild-type hNMD3 in the nucleus did not affect the rate of export of 60S subunits but it did accelerate the production of mature length 28S rRNA (Figure 8A). Thus the rate-limiting step in 60S subunit export is likely to be a step that occurs after formation of the 28S rRNA but before formation of export complexes, perhaps one that involves a conformational change or protein rearrangement in the subunit. hNMD3 binds to nascent 60S subunits very late in their maturation (Figure 9B), so its effects on rRNA processing may be indirect, through the release of subunit-bound accessory factors that are limiting for early processing events.
It is likely that NMD3 normally interacts with 60S subunits within the nucleolus, since the NoLS can direct GFP–hNMD3 to that compartment and the protein accumulates there when rRNA synthesis is inhibited by ActD. However, the binding apparently can also occur in the nucleoplasm, as a mutant of hNMD3 that is deficient in both NoLS and NES function is still a potent inhibitor of export of 60S subunits (Figure 8A).
The maturity of a 60S subunit within the nucleus may be monitored by ‘structural proofreading’ through its ability to bind NMD3 in a productive manner that ultimately allows for export. We propose that this constitutes a major check for subunit integrity before export. ‘Functional proofreading’ of newly made 60S subunits by translation within the nucleus is unlikely because 60S subunit export is not blocked by treatment of oocytes with translation inhibitors, such as cycloheximide (E.Lund and J.E.Dahlberg, unpublished data), and newly made subunits were not found in 80S ribosomes in nuclear extracts (Figure 9A). Moreover, no compelling evidence has been published showing that translation occurs within the nucleus (Dahlberg et al., 2003).
Attempts to reconstitute an in vitro export complex containing CRM1 and Ran·GTP, with cytoplasmic 60S subunits bound to NMD3, have not been successful to date (Thomas and Kutay, 2003; G.Kallstrom and A.Johnson, unpublished data). This may indicate that only nascent nuclear 60S pre-export particles can expose the NES of bound NMD3, and thereby become substrates for the CRM1 export complex. Nuclear 60S particles differ from their cytoplasmic counterparts in the complement of associated proteins (Nissan et al., 2002) and in the degree of rRNA maturation (Figure 8). For example, in oocytes, nuclear 60S subunits contain 6S rRNA (a 3′-extended precursor form of 5.8S) (Figures 8B and 9), which undergoes maturation only upon export of 60S subunits to the cytoplasm (E.Lund, C.R.Trotta and J.E.Dahlberg, in preparation; compare with Figure 6A). Hence, completion of 60S subunit export may involve several steps in the cytoplasm (discussed in Johnson et al., 2002). It remains to be determined how, or if, any of these additional proteins or rRNA sequences might support the assembly of 60S subunit export complexes.
The work presented here indicates that NMD3 furnishes an essential—perhaps the only—NES used for export of 60S subunits. The necessity for NMD3 in export of 60S subunits means that this protein could be used both to control export and to monitor the maturity of nuclear 60S subunits. Since NMD3 binds free subunits in the cytoplasm (Figure 9; Belk et al., 1999; Ho and Johnson, 1999), an excess of such subunits could lower the amount of this shuttling adaptor that is available to export yet more subunits, thereby slowing the rate at which even more subunits appear in the cytoplasm. Furthermore, the ability of NMD3 to discriminate between newly assembled nuclear subunits at different stages of maturation (Figure 9B) would prevent export of immature, non-functional subunits (discussed in Johnson et al., 2002). Thus NMD3, as a shuttling export adaptor, may have several roles in ensuring that functional 60S subunits are exported when needed in the cytoplasm.
Materials and methods
Cloning, mutagenesis and expression of hNMD3
All DNA manipulations were performed as described by Sambrook and Russell (2000). The coding region of the gene for hNMD3 (accession No. AF132941) was obtained by PCR amplification from a HeLa cDNA library, using oligonucleotides 1 and 2 (Supplementary table S-II) and Pfu polymerase (Stratagene). Two nucleotide sequence discrepancies (relative to the hNMD3 sequences in DDBJ/EMBL/GenBank) were noted in all PCR amplifications of hNMD3 from several different cDNA sources: A565G, resulting in a silent mutation; and C775G, resulting in a change from isoleucine to methionine. A mutant of hNMD3 lacking the C-terminal 79 amino acids (equivalent to yNmd3Δ100p; Belk et al., 1999; Ho et al., 2000) was obtained by PCR amplification using oligonucleotides 2 and 3.
For expression in HeLa cells, the PCR products of wild-type and C-terminally truncated hNMD3 were cloned in-frame into the BglII–EcoRI sites of pEGFP-C1 (Clontech). The resulting clones, pEGFP-C1-hNMD3WT and pEGFP-C1-hNMD3Δ79, were used to generate hNMD3 mutants (see text and Supplementary table S-I) via mutagenesis (QuikChange Mutagenesis Kit, Stratagene). The nucleotide sequences of all clones were confirmed by DNA sequencing.
For expression in Xenopus oocytes, hNMD3 coding regions were cloned into pSP64 poly(A) (Promega). A BspEII (blunt ended)–BamHI fragment of hNMD3Δ79 was excised from pEGFP-C1-hNMD3Δ79 and cloned into the HincII–BamHI sites of pSP64 poly(A), generating pSP64 poly(A)-hNMD3Δ79. Other pSP64 poly(A) clones were obtained by replacement of the NcoI–BamHI hNMD3Δ79-fragment with NcoI–BamHI fragments of various pEGFP-C1-hNMD3 DNAs. For synthesis of m7G-capped mRNAs, BsrBI-linearized pSP64 poly(A)-hNMD3 DNAs were used as templates for in vitro transcription with SP6 RNA polymerase (Promega), as detailed elsewhere (Pasquinelli et al., 1995; Petersen et al., 2000).
Plasmids encoding GST fusion proteins were generated by PCR amplification of the C-terminal sequence of hNMD3 encoding amino acids 426–503 using oligonucleotides 4 and 5 (Supplemetary table S-II) and cloning in-frame into the BamHI–EcoRI sites of pGEX-2T (Pharmacia). Mutations were introduced as above using the QuikChange Mutagenesis Kit (Stratagene). GST fusion proteins were expressed in Escherichia coli strain BL21(DE3) (Novagen) carrying plasmid pTR99 (Del Tito et al., 1995) by induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 37°C. Cells were broken by sonication in buffer containing 50 mM Tris–HCl pH 7.5, 200 mM NaCl, 1% Triton X-100, 5 mM β-mercaptoethanol, 5% glycerol, and purified on glutathione–Sepharose (Amersham).
For generation of MBP fusion proteins, wild-type or NESmut hNMD3 coding region fragments were excised from pSP64 poly(A) plasmids by cleavage with NcoI (end-filled) and BamHI and cloned in-frame into EcoRI (end-filled)–BamHI sites of pMAL-c2G (New England Biolabs). MBP–hNMD3 fusion proteins were expressed in E.coli DH5α and affinity purified on amylose resin (New England Biolabs).
Transfection of HeLa cells
Transient transfection of HeLa cells grown on coverslips was carried out with Lipofectamine (Gibco-BRL). Cells were allowed to express GFP–hNMD3 fusion proteins overnight (12–18 h) and then either fixed and mounted for visualization, or treated with inhibitors (see figure legends) prior to fixation. LMB (a kind gift from S.Blair and M.Yoshida) was used at 3 ng/ml. Actinomycin D (Sigma) was used at 0.04 µg/ml. For visualization of GFP, cells were fixed with 3% paraformaldehyde for 20 min, washed with phosphate-buffered saline (PBS) and mounted for direct fluorescence microscopy using an Axioplan II Microscope (Zeiss).
Injection of Xenopus laevis oocytes
Stage V and VI oocytes were obtained from X.laevis frog ovary (Petersen et al., 2000). GST or MBP fusion proteins or hNMD3 mRNAs (at 0.5–1.0 µg/µl) were injected into oocyte nuclei (10–15 nl) or cytoplasm (20–50 nl), using blue dextran to control for the accuracy of nuclear injection and dissection (Terns and Goldfarb, 1998). For inhibition of export, oocytes were incubated in medium with LMB (400 ng/ml) or injected with PKI NES peptide conjugates (Pasquinelli et al., 1997), RanT24N (a gift of I.G.Macara) or mRNA encoding VSV M protein (Petersen et al., 2000).
For labeling of newly made hNMD3 proteins, oocytes were incubated in medium containing 20–40 µCi/ml of [35S]methionine + cysteine (Amersham). Nuclear and cytoplasmic extracts were prepared by homogenization in buffer A (10 mM HEPES pH 7.5, 83 mM KCl, 17 mM NaCl, 2 mM MgCl2, 0.8 mM EGTA, 2 mM dithiothreitol (DTT), 0.2 U of RNasin (Promega) and protease inhibitors (Roche).
For labeling of endogenous rRNAs, oocytes were injected in the cytoplasm with 0.5–1.0 µCi of [α-32P]GTP (NEN Life Science). Total nuclear and cytoplasmic RNAs were isolated by proteinase K digestion, phenol/chloroform extraction and ethanol precipitation, and resuspended in H2O.
Gel analysis of proteins and RNAs
Protein extracts were fractionated on SDS-containing 8 or 10% (29:1) polyacrylamide gels which were either dried for autoradiography of 35S-labeled proteins or transferred to Immobilon-P membranes (Millipore) for western blot analysis. Anti-GST (Santa Cruz Biotechnology) and anti-MBP (New England Biolabs) antibodies were used as directed.
32P-labeled RNAs (equivalent to 0.5 oocytes) were separated in thin 8% (30:0.8) polyacrylamide gels containing 7 M urea (for small rRNAs) or following denaturation by treatment with glyoxal (Sambrook and Russell, 2000) in 1.2% agarose gels (for large rRNAs). For complete recovery of all large rRNAs, agarose gels were dried down on Zeta-Probe Blotting membranes (Bio-Rad), prior to autoradiography and quantitation by phosphorImager (Molecular Dynamics) analysis.
Sucrose gradient and immunoprecipitation analyses
Nucleoplasmic and cytoplasmic extracts (corresponding to 15–25 or 5–10 oocytes, respectively) prepared from 32P-labeled oocytes injected with MPB–hNMD3 fusion proteins were fractionated on 5 ml of 10–40% sucrose gradients (made in buffer A containing 0.05% deoxycholate) in an SW55 rotor for 3 h at 45 000 r.p.m. and 4°C. Fractions (0.25 ml) were collected and split for analyses of MPB fusion proteins (by western blotting) and large rRNAs (by agarose gel electrophoresis).
For immunoprecipitation, similar nuclear extracts (3–5 oocyte equivalents) were incubated (1–2 h at 4°C) with anti-MBP antibodies (New England Biolabs) coupled to protein A–Sepharose beads (Sigma). The beads were washed with buffer A containing 0.05% Triton X-100, and RNAs in the precipitate and supernatant fractions were recovered by proteinase K digestion as above and analyzed by gel electrophoresis.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr U.Kutay for communicating unpublished results, and C.Slominski and K.Hollister for technical help. Supported by NIH grants GM30220 to J.E.D., GM53655 to A.W.J., a University of Wisconsin Faculty Development grant to L.K. and a Damon Runyon–Walter Winchell Postdoctoral Fellowship DRG 1546 to C.R.T.
References
- Allison L.A., Romaniuk,P.J. and Bakken,A.H. (1991) RNA–protein interactions of stored 5S RNA with TFIIIA and ribosomal protein L5 during Xenopus oogenesis. Dev. Biol., 144, 129–144. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Askjaer P. et al. (1999) RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol., 19, 6276–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille N., Helser,T. and Fried,H.M. (1990) Cytoplasmic transport of ribosomal subunits microinjected into the Xenopus laevis oocyte nucleus: a generalized, facilitated process. J. Cell Biol., 111, 1571–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belk J.P., He,F. and Jacobson,A. (1999) Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA, 5, 1055–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. (1993) Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr., 3, 193–227. [PubMed] [Google Scholar]
- Dahlberg J.E. and Lund,E. (1998) Functions of the GTPase Ran in RNA export from the nucleus. Curr. Opin. Cell Biol., 10, 400–408. [DOI] [PubMed] [Google Scholar]
- Dahlberg J.E., Lund,E. and Goodwin E. (2003) Nuclear translation: what’s the evidence? RNA, 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Tito B.J., Ward,J.M., Hodgson,J., Gershater,C.J., Edwards,H., Wysocki,L.A., Watson,F.A., Sathe,G. and Kane,J.F. (1995) Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol., 177, 7086–7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.K. and Ford,P.J. (1982) Persistence of nonribosome bound 5S RNA in full-grown oocytes of Xenopus laevis. Dev. Biol., 91, 474–477. [DOI] [PubMed] [Google Scholar]
- Dunbar D.A. and Baserga,S.J. (1998) The U14 snoRNA is required for 2′-O-methylation of the pre-18S rRNA in Xenopus oocytes. RNA, 4, 195–204. [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Leno,G.H., Hammarskjold,M.-L., Rekosh,D., Helga-Maria,C. and Olson,M.O. (1995) The roles of nucleolar structure and function in the subcellular location of HIV-1 Rev protein. J. Cell Sci., 108, 2811–2823. [DOI] [PubMed] [Google Scholar]
- Fatica A. and Tollervey,D. (2002) Making ribosomes. Curr. Opin. Cell Biol., 14, 313–318. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Luhrmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001) Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. (1985) The Nucleolus and Ribosome Biogenesis. Springer Verlag, New York, NY.
- Her L.S., Lund,E. and Dahlberg,J.E. (1997) Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science, 276, 1845–1848. [DOI] [PubMed] [Google Scholar]
- Ho J.H. and Johnson,A.W. (1999) NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2389–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom,G. and Johnson,A.W. (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol., 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E. and Adam,S. (1998) Transport of macromolecules between the nucleus and the cytoplasm. RNA, 4, 351–364. [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Kutay,U., Von Kobbe,C., Mattaj,I.W. and Gorlich,D. (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J., 16, 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N. (2002) Human ribonuclease P: subunits, function and intranuclear localization. RNA, 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.W., Lund,E. and Dahlberg,J.E. (2002) Nuclear export of ribosomal subunits. Trends Biochem. Sci., 27, 580–585. [DOI] [PubMed] [Google Scholar]
- Kaffman A. and O’Shea,E.K. (1999) Regulation of nuclear localization: a key to the door. Annu. Rev. Cell. Dev. Biol., 15, 291–339. [DOI] [PubMed] [Google Scholar]
- Kudo N., Wolff,B., Sekimoto,T., Schreiner,E.P., Yoneda,Y., Yanagida,M., Horinouchi,S. and Yoshida,M. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res., 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Leary D.J. and Huang,S. (2001) Regulation of ribosome biogenesis within the nucleolus. FEBS Lett., 509, 145–150. [DOI] [PubMed] [Google Scholar]
- Lund E. and Dahlberg,J.E. (2001) Direct and Indirect roles of Ran·GTP in nuclear export of RNAs in higher eukaryotes. In Rush,M. and D’Eustachio,P. (eds), The Small GTPase Ran. Kluwer Academic Publishers, Boston, MA, pp. 59–77.
- Lund E. and Paine,P.L. (1990) Non-aqueous isolation of transcriptionally active nuclei from Xenopus oocytes. Methods Enzymol., 181, 36–43. [DOI] [PubMed] [Google Scholar]
- Macara I.G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev., 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleo-cytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Nissan T.A., Bassler,J., Petfalski,E., Tollervey,D. and Hurt,E. (2002) 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J., 21, 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.O., Dundr,M. and Szebeni,A. (2000) The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol., 10, 189–196. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Dahlberg,J.E. and Lund,E. (1995) Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA, 1, 957–967. [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E., Powers,M.A., Lund,E., Forbes,D. and Dahlberg,J.E. (1997) Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc. Natl Acad. Sci. USA, 94, 14394–14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peculis B.A. and Steitz,J.A. (1993) Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell, 73, 1233–1245. [DOI] [PubMed] [Google Scholar]
- Petersen J.M., Her,L.S., Varvel,V., Lund,E. and Dahlberg,J.E. (2000) The matrix protein of vesicular stomatitis virus inhibits nucleo-cytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Cell. Biol., 20, 8590–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J., Dilworth,S.M., Laskey,R.A. and Dingwall,C. (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell, 64, 615–623. [DOI] [PubMed] [Google Scholar]
- Sambrook J. and Russell,D.W. (2000) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Savino R. and Gerbi,S.A. (1990) In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J., 9, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tycowski,K.T. and Steitz,J.A. (1996) Ribosomal RNA processing in eukaryotes. In Dahlberg,A.E. and Zimmermann,R.A. (eds), Ribosomal RNA: Structure, Evolution, Gene Expression and Function in Protein Synthesis. CRC Press, Boca Raton, FL, pp. 469–490.
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T., Schmidt,U. and Silver,P.A. (2000) Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell., 11, 3777–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M.P. and Goldfarb,D.S. (1998) Nuclear transport of RNAs in microinjected Xenopus oocytes. Methods Cell Biol., 53, 559–589. [DOI] [PubMed] [Google Scholar]
- Thomas F. and Kutay,U. (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci., in press. [DOI] [PubMed] [Google Scholar]
- van Eenennaam H., van der Heijden,A., Janssen,R.J., van Venrooij,W.J. and Pruijn,G.J. (2001) Basic domains target protein subunits of the RNase MRP complex to the nucleolus independently of complex association. Mol. Biol. Cell, 12, 3680–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J.R. (1990) The nucleolus and ribosome formation. Curr. Opin. Cell Biol., 2, 521–527. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Wolff B., Sanglier,J.J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]
- Wong J.M., Kusdra,L. and Collins,K. (2002) Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol., 4, 731–736. [DOI] [PubMed] [Google Scholar]
- Wool I.G., Chan,Y.L. and Gluck,A. (1995) Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol., 73, 933–947. [DOI] [PubMed] [Google Scholar]
- Yang Y., Chen,Y., Zhang,C., Huang,H. and Weissman,S.M. (2002) Nucleolar localization of hTERT protein is associated with telomerase function. Exp. Cell Res., 277, 201–209. [DOI] [PubMed] [Google Scholar]
- Zirwes R.F., Eilbracht,J., Kneissel,S. and Schmidt-Zachmann,M.S. (2000) A novel helicase-type protein in the nucleolus: protein NOH61. Mol. Biol. Cell., 11, 1153–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]