Abstract
Candida albicans is the most prevalent human fungal pathogen. Here, we take advantage of haploinsufficiency and transposon mutagenesis to perform large-scale loss-of-function genetic screen in this organism. We identified mutations in 146 genes that affect the switch between its single-cell (yeast) form and filamentous forms of growth; this switch appears central to the virulence of C.albicans. The encoded proteins include those involved in nutrient sensing, signal transduction, transcriptional control, cytoskeletal organization and cell wall construction. Approxim ately one-third of the genes identified in the screen lack homologs in Saccharomyces cerevisiae and other model organisms and thus constitute candidate antifungal drug targets. These results illustrate the value of performing forward genetic studies in bona fide pathogens.
Keywords: Candida albicans/filamentous growth/haploinsufficiency
Introduction
Candida albicans is a commensal of humans and other warm-blooded animals that can cause mucosal infections in immunocompetent individuals as well as a broad spectrum of symptoms in immunocompromised patients, including serious disseminated infections (for reviews see Odds, 1988; San-Blas et al., 2000; Calderone and Fonzi, 2001; Haynes, 2001). Progress in understanding many aspects of the biology of C.albicans has been hindered by the inability to carry out simple, large-scale genetic screens. Such screens are highly effective ways of gaining access to and ultimately understanding biological problems, as evidenced by their widespread utility in the ‘model’ yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. The lack of traditional genetic approaches in C.albicans has been due largely to the absence of a well characterized sexual cycle in this organism. The lack of a well-behaved plasmid system and the fact that C.albicans is diploid have also been impediments to this type of approach.
A critical feature of C.albicans—one that has attracted researchers for decades—is its ability to switch between different morphological forms. Candida albicans can grow as single-celled, budding yeast forms (blastospores) or as filamentous forms (including both pseudohyphae and true hyphae) in which cells remain joined end-to-end following cell division (Odds, 1988). The three primary morphological forms, blastospores, pseudohyphae and hyphae, are all found in infected tissues, and work to date indicates that the transition between these forms is critical for pathogenesis. The transition between the morphological forms can also be manipulated in the laboratory, by altering the growth medium. For example, on rich media [such as yeast extract with peptone (YEP) supplemented with 2% glucose (YEPD)] at 30°C, C.albicans grows primarily in the blastospore form; addition of fetal calf serum (FCS) rapidly induces filamentous (pseudohyphal and hyphal) growth of nearly every cell. Other environmental factors, including pH, temperature, oxygen availability, nitrogen availability and carbon source, also affect the distribution of cells among the three primary morphological forms.
To date, molecular genetic studies of the C.albicans morphological transitions have largely relied on S.cerevisiae as an experimental and conceptual model. In response to limitation of specific nutrients, S.cerevisiae undergoes a transition from the single-celled budding form to a pseudohyphal form (Gimeno et al., 1992; for a recent review see Palecek et al., 2002). Based on this overall resemblance to the transition in C.albicans, S.cerevisiae has been used as a recipient to screen gene libraries from C.albicans for their ability to affect pseudohyphal growth (see for example Stoldt et al., 1997; Feng et al., 1999; Kadosh and Johnson, 2001). Alternatively, C.albicans genes with close similarity to S.cerevisiae genes involved in pseudohyphal growth have been identified and disrupted in Candida and the resultant phenotype studied (see for example Liu et al., 1994; Leberer et al., 1996). While these strategies have been successful for identifying certain aspects of filamentous growth regulation in C.albicans, there are important differences between S.cerevisiae and C.albicans with regard to filamentous growth. For example, S.cerevisiae does not exhibit true hyphal growth (in which cells are joined end-to-end with no visible constrictions at cell junctions), whereas this is the predominant morphological form of C.albicans under a number of growth conditions (Odds, 1988). Moreover, certain environmental signals, such as serum, are very strong inducers of filamentous growth in C.albicans but have little or no effect on S.cerevisiae. In addition, a comparison of their genome sequences has revealed that only about two-thirds of Candida genes appear to have clear orthologs in S.cerevisiae. All of these considerations raise the possibility that strategies that rely strictly on similarities between S.cerevisiae and C.albicans are likely to miss crucial features of Candida biology, especially those specific to its pathogenesis.
Here, we describe a large-scale genetic screen in C.albicans designed to identify genes that affect the transition between blastospore and filamentous forms of the yeast. Of particular significance, this strategy makes no prior assumptions about the similarities or differences between C.albicans and S.cerevisiae. This effort led to the identification of 146 different genes that affect the blastospore–filament transition. Only six of these genes had been identified from previous work; the majority, including 39 genes that lack close relatives in S.cerevisiae, were not predicted from previous studies and provide new insights into the mechanism of the blastospore– filament transition. The results of this screen provide a framework for understanding the complex control of this morphological transition.
Results
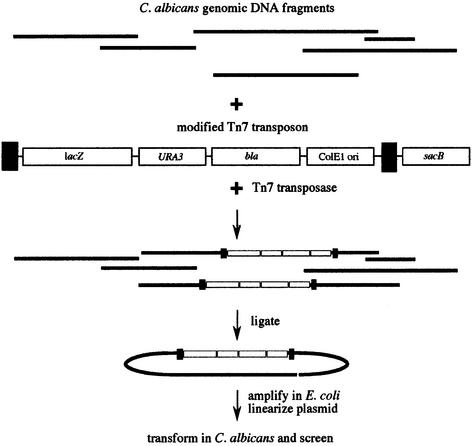
Transposon mutagenesis has been used in many bacterial and yeast genetic screens (Berg and Howe, 1989; Cormack et al., 1999; Ross-Macdonald et al., 1999). To carry out a large-scale transposon-based screen in C.albicans, we constructed a library of 18 000 strains, each containing an independent Tn7-based transposon insertion. These insertion strains were constructed by first transposing Tn7 into Candida genomic DNA in vitro and then transforming a large population of C.albicans with this DNA (Figure 1). Individual transformants were picked and arranged in microtiter dishes; this library of 18 000 strains represents an insertion, on average, every 2.5 kb per haploid genome. Because Candida is diploid, we relied on haploinsufficiency combined with sensitive indicator plates (discussed below) to identify insertion mutants altered in the blastospore–filament transition. Since the transposon introduces unique DNA sequences into the C.albicans genome, the sites of insertion of identified mutants could readily be determined by direct DNA sequencing and comparison with the Stanford genome sequence (http://www-sequence.stanford.edu/group/candida). Details of the library construction and screening conditions are given in Materials and methods. Here, we discuss several general issues that pertain to the screen.
Fig. 1. Method for transposon mutagenesis of C.albicans. Linearized C.albicans genomic DNA fragments generated by restriction enzyme digestion were added to the donor plasmid containing a modified Tn7 transposon and Tn7 transposase. The modified transposon contains a promoterless Streptococcus thermophilus lacZ (Uhl and Johnson, 2001), C.albicans URA3 (Gillum et al., 1984) and the ampicillin resistance gene (bla) and origin of replication from pBluescriptKS+ (Stratagene). The sacB gene located on the donor plasmid external to the Tn7 repeats allows for selection against the donor plasmid (see Materials and methods). Following the transposition reaction (Biery et al., 2000), mutagenized genomic DNA was ligated and transformed into E.coli. The library was amplified, linearized by digestion with BsrGI and transformed in batch into C.albicans strain CAI4 (ura3/ura3). The transformed DNA was allowed to integrate into the C.albicans genome by homologous recombination, and successful integrants were selected as URA+ transformants.
First, we consider the use of haploinsufficiency to screen directly for loss-of-function mutations in a diploid organism. Although C.albicans is indeed diploid, loss of one functional copy of a gene often produces a noticeable phenotype. For example, haploinsufficiency has been reported for a number of genes, including CPH1, INT1, TUP1, CLN1, STE7 and STE20, involved in the regulation of filamentous growth (Liu et al., 1994; Köhler and Fink, 1996; Leberer et al., 1996; Braun and Johnson, 1997; Gale et al., 1998; Loeb et al., 1999). In these studies, one allele of a gene of interest was specifically disrupted by transformation and homologous recombination, and the resultant phenotype was analyzed. In most cases, loss of one allele caused a less severe form of the phenotype produced by disrupting both alleles. To test the feasibility of utilizing such haploinsufficiency for a general genetic screen, we experimented with three C.albicans strains hemizygous for disruptions of genes known to be involved in filamentous growth: TUP1 (a negative regulator of filamentous growth; Braun and Johnson, 1997), CLN1 (a positive regulator of filamentous growth; Loeb et al., 1999) and EFG1 (a positive regulator of filamentous growth; Stoldt et al., 1997). Consistent with published reports, the TUP1/tup1, CLN1/cln1 and EFG1/efg1 strains each showed a clear difference in colony appearance when compared with the ‘wild-type’ parent strain, CAF2-1 (CAF2-1 is a clinical isolate of C.albicans with one copy of URA3 disrupted; Fonzi and Irwin, 1993). Moreover, microscopic examination of the cells in these colonies confirmed published reports that macroscopic differences in colony morphology could be used to monitor microscopic differences in filamentous growth (see Materials and methods).
We used these three strains to choose two screening conditions, one based on serum induction of filamentous growth and one based on nutrient limitation as the inducer. Serum and starvation are both potent inducers of filamentous growth and appear to represent different but overlapping responses. For example, some gene knockouts affect induction by one environmental condition but not the other, whereas others affect the response to both (for reviews see Brown and Gow, 1999; Ernst, 2000; Whiteway, 2000). Although 10–15% FCS is most commonly utilized for induction of filamentous growth, exposure of the EFG1/efg1, CLN1/cln1 and TUP1/tup1 test strains to a range of serum concentrations indicated that 1% FCS was a more sensitive condition for detecting changes in filamentous growth caused by deletion of a single allele. For nutrient limitation, we used Spider medium, as it revealed clear differences among the three test strains and between each test strain and the parent strain (Liu et al., 1994). Based on these preliminary experiments, the mutant screen was performed twice, once on YEPD + 1% FCS and once on Spider medium.
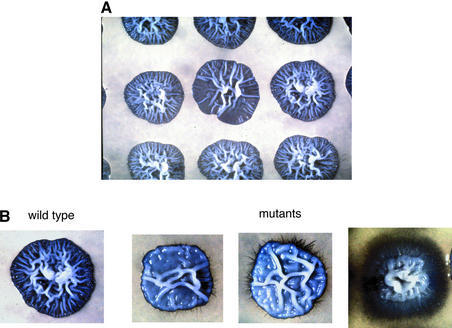
We next consider the overall number of mutants obtained in the screen. We replica-plated 18 000 independent insertion mutants on each type of medium and examined them over the course of 2 weeks. Colonies that displayed altered filamentous growth (either increased or decreased) were rescreened, and only those with reproducible phenotypes were considered further. A total of 340 (∼2%) of the insertion strains exhibited altered patterns of filamentous growth as deduced from changes in the pattern of wrinkling of the central portion of the patch or from differences in the appearance of peripheral hyphae (Figure 2; see also Figure 3). Some mutant strains displayed altered filamentous growth on both types of medium, but many showed differences from the parent strain on only one medium (Table I). Overall, roughly half the mutants showed enhanced filamentous growth on at least one type of medium, and half showed reduced filamentous growth.
Fig. 2. Examples of mutants identified in the screen. (A) A portion of a replica plate containing nine independent members of the insertion library grown on Spider medium for 4 days at 30°C. The center patch has a significantly less wrinkled appearance relative to its neighbors and was scored as a mutant reduced for filamentous growth. At this time point, the patches have not yet produced peripheral hyphae. (B) Examples of three other types of mutants obtained from the screen. From left to right, wild-type strain CAF2-1 (URA3/ura3:: imm434) (Fonzi and Irwin, 1993), a mutant reduced for filamentous growth, a mutant reduced for filamentous growth in the center of the patch but increased for production of peripheral hyphae, and a strongly hyperfilamentous mutant. Strains were grown for 4 days on Spider medium.
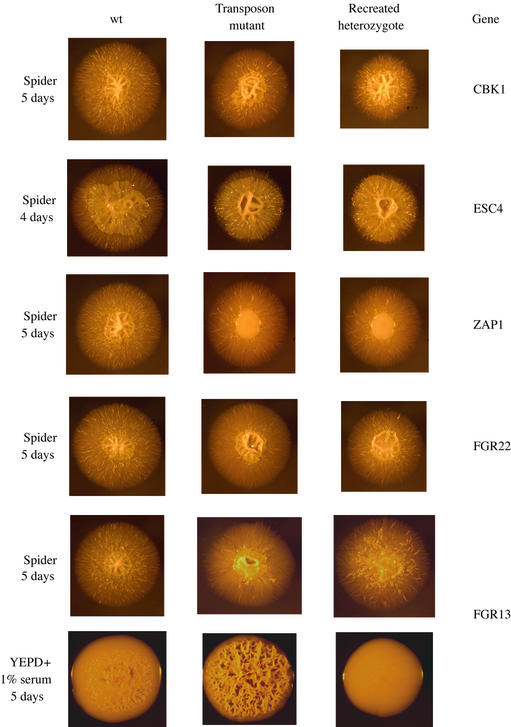
Fig. 3. Comparison of phenotypes of original transposon insertion mutants with those of reconstructed hemizygous deletion mutants. In each case, the deletion mutant was constructed in a fresh strain (RM1000) by deleting one copy of the indicated gene. These genes were identified by the site of transposon insertion in the original mutants. Colonies were grown on the indicated media for the indicated length of time and photographed at 2.5× magnification. Note the complex and individualistic colony morphologies, particularly those of the ZAP1/zap1 and FGR22/fgr22 mutants.
Table I. Categories of mutants obtained from the screen.
| Category | Description of mutant | Total obtained |
|---|---|---|
| I | Less filamentous only on Spider | 117 |
| II | Less filamentous only on serum | 1 |
| III | Less filamentous on both media | 12 |
| IV | More filamentous only on Spider | 57 |
| V | More filamentous only on serum | 44 |
| VI | More filamentous on both media | 49 |
| VII | Hyperfilamentous with growth defect | 45 |
Seven general categories are defined, based on the phenotypes on Spider medium (Liu et al., 1994) and YEPD (yeast extract with peptone and 2% glucose) with 1% FCS.
After restreaking and verifying the original mutant phenotypes, the transposon insertion points were determined for a subset of the mutants; this analysis revealed 163 unambiguous insertion points (see Materials and methods). Eighty-eight insertion points were within structural genes, 47 were within the upstream regions of structural genes (within 500 nucleotide pairs of the presumptive translation startpoint) and 28 were downstream of structural genes (within 500 nucleotide pairs of the presumptive stop codon). Ten insertions were in unique sequence but were located >500 nucleotide pairs from the nearest structural gene. We have assigned the transposon insertion point to the nearest open reading frame (ORF), with the understanding that these assignments become less certain the further the insertion point is from the assigned ORF (Supplementary table 1 available at The EMBO Journal Online). When an identified gene had a close relative in S.cerevisiae, we used the same name for the C.albicans gene. The C.albicans genes identified in the screen that did not have close relatives in S.cerevisiae or other model organisms (see Materials and methods) were denoted FGR (filamentous growth regulator) and arbitrarily designated FGR1, FGR2, etc.
A critical issue in the study of filamentous growth in C.albicans concerns position effects on URA3 expression. Diminished URA3 expression (caused by, for example, insertion of URA3 into a transcriptionally ‘cold’ region of the genome) can have a direct effect on filamentous growth, as recently described by Sundstrom et al. (2002) for a URA3 insertion in the HWP1 gene. In this case, the filamentous defect could be reversed by adding uridine to the growth medium. In our library of transposon mutants, the only source of URA3 is the transposon; therefore, it was important to demonstrate that the filamentous growth effects exhibited by the different mutants were not due simply to differences in URA3 expression that might arise from different genomic positions of the transposon. To this end, the entire collection of mutants was re-tested on plates containing uridine; all of the mutant phenotypes were reproduced on this medium, and it is therefore highly unlikely that differences in URA3 expression could account for the mutant phenotypes. In this regard, we note that the URA3 gene in our transposon is flanked by several kilobases of DNA, and this may serve to buffer URA3 from position effects.
We next addressed whether the transposon insertions themselves (and not additional, unlinked genetic changes caused by the transformation or other manipulations) were the causes of the phenotypes. This question can be answered easily by simple backcrosses in organisms with well characterized sexual cycles, but is problematic in Candida. This issue is particularly worrisome since C.albicans is known to undergo spontaneous chromosomal rearrangements at relatively high frequencies. Although we cannot yet be certain for every one of the insertion mutants, we believe that the transposon insertion is nearly always the direct cause of the mutant phenotype. This belief is supported by several lines of evidence. First, six genes previously known to affect filamentous growth in C.albicans were identified in the screen (Table II), and the phenotypes of the newly isolated insertion mutations matched those described in the published studies. Secondly, genes closely related to those that affect filamentous growth in other organisms [for example, kin1 in Ustilago maydis (Lehmler et al., 1997) and PDE2 in S.cerevisiae (Lorenz and Heitman, 1997)] were identified in the screen. Thirdly, for 18 genes identified in the screen, we isolated at least two independent mutants; we know they are independent because the transposon insertion points within the ORF are different. In all 18 cases, the phenotypes produced by independent insertions in the same coding region were identical. Considering that 98% of the transposon insertion mutations did not show any filamentous phenotype, the isolation of independent insertion mutations in the same gene that cause the same phenotype argues very strongly that the screen is converging on a specific set of genes, and that the phenotypes are caused directly by the insertions.
Table II. Known regulators of filamentous growth identified by the screen.
| Name | Nearest homolog | Protein function | Organism | Reference |
|---|---|---|---|---|
| TUP1 | TUP1 | General repressor of transcription | S.cerevisiae | Braun and Johnson (1997) |
| PDE2 | PDE2 | cAMP phosphodiesterase | S.cerevisiae | Lorenz and Heitman (1997) |
| CBK1 | cot-1 | Ser/thr kinase | N.crassa | Yarden et al. (1992) |
| NRG1 | NRG1 | DNA-binding protein | S.cerevisiae | Braun et al. (2001); Murad et al. (2001) |
| RFG1 | ROX1 | DNA-binding protein | S.cerevisiae | Kadosh and Johnson (2001) |
| CZF1 | DNA-binding protein | C.albicans | Brown et al. (1999) |
Names of genes are indicated, along with their closest homolog as defined by BLAST searches (Altschul et al., 1997).
Finally, we performed a direct test of the idea that the transposon insertions were the cause of the mutant phenotypes. For this analysis, we chose 18 genes representing a variety of different phenotypes observed among the original insertion mutants. Using a PCR-based knockout strategy (Wilson et al., 2000), we directly inactivated one copy of each of the 18 genes in a wild-type strain and compared them, on several types of media, with the original transposon insertion strains. Of the 18 genes tested, disruption of 16 exactly reproduced the phenotype of the original transposon insertion, despite the fact that the phenotypes were often complex and highly specific to each mutant (Figure 3; Table III). Although this correlation is very strong, it is instructive to consider the two exceptions. A knockout of one allele of FGR32 gave a similar but more severe phenotype (hyperfilamentous) than that displayed by the original transposon insertion. Because the original transposon insertion is located in the C-terminal third of the predicted ORF, the transposon insertion may have generated an allele with a partial function. In the case of ERG5, the recreated deletion strain had a wild-type appearance. The transposon insertion in this mutant was ∼400 bases downstream of the ORF, and may affect ERG5 expression or localization or even influence a neighboring gene. The overall analysis indicates there is a very high probability that the insertion is the cause of the phenotype, and suggests that the majority of the phenotypes observed in the original mutants are due to the loss of function of one allele caused by the transposon insertion.
Table III. Recreation of phenotypes.
| Gene | Function | Tn location | Transposon phenotype | Recreated phenotype | Phenotype match |
|---|---|---|---|---|---|
| FGR3 | ??? | ORF | LF Spider | LF Spider | Yes |
| LIP2-3 | Lipase | Upstream | LF Spider | LF Spider | Yes |
| PDE2 | cAMP response | ORF | HF Spider/serum | HF Spider/serum | Yes |
| CBK1 | Ser/thr kinase | ORF | LF Spider | LF Spider | Yes |
| FGR13 | ??? | ORF | Variable morphology | Variable morphology | Yes |
| ERG5 | Lipid synthesis | Downstream | HF Spider/serum | Wild type | No |
| FGR5 | ??? | ORF | LF Spider | LF Spider | Yes |
| ZAP1 | DNA binding | ORF | LF Spider | LF Spider | Yes |
| FGR32 | Transport | ORF | HF serum | HF Spider/LF serum | No |
| ESC4 | Silencing | ORF | LF Spider | LF Spider | Yes |
| FGR22 | ??? | Downstream | HF Spider/LF serum | HF Spider/LF serum | Yes |
| FGR17 | DNA binding | ORF | LF Spider | LF Spider | Yes |
| SHE3 | ORF | HF Spider/serum | HF Spider/serum | Yes | |
| FGR27 | DNA binding | Upstream | HF Spider | HF Spider/HF serum | Yes |
| RDI1 | Cell polarity | ORF | HF Spider/serum | HF Spider/serum | Yes |
| TUP1 | Repression | ORF | HF Spider/serum | HF Spider/serum | Yes |
| NRG2 | DNA binding | ORF | HF Spider/serum | HF Spider | Yes |
| RFG1 | DNA binding | ORF | HF Spider/serum | HF Spider/serum | Yes |
Phenotypes are shown for transposon insertions and the recreated strains containing a deletion of the respective ORF. The location of the transposon insertion (upstream of an ORF, in an ORF and downstream of an ORF) is shown, as well as the phenotypes on Spider medium and YEPD with 1% FCS (HF, hyperfilamentous; LF, less filamentous). Of the 18 genes tested, 16 of the deletion strains matched the original transposon mutant phenotype.
Discussion
We identified 146 genes that affect the blastospore– filament transition in C.albicans. As discussed in the Introduction, this transition is believed to be central to the virulence of this human fungal pathogen.
General features of the blastospore–filament transition
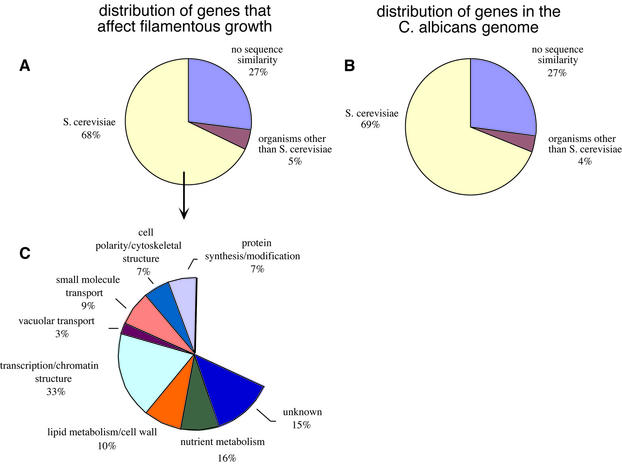
In this section, we discuss several broad generalizations based on the results of the screen. First, 40% of the genes identified do not have close relatives in S.cerevisiae and other well-studied organisms (Figure 4A). Because they lack close relatives in S.cerevisiae, it seems likely that these genes control aspects of filamentous growth that are specific to the ability of C.albicans to colonize and proliferate in warm-blooded animals, the only known hosts for this organism. Because these genes also lack close relatives in humans, they may identify useful targets for antifungal drugs. Some of these genes are members of much larger gene families in the Candida genome: for example, the FGR6 family has ∼10 members, and the FGR33/FGR38 family has >20 members. If the C.albicans genome as a whole is considered, approximately one-third of all C.albicans genes do not have close relatives in S.cerevisiae (Figure 4B). Thus the filamentous growth program in C.albicans is made up of both conserved and non-conserved genes in approximately the same proportion as those of the genome as a whole.
Fig. 4. Similarity of C.albicans genes that affect filamentous growth to genes in S.cerevisiae. (A) Roughly two-thirds of the C.albicans genes identified in the screen for filamentous growth mutants have closely related genes in S.cerevisiae. Nearly 30% lack significant similarity (as indicated by a BLAST score less than –15) to genes in any available databases. (B) The overall distribution of C.albicans genes relative to the S. cerevisiae genome. (C) Putative functions for the C.albicans genes that affect filamentous growth and have a close relative in S.cerevisiae. Each of these C.albicans genes was assigned a function based on the S.cerevisiae counterpart (see Supplementary table 1 for individual assignments). Categories are derived from the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/).
Approximately 60% of the 146 genes identified in the screen have close relatives in S.cerevisiae, and most of these can be assigned a general function (see Figure 4C; Tables II and IV) The largest class of these genes is involved with transcription and its regulation, indicating that the blastospore–filament transition is based on a large transcriptional program. In this class are 11 putative sequence-specific DNA-binding proteins (encoded by ADR1, CZF1, FCR1, FGR17, FGR27, GAT2, RFG1, NRG1, TEA1, YBR239c and ZAP1), three of which were known from previous work. It is known that filamentous growth can be induced by a variety of different environmental signals, including serum, starvation, contact, pH and temperature, and it seems likely that these gene-regulatory proteins are involved in coordinating these different responses. Consistent with this idea, mutations in five of the genes encoding DNA-binding proteins affect filamentous growth only on Spider (nutrient-poor) medium, one affects filamentous growth only on serum, and five affect filamentous growth on both types of media.
Table IV. Categories of genes identified by screen.
| Category | Examples |
|---|---|
| Transcription/chromatin structure | RPO21, SIN3, SPT5, SPT6, SRB9, ESC4, CDC39, CDC73, POB3, RPC40, TFG1, YTA6 |
| Cell wall biosynthesis, lipid metabolism | ECM4, ECM17, CDS1, ECM29, ECI1, ERG5, CHO1, FAS1, CSI2, SUR1 |
| Cell polarity/cytoskeletal structure | SHE3, RSR1, RDI1, ARC40, CDC25, BNI4 |
| Nutrient acquisition/metabolism | CDC19, FGR2, FCY2, CHA1, CDC25, ZAP1, RNR1, TFI1, ADR1, QNS1 |
| Filamentous growth | TUP1, RFG1, PDE2, CZF1, NRG1, NOT1 |
| Other signaling pathways | FUS3, STE13, STE23, CBK1 |
| Unknown function without related genes | FGR1–FGR51 |
Categories are defined based upon examples from S.cerevisiae and the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces).
Genes involved in more general aspects of transcriptional regulation were also identified in the screen. For example, we isolated mutations in TUP1 (which encodes a general transcriptional repressor) and mutations in components and associated proteins of the large CCR4/NOT1 complex (including CCR4, CDC73, CDC39, SRB9 and RBP4) (Liu et al., 1998). We also identified homologs of two members of the SET3 histone deacetylase complex of S.cerevisiae, Set3 and Snt1 (Pijnappel et al., 2001). These results indicate that these general regulators of transcription may have particularly prominent roles in the program of induction of filamentous growth.
Another major class of conserved genes identified in the screen is concerned with nutrient acquisition and metabolism. This observation makes intuitive sense, since nutrient composition of the medium—especially the presence of certain sugars (N-acetylglucosamine and glucose, for example) and amino acids (proline, for example)—is a major determinant of filamentous growth. It seems plausible that at least some of these mutations skew the metabolism of the cell, making it either more or less sensitive to the normal signals for the blastospore– filamentous growth transitions. Included in this group are nine genes encoding small molecule transporters; these may well have roles in the acquisition or sensing of particular nutrients that regulate filamentous growth.
The next largest class of conserved genes identified in the screen are involved with lipid and cell wall biosynthesis. Again, this finding makes intuitive sense, as it is known that the C.albicans plasma membrane and cell wall are remodeled extensively during the transition from the blastospore to filamentous forms of growth. The genetic screen identifies specific enzymes and pathways that probably function in this remodeling. These include several genes related to the ECM genes of S.cerevisiae (identified in that organism by a screen for sensitivity to calcofluor white) (Lussier et al., 1997), genes involved in chitin synthesis (CSI2; Roncero et al., 1988) and genes required for synthesis of membrane lipids or cell wall components (CDS1, CHO1, FAS1 and ERG5; Daum et al., 1998)
Other classes of genes identified in the screen include those encoding cell polarity and cytoskeletal components (13 genes) and those involved in protein synthesis and modification (10 genes). Again, all these findings make conceptual sense, as blastospore and filamentous cells differ dramatically in both size and shape.
Specific insights into the regulation of filamentous growth in C.albicans
The previous discussion outlines generalizations and broad trends amongst the mutations that affect the control of filamentous growth in C.albicans. However, each individual mutant typically suggests a specific model or reveals a previously unrecognized feature of filamentous growth. Obviously, space does not permit a discussion for all 146 mutants, but two examples will be given where a mutant or group of mutants suggests a specific idea.
Analysis of the C.albicans ZAP1 mutant (which encodes a Zn-finger transcriptional regulator) illustrates the idea that filamentous growth is not simply an on–off switch, but can result from distinct transcriptional pathways. On Spider (nutrient poor) medium, colonies of wild-type C.albicans strains (CAF2-1) exhibit two types of filamentous growth: a wrinkling of the central colony (indicative of a high proportion of hyphae and pseudohyphae) and peripheral hyphae that emanate from the colony (see Figure 3, third row). Analysis of the ZAP1/zap1 mutant clearly shows that these two types of filamentous growth are induced by distinct pathways: the ZAP1/zap1 mutant exhibits a smooth central colony (indicative of blastospores), but normal numbers of peripheral hyphae (see Figure 3, third row). This observation also shows that Zap1 is required for the formation of filaments in the central portion of colonies but is relatively unimportant for peripheral hyphae. Environmental conditions in the center of a colony differ dramatically from those on the periphery, and the analysis of this mutant reveals that C.albicans recognizes and responds to this difference. Candida albicans Zap1 is very closely related to S.cerevisiae Zap1, which regulates zinc homeostasis (Zhao and Eide, 1997). In C.albicans, it is likely that Zap1 activates a specialized program of filamentous growth that is specific to the environmental conditions (perhaps severe deprivation of one or more critical nutrients, maybe even zinc) present at the center of the colony but not at the colony periphery. Consistent with this idea, Bedell and Soll (1979) showed that zinc levels in the media can affect filamentous growth in C.albicans. In any case, analysis of the ZAP1/zap1 mutant clearly distinguishes peripheral hyphae from those at the colony center and has thus revealed a new aspect of filamentous growth and suggests a specific set of models that can be tested experimentally.
As a second example of a specific insight arising from the screen, STE13 and STE23 were identified as mutants reduced in filamentous growth. In S.cerevisiae, the products of these genes are proteases involved in processing of the mating pheromones α-factor (STE13) and a-factor (STE23). Mating pheromones are involved in filamentous growth of other fungi, including Cryptococcus neoformans, Ustilago maydis and, under special circumstances, S.cerevisiae (Spellig et al., 1994; Wang and Heitman, 1999; Yue et al., 1999; Erdman and Snyder, 2001). Additionally, C.albicans mutants lacking Kex2, another protease necessary for processing of the α-factor precursor in S.cerevisiae, are deficient in filamentous growth and mating of α-mating type (Newport and Agabian, 1997; Magee et al., 2002). These results taken together suggest that mating factors (or other peptides that are similarly processed) may be required for filamentous growth.
The utility of haploinsufficiency and its comparison with other genetic screening strategies in C.albicans
Based on the number of genes (18) for which we obtained multiple insertions, we estimate from the Poisson distribution that we have identified ∼30% of the genes that would, in principle, come from this screen if it were carried out on a significantly larger scale. In addition, we do not know whether our reliance on haploinsufficiency or whether any details of the transposon mutagenesis scheme systematically excluded additional genes important for the blastospore–filament transition. However, since we did obtain a relatively large number of mutants, it is perhaps worth considering why the reliance on haploinsufficiency worked as well as it did. Three possibilities could explain why haploinsufficiency seems so prevalent in C.albicans: (i) Since C.albicans exists almost exclusively in the diploid state, gene dosage may have become so finely tuned that the organism is highly sensitive to a 50% loss of a gene product. (ii) A transvection effect could be responsible for depressing gene expression from the intact copy of the gene when the other copy is deleted or has suffered a transposon insertion. (iii) The two alleles of a given gene could have differentiated, encoding proteins that have subtly different functions. Inactivating one allele might thereby be expected to produce a phenotype. We believe the first idea is generally correct: an examination of five different hemizygous knockout strains revealed that the transcriptional level of the remaining gene—as determined by northern analysis—is at least 50% the level of that of the parent strain (see Supplementary data). This observation rules out transvection effects as an overall explanation for haploinsufficiency and suggests that gene dosage can account for it in many cases. Consistent with this idea is the observation reported by many laboratories that the phenotype of a hemizygous deletion strain is simply a less severe version of that seen in the homozygous mutation (see Results). This idea was tested explicitly for eight genes identified in our screen: FGR17, YKR029C, ZAP1, YBR239C, TUP1, NRG1, RFG1, CZF1 and CBK1. For all cases except ZAP1, the homozygous deletion mutants showed a more severe phenotype than did the hemizygous mutants; for ZAP1, the homozygous and hemizygous mutants were identical in phenotype, as determined by colony morphology on a variety of media (M.A.Uhl and A.D.Johnson, unpublished observations). Thus, although the general trend holds for our mutants, there is at least one exception. For some specific instances of haploinsufficiency (perhaps that of ZAP1), allele diversification could be pertinent. Allele differences in C.albicans range from the extreme case of the MTL (Hull and Johnson, 1999) to more limited examples where the products of the two alleles differ by several amino acid substitutions (see Yesland and Fonzi, 2000). It is therefore possible that for some of our insertion mutations, haploinsufficiency is caused by the inactivation of the more active of a pair of alleles, or of an allele that has diversified from its partner.
Two previous genetic screens have been carried out directly in C.albicans, and both relied on dominant mutations rather than haploinsufficiency. In one, a collection of strains was constructed by the restriction enzyme-mediated integration of a gene library under the control of the C.albicans MAL2 promoter. This collection was then screened for maltose-dependent filamentous growth on medium that does not ordinarily promote filamentous growth; the strategy thus relied on a phenotype dependent on overexpression of a gene. Of 2000 integrants screened, a single gene (CZF1) was identified that is an activator of filamentous growth (Brown et al., 1999); an insertion in CZF1 was also identified in our screen as being reduced in filamentous growth (Table II). A second C.albicans screen was based on inhibitory RNA. Here a cDNA library in reverse orientation from a galactose-inducible promoter was integrated in the genome and 2000 transformants were screened for galactose-inducible growth defects. Eighty-six genes (whose antisense transcription caused growth defects) were identified by this screen, including a number whose orthologs are known to be essential for growth in S.cerevisiae. Phenotypes of six of these genes were tested by making a deletion of one copy of the gene and scoring for growth defects; four of the six heterozygous strains had growth defects similar to the original (De Backer et al., 2001). Given the relatively non-specific nature of the phenotype tested, it is not yet known how many of the antisense constructs produced their phenotypes by specifically inactivating the homologous gene.
The results from our transposon mutagenesis indicate that a forward screen utilizing haploinsufficiency is feasible for identifying genes with recessive phenotypes in C.albicans. As described, we screened 18 000 insertion mutants and uncovered 146 genes involved in filamentous growth, only six of which had been identified previously as having a role in this process. This same approach should also be readily adaptable to other genetic screens in C.albicans, provided that sensitive laboratory conditions are available to monitor the phenotype in question. Haploinsufficiency can also be used to identify targets for drugs, as mutations in one copy of the target gene can render a diploid more sensitive to the drug; this strategy was used recently in S.cerevisiae (Giaever et al., 1999). Finally, haploinsuffiency has also been noted for genes required for virulence in systemic models of candidosis (e.g. Lo et al., 1997; Gale et al., 1998; Braun et al., 2000), suggesting that our library can also be utilized for general screens in vivo for virulence factors in C.albicans.
Materials and methods
Strains and media
Candida albicans CAI4 (ura3/ura3) was used as the parental strain for transposon mutagenesis (Fonzi and Irwin, 1993). RM1000 (Negredo et al., 1997) (ura3/ura3 his1/his1) was used for PCR-based disruption of genes analyzed in the experiment in Figure 3. For routine growth of C.albicans, YEPD was used. URA+ transformants were selected on media lacking uridine. Screens of the transposon mutants were carried out on Spider medium and YEPD + 1% FCS.
Transposon mutagenesis
Large-scale transposon mutagenesis of C.albicans was carried out as schematized in Figure 1. Details of the procedure are described in the Supplementary data.
Choice of media for haploinsufficiency screens
As described above, Spider medium (a nutrient-poor medium; Liu et al., 1994) could be used to distinguish known hemizygous deletion strains. For example, when grown for 4 days on Spider medium (Liu et al., 1994), colonies of the CLN1/cln1 and EFG1/efg1 were significantly less wrinkled and had fewer peripheral hyphae penetrating the agar when compared with the parent strain. Microscopic examination of cells isolated from the wrinkled portions of the colonies revealed a significantly lower fraction of filamentous cells in the two mutant strains than in the parent strain. These observations are consistent with prior studies implicating CLN1 and EFG1 as activators of filamentous growth (Stoldt et al., 1997; Loeb et al., 1999). In contrast, TUP1/tup1 colonies were more wrinkled than those of CAF2-1, and the colonies had pronounced ‘domes’. Examination of these cells showed a much higher proportion of filamentous cells in the TUP1/tup1 strain, verifying previous reports that TUP1 is formally a negative regulator of filamentous growth (Braun and Johnson, 1997). These results support the idea that differences in colony appearance on Spider medium could be used to monitor differences in filamentous growth and that a genetic screen based on haploinsufficiency might be feasible. Similar observations revealed that YPD + 1% FCS could also be used to distinguish the colony morphologies of these three strains easily.
Library screens
Plasmids containing the transposon insertions were digested with BsrGI, which should recreate the original linear genomic DNA fragment. Linear DNA was transformed into C.albicans by the Fast-Track method (Geno Technology, St Louis, MO). Colonies were patched onto medium without uridine and then replicated onto Spider medium and YEP with 1% FCS. Plates were incubated at 30°C and colonies were observed every day over the course of 2 weeks. Colonies were scored for appearance of wrinkles and peripheral hyphae; any colony that demonstrated a difference compared with CAF2-1 (wild-type control) was rescreened on Spider and YEP with 1% FCS.
Identification of transposon junctions
Following confirmation of mutant phenotypes, the transposon insertion points were determined for 228 mutant strains using a nested vectorette PCR methodology (Vectorette system; Sigma/Genosys, The Woodlands, TX). Details are described in the Supplementary data. Roughly 20% (45/228) of the insertions were found to occur in the major repeat sequence of C.albicans, RPS1, which comprises ∼10% of the Candida genome and occurs in all but one of the chromosomes. Seventy-three percent of the 45 RPS1 insertions obtained in the screen gave a hyperfilamentous phenotype. No discernible ORFs lie near the RPS1 sequences, and the basis of the phenotype arising from these insertions currently is under investigation. One possibility, suggested by the work of Barton and Scherer (1994), is that insertion into RPS1 sequences causes chromosomal rearrangements, and that those subsequent rearrangements are the cause of the mutant phenotypes. CHEF gel analysis was performed on 10 RPS1 insertion mutants that displayed a variety of phenotypes. No gross chromosomal rearrangements were detected, suggesting either that rearrangements occurred and were too small to detect by this method or that the phenotypes were due to some other effect of insertion at the RPS1 locus. In any case, all the RPS1 insertions were not studied further and are not included in the analysis presented herein.
BLAST searches
Candida albicans gene sequences were conceptually translated into protein and compared with the S.cerevisiae genome using BLAST (Altschul et al., 1997). Predicted protein sequences with a BLAST score of less than E-15 were assigned the name of the corresponding S.cerevisiae gene. Genes whose predicted products fell under that cut-off were assigned an FGR name. In one instance, a gene was assigned an FGR name (FGR22), although it had high similarity to a predicted phospholipase C from Bacillus subtilis. In this case, the name from B.subtilis did not match the naming conventions from C.albicans.
Disruption of genes
The C.albicans strain RM1000 (ura3/ura3 his1/his1) (Negredo et al., 1997) was used for gene knockouts. Although the strain background for transposon mutagenesis (strain CAI4) and reconstruction of the disruptions (strain RM1000) differed slightly, there was no apparent difference in filamentous growth between RM1000 and CAI4 or URA+ derivatives of the two strains on Spider medium or YEP with 1% FCS (data not shown). Genes were disrupted using a PCR-based strategy (Wilson et al., 1999, 2000) by a lithium acetate transformation protocol (Gietz et al., 1995). Transformants were selected on media lacking uridine, and disruptions were confirmed by PCR utilizing a primer hybridizing upstream of the disrupted gene and a primer hybridizing to URA3 (Braun and Johnson, 2000).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
Sequence data for C.albicans were obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida. Sequencing of C.albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. We are particularly grateful to Richard Bennett for carrying out the haploinsufficiency experiment described in the Supplementary data. We thank Peggy Cotter for the gift of pEGBR, Anita Sil for advice on vectorette PCR, Makoto Tsunozaki and Jen Ramond for strain construction, members of the Johnson laboratory for input, Ira Herskowitz, Anita Sil, Jeff Cox and Hiten Madhani for valuable comments on the manuscript, and Burk Braun for construction of a C.albicans ORF database that greatly facilitated analysis of this work. This work was supported by a Damon Runyon–Walter Winchell post-doctoral research grant to M.A.U. and NIH grant GM37049 to A.D.J. M.B. and N.C. are employees of HHMI.
Note added in proof
Recently Davis et al. (2002) developed a different type of transposon-based mutagenesis scheme for C.albicans. Using this methodology, they screened 253 C.albicans strains (each of which had both copies of a known gene disrupted) and found three genes that affected pH-dependent filamentous growth.
References
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton R.C. and Scherer,S. (1994) Induced chromosome rearrangements and morphologic variation in Candida albicans. J. Bacteriol., 176, 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell G.W. and Soll,D.R. (1979) Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect. Immun., 26, 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D.E. and Howe,M.M. (1989) Mobile DNA. American Society for Microbiology Press, Washington, DC.
- Biery M.C., Stewart,F.J., Stellwagen,A.E., Raleigh,E.A. and Craig,N.L. (2000) A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res., 28, 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B.R. and Johnson,A.D. (1997) Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science, 277, 105–109. [DOI] [PubMed] [Google Scholar]
- Braun B.R. and Johnson,A.D. (2000) TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics, 155, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B.R., Head,W.S., Wang,M.X. and Johnson,A.D. (2000) Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics, 156, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J. and Gow,N.A. (1999) Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol., 7, 333–338. [DOI] [PubMed] [Google Scholar]
- Brown D.H. Jr, Giusani,A.D., Chen,X. and Kumamoto,C.A. (1999) Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol., 34, 651–662. [DOI] [PubMed] [Google Scholar]
- Calderone R.A. and Fonzi,W.A. (2001) Virulence factors of Candida albicans. Trends Microbiol., 9, 327–335. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Ghori,N. and Falkow,S. (1999) An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science, 285, 578–582. [DOI] [PubMed] [Google Scholar]
- Daum G., Lees,N.D., Bard,M. and Dickson,R. (1998) Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast, 14, 1471–1510. [DOI] [PubMed] [Google Scholar]
- Davis D.A., Bruno,V.M., Loza,L., Filler,S.G. and Mitchell,A.P. (2002) Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics, 162, 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer M.D. et al. (2001) An antisense-based functional genomics approach for identification of genes critical for growth of Candida albicans. Nat. Biotechnol., 19, 235–241. [DOI] [PubMed] [Google Scholar]
- Erdman S. and Snyder,M. (2001) A filamentous growth response mediated by the yeast mating pathway. Genetics, 159, 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J.F. (2000) Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology, 146, 1763–1774. [DOI] [PubMed] [Google Scholar]
- Feng Q., Summers,E., Guo,B. and Fink,G. (1999) Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol., 181, 6339–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi W.A. and Irwin,M.Y. (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics, 134, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale C.A., Bendel,C.M., McClellan,M., Hauser,M., Becker,J.M., Berman,J. and Hostetter,M.K. (1998) Linkage of adhesion, filamentous growth and virulence in Candida albicans to a single gene, INT1. Science, 279, 1355–1358. [DOI] [PubMed] [Google Scholar]
- Giaever G., Shoemaker,D.D., Jones,T.W., Liang,H., Winzeler,E.A., Astromoff,A. and Davis,R.W. (1999) Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet., 21, 278–283. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl,R.H., Willems,A.R. and Woods,R.A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast, 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Gillum A.M., Tsay,E.Y. and Kirsch,D.R. (1984) Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S.cerevisiae ura3 and E.coli pyrF mutations. Mol. Gen. Genet., 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Gimeno C.J., Ljungdahl,P.O., Styles,C.A. and Fink,G.R. (1992) Unipolar cell divisions in the yeast S.cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell, 68, 1077–1090. [DOI] [PubMed] [Google Scholar]
- Haynes K. (2001) Virulence in Candida species. Trends Microbiol., 9, 591–596. [DOI] [PubMed] [Google Scholar]
- Hull C.M. and Johnson,A.D. (1999) Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science, 285, 1271–1275. [DOI] [PubMed] [Google Scholar]
- Kadosh D. and Johnson,A.D. (2001) Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol., 21, 2496–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler J.R. and Fink,G.R. (1996) Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl Acad. Sci. USA, 93, 13223–13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E. et al. (1996) Signal transduction through homologues of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl Acad. Sci. USA, 93, 13217–11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmler C., Steinberg,G., Snetselaar,K.M., Schliwa,M., Kahmann,R. and Bolker,M. (1997) Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J., 16, 3464–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kohler,J. and Fink,G.R. (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science, 266, 1723–1726. [DOI] [PubMed] [Google Scholar]
- Liu H.Y., Badarinarayana,V., Audino,D.C., Rappsilber,J., Mann,M. and Denis,C.L. (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J., 17, 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.J., Kohler,J.R., DiDomenico,B., Loebenberg,D., Cacciapuoti,A. and Fink,G.R. (1997) Nonfilamentous C.albicans mutants are avirulent. Cell, 90, 939–949. [DOI] [PubMed] [Google Scholar]
- Loeb J.D., Sepulveda-Becerra,M., Hazan,I. and Liu,H. (1999) A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol., 19, 4019–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M.C. and Heitman,J. (1997) Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J., 16, 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M. et al. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cere visiae. Genetics, 147, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B.B, Legrand,M., Alarco,A.M., Raymond,M. and Magee,P.T. (2002) Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol., 46, 1345–1351. [DOI] [PubMed] [Google Scholar]
- Negredo A., Monteoliva,L., Gil,C., Pla,J. and Nombela,C. (1997) Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology, 143, 297–302. [DOI] [PubMed] [Google Scholar]
- Newport G. and Agabian,N. (1997) KEX2 influences Candida albicans proteinase secretion and hyphal formation. J. Biol. Chem., 272, 28954–28961. [DOI] [PubMed] [Google Scholar]
- Odds F.C. (1988) Candida and Candidosis. Baillière Tindall, London, UK.
- Palecek S.P., Parikh,A.S. and Kron,S.J. (2002) Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology, 148, 893–907. [DOI] [PubMed] [Google Scholar]
- Pijnappel W.W. et al. (2001) The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1 and is a meiotic-specific repressor of the sporulation gene program. Genes Dev., 15, 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried J.L. and Collmer,A. (1987) An nptI–sacB–sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene, 57, 239–246. [DOI] [PubMed] [Google Scholar]
- Roncero C., Valdivieso,M.H., Ribas,J.C. and Duran,A. (1988) Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J. Bacteriol., 170, 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- San-Blas G. et al. (2000) Fungal morphogenesis and virulence. Med. Mycol., 38, 79–86. [PubMed] [Google Scholar]
- Spellig T., Bolker,M., Lottspeich,F., Frank,R.W. and Kahmann,R. (1994) Pheromones trigger filamentous growth in Ustilago maydis. EMBO J., 13, 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A.E. and Craig,N.L. (1997) Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. Genetics, 145, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt V.R., Sonneborn,A., Leuker,C.E. and Ernst,J.F. (1997) Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J., 16, 1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom P., Cutler,J.E. and Staab,J.F. (2002) Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect. Immun., 70, 3281–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl M.A. and Johnson,A.D. (2001) Development of Streptococcus thermophilus LacZ as a reporter gene for Candida albicans. Microbiology, 147, 1189–1195. [DOI] [PubMed] [Google Scholar]
- Wang P. and Heitman,J. (1999) Signal transduction cascades regulating mating, filamentation and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol., 2, 358–362. [DOI] [PubMed] [Google Scholar]
- Whiteway M. (2000) Transcriptional control of cell type and morphogenesis in Candida albicans. Curr. Opin. Microbiol., 3, 582–588. [DOI] [PubMed] [Google Scholar]
- Wilson R.B., Davis,D. and Mitchell,A.P. (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol., 181, 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.B., Davis,D., Enloe,B.M. and Mitchell,A.P. (2000) A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast, 16, 65–70. [DOI] [PubMed] [Google Scholar]
- Yesland K. and Fonzi,W.A. (2000) Allele-specific gene targeting in Candida albicans results from heterology between alleles. Microbiology, 146, 2097–2104. [DOI] [PubMed] [Google Scholar]
- Yue C., Cavallo,L.M., Alspaugh,J.A., Wang,P., Cox,G.M., Perfect,J.R. and Heitman,J. (1999) The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics, 153, 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. and Eide,D.J. (1997) Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]