Abstract
Mis12 is a kinetochore protein essential for equal chromosome segregation and is evolutionarily conserved from yeast to human. In this study, we report the isolation and characterization of suppressors of the mis12 mutant in fission yeast. Our results indicate that Mis12 is negatively regulated by a highly conserved protein phosphatase Ppe1 (scSit4/dmPPV/hPP6) or its bound partner Ekc1 (scSAP), and it is positively regulated by a counteracting kinase Gsk3. Mass spectrometry analysis shows that at least two sites in Mis12 are phosphorylated. This mechanism of suppression occurs at the level of localization recovery of Mis12 to the kinetochore chromatin. Consistently, Mis12 and a subpopulation of Ppe1/Ekc1 were found to behave like non-histone-type chromatin-associating proteins in the chromatin fractionation assay. Mutant analysis of Ppe1 and Ekc1 revealed that they are important for faithful chromosome segregation, as the mutants exhibited unequal chromosome segregation similar to mis12 in the presence of a low concentration of tubulin poison. Ppe1/PP6 directly or indirectly modulates kinetochore chromatin protein Mis12 to ensure progression into normal anaphase.
Keywords: chromatin/kinetochore/phosphatase/S.pombe
Introduction
Equal segregation of chromosomes to two daughter cells is a process fundamental for genome inheritance. Centromeres are the chromosomal attachment sites for spindle microtubules during mitosis and meiosis. Proteins are located on the centromere DNAs to form a highly ordered complex structure, the kinetochore. The kinetochore has vital roles in ensuring the fidelity of chromosome segregation.
We have employed the fission yeast Schizosaccharo myces pombe as an excellent model organism for the analyses of centromere/kinetochore structure and function. Centromeres of fission yeast (35–110 kb) are composed of two types of chromatin domains, specialized central chromatin and outer heterochromatin (e.g. Takahashi et al., 1992). The central domains contain 15 kb unique sequences and form the specialized chromatin, which displays a smeared nucleosome ladder after micrococcal nuclease (MNase) digestion. The outer regions produced the patterns of regular nucleosomal ladders (Polizzi and Clarke, 1991; Takahashi et al., 1992). This two-domain structure bears some resemblance to the higher eukaryotic centromeres (Blower and Karpen, 2001; Oegema et al., 2001). Stability tests of various artificial minichromosomes of fission yeast established that the central regions are essential for centromere function in both mitosis and meiosis, whereas outer repetitious regions make minor contributions in mitosis but are indispensable for meiotic chromosome segregation (e.g. Takahashi et al., 1992). Kinetochore microtubules as well as microtubule-associating proteins and spindle checkpoint proteins are likely to interact at the surface of central centromere regions during mitosis (Garcia et al., 2001; Nakaseko et al., 2001; Toyoda et al., 2002). Identification of proteins that bind to the central regions and form the structure for higher ordered kinetochore chromatin is, therefore, important for understanding mitotic kinetochore functions.
Fission yeast mis6-302 and mis12-537 mutants exhibit unequal segregation of regular chromosomes when shifted to a restrictive temperature. In these mutants, sister chromatids were segregated to the daughter cells at anaphase without a long delay, but the segregation patterns of sister chromatids were random. Consequently, these mutations produced aneuploid cells with a high frequency. Localization studies using green fluorescent protein (GFP) and chromatin immunoprecipitation (ChIP) assays established that Mis6 and Mis12 are bound to the central centromere regions throughout the cell cycle. MNase digestion experiments indicated that these proteins are essential for forming the specialized chromatin of the central centromere regions (Saitoh et al., 1997; Goshima et al., 1999). However, no genetic or physical interactions have been found between Mis6 and Mis12.
CENP-A, a histone H3 variant protein, is localized exclusively at centromere regions and plays an essential role in chromosome segregation from yeast to humans (Stoler et al., 1995; Howman et al., 2000; Takahashi et al., 2000; Blower and Karpen, 2001; Oegema et al., 2001). In fission yeast, spCENP-A is located to central centromere regions in a Mis6-dependent manner, and an spCENP-A mutant shows missegregation and central chromatin disruption phenotypes identical to mis6/mis12 (Takahashi et al., 2000). spCENP-A, therefore, appears to confer specialized nucleosomes to central DNAs. In other organisms so far investigated, Mis6 homologue depletion is unlikely to affect CENP-A localization at centromeres and, instead, CENP-A is indispensable for Mis6 recruitment to centromeres (Measday et al., 2002; Nishihashi et al., 2002; Goshima et al., 2003). CENP-A is also essential for kinetochore localization of other components in higher eukaryotes. These results lead to the model that CENP-A acts as a core nucleosome for the whole kinetochore chromatin architecture (reviewed by Smith, 2002).
On the other hand, there has been little evidence that shows the interaction and obvious localization dependence between Mis12 and CENP-A, both of which locate in the central centromere regions. Localization of Mis12 was unaffected in the absence of CENP-A, and vice versa in fission yeast [temperature-sensitive (ts) mutant analyses] and HeLa cells [RNA interference (RNAi) analyses], indicating that the loading pathway for Mis12 is independent of CENP-A (Takahashi et al., 2000; Goshima et al., 2003). The reduction of human hMis12 or CENP-A by RNAi led to chromosome misalignment at metaphase, chromosome lagging in anaphase and micronuclei formation in interphase. These genetic and cytological results established that Mis12 is an important conserved kinetochore protein in an independent pathway forming kinetochore chromatin (Goshima et al., 2003).
Although CENP-A’s molecular characterization as well as its relationships to other kinetochore components have been well investigated, such knowledge was scarce for Mis12. The initial aim of this study was to identify proteins that interact directly or indirectly with Mis12 through genetic screening. To this end, extragenic and high copy suppressors were isolated for the fission yeast mis12 mutant. Through the analyses of extragenic suppressors, Mis12 was found to be under the control of protein phosphorylation–dephosphorylation by a counteracting kinase and phosphatase.
Results
Suppression of mis12-537 by mutations in conserved proteins Ppe1 and Ekc1
We attempted to isolate extragenic or high dosage suppressors for mis12-537 in the hope that identification of suppressor gene products would produce an understanding of the regulation of Mis12 kinetochore function. Among 500 spontaneously isolated pseudo-revertants of the mis12-537 mutant, 24 strains were found to be cold sensitive (cs) and could grow at 36°C but failed at 22°C (Goshima et al., 1999). These extragenic suppressors are designated ekc (extragenic suppressor of kinetochore) mutants. Genetic analysis by crossing (Materials and methods) established that these 24 mutant alleles were derived from only two genetic loci (designated ekc1 and ekc2).
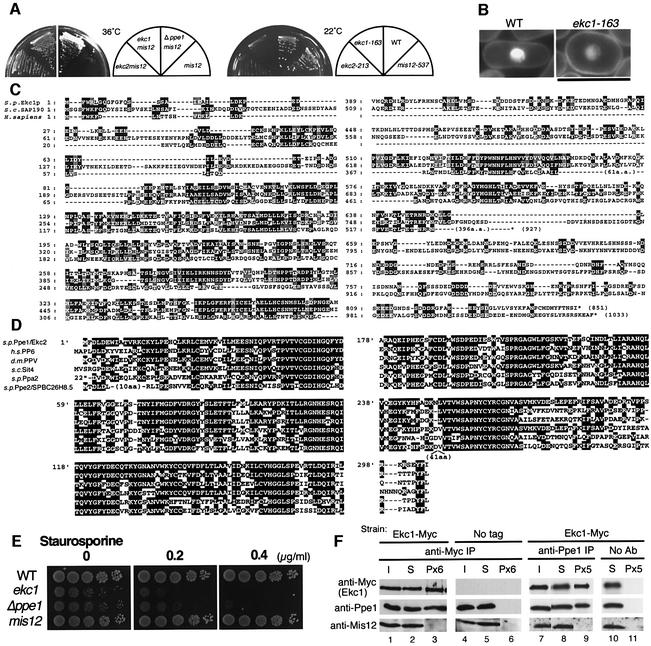
As shown in Figure 1A, the double mutants ekc1 mis12 and ekc2 mis12 formed colonies at 36°C, while single segregants ekc1 and ekc2 failed to form colonies at 22°C. Curiously, all the ekc mutant strains invariably revealed pear-shaped cells at any temperature (Figure 1B). Regulation of cell morphology may be defective in ekc mutant cells.
Fig. 1. Extragenic suppression of mis12-537 by ppe1 and ekc1 mutants. (A) Left: double mutants mis12-537 ekc1-163, mis12-537 ekc2-213 and mis12-537 Δppe1 could form colonies at 36°C, whereas single mis12-537 did not. Right: single ekc1 and ekc2 mutants were cold sensitive and unable to form colonies at 22°C. (B) ekc1-163 is pear-shaped even at the permissive temperature. Bar, 10 µm. (C) Amino acid sequence alignment of S.pombe Ekc1, S.cerevisiae SAP190 and Homo sapiens KIAA0685. Identical residues are boxed, and similar residues are hatched. Saccharomyces cerevisiae has four SAPs (SAP4, 190, 185 and 155), while human has three similar sequences (KIAA0685, KIAA1115 and KIAA1558). (D) Sequence alignment of S.pombe Ppe1 phosphatase, S.cerevisiae Sit4, H.sapiens PP6, D.melanogaster PPV and S.pombe Ppa2 and Ppe2/SPBC26H8.05. (E) Exponentially growing wild-type, ekc1, ppe1 and mis12 cells were spotted after dilution onto YPD plate that contained 0.2 or 0.4 µg/ml staurosporine, and incubated at 33°C. (F) The strain containing the integrated Ekc1-Myc was immunoprecipitated by anti-Myc antibodies (lanes 1–3) and anti-Ppe1 antibodies (lanes 7–9), and immunoblotting was used to detect the three proteins Ekc1, Ppe1 and Mis12 in the resulting precipitates. Beads without antibodies (lanes 10 and 11) or the strain without myc epitopes (lanes 4–6) were used as control. I, input; S, supernatant; P, immunoprecipitate. Equal amounts of I and S were loaded, whereas immunoprecipitates were 6- (Px6) or 5-fold (Px5) concentrated.
The ekc1+ gene was cloned from a genomic DNA library using transformation of the cs phenotype of the ekc1 mutant. Ekc1 was identical to SPCC663.01, an uncharacterized gene. The gene product contains 851 amino acids and is 30–37% identical to the budding yeast SAPs (SAP190, SAP185, SAP155 and SAP4), which are phosphatase modulators and bind to Sit4 phosphatase as a positive regulator (Luke et al., 1996). The SAP proteins are conserved from yeast to humans (Figure 1C), and although Saccharomyces cerevisiae and human have four and three SAPs, respectively, the genome of Schizosaccharomyces pombe has only one.
We speculated that Ekc2 might be identical to Ppe1 phosphatase (Matsumoto and Beach, 1993; Shimanuki et al., 1993), the fission yeast orthologue of scSit4/hPP6/dmPPV (Mann et al., 1993; Bastians and Ponstingl, 1996; Cohen, 1997). The cs phenotype of the ekc2-213 mutant was indeed rescued by a multicopy plasmid carrying the authentic ppe1+ gene. Subsequent crossing between ekc2-213 and the Δppe1 deletion mutant confirmed that ekc2 was allelic to ppe1. Consistently, the double mutant Δppe1 mis12 could grow at 36°C (Figure 1A, left panel). The deletion of ppe1+ is known to cause the cs phenotype and pear-shaped or round cells at both permissive and restrictive temperatures (Shimanuki et al., 1993).
Ppe1 is a protein phosphatase that is highly conserved from yeast to human (Figure 1D). Indeed, the human orthologue PP6 could suppress the phenotype of budding yeast sit4 and fission yeast ppe1 mutants (Bastians and Ponstingl, 1996). The amino acid sequence of Ppe1 is similar (51% identical) to that of type 2A phosphatase Ppa2 (Kinoshita et al., 1990), but Δppa2, the deletion mutant of Ppa2, did not suppress the ts phenotype of mis12-537 (data not shown). Schizosaccharomyces pombe has another highly conserved protein phosphatase designated Ppe2 (SPBC26H8.05), the orthologue of human and fly PP4 (Helps et al., 1998), and budding yeast Pph3 (Ronne et al., 1991). It is highly similar (53% identity) to Ppe1, but the deletion mutant Δppe2 that was viable did not suppress the ts phenotype of mis12-537 (data not shown). The ts phenotype of mis6-302 was not suppressed by Δppe1. The suppression of mis12-537 by the Δppe1 mutant was thus quite specific.
While Δppe1 was hypersensitive to staurosporine, a potent kinase inhibitor (Toda et al., 1991; Shimanuki et al., 1993), ekc1 mutants were similarly sensitive to the drug (Figure 1E). To determine if there was a physical interaction between Ekc1 and Ppe1, we performed immunoprecipitation using polyclonal anti-Ppe1 antibodies and monoclonal anti-myc antibodies for a strain chromosomally integrated with the Ekc1-Myc gene (Figure 1F). Quantitative analysis indicated that 20% of the total Ekc1-Myc was precipitated by anti-myc antibody (comparison between lanes 1 and 3). In the precipitates, 20% of Ppe1 was co-precipitated (lanes 1 and 3). Similar levels of Ppe1 and Ekc1-Myc co-precipitation were obtained for the precipitates made by anti-Ppe1 antibodies (lanes 7 and 9). However, Mis12 protein was not detected in the precipitates with either anti-Ppe1 or anti-Ekc1-Myc antibodies under the experimental conditions used (lanes 3 and 9). Ekc1 thus stably binds to Ppe1, but not to Mis12. As the functional link between Ppe1–Ekc1 and kinetochore is a novel aspect, we investigated the mitotic roles of Ppe1–Ekc1.
Ekc1 is enriched in the nucleus
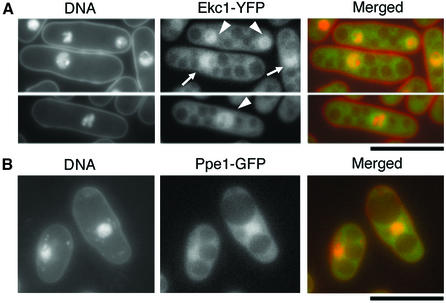
To determine its localization, Ekc1 tagged with yellow fluorescent protein (YFP) at the C-terminus was chromosomally integrated in wild type and observed (Figure 2A). The Ekc1–YFP signal was enriched in the nucleus in both interphase and mitosis. The signal was also found in the cytoplasm, except for vacuole-like structures. The nuclear signals of Ekc1 closely resembled that of nuclear chromatin in interphase (indicated by the arrow), whereas the mitotic nuclear signals (indicated by the arrowhead) were more diffuse in the whole nucleus, suggesting that a pool of Ekc1 might be dissociated from nuclear chromatin during mitosis.
Fig. 2. Nuclear-enriched localization of Ekc1–YFP and Ppe1–GFP. Chromosomally integrated Ekc1–YFP (A, green) and Ppe1–GFP (phosphatase-dead) (B, green) showed nuclear chromatin-enriched localization. 4′,6-diamidino-2-phenylindole (DAPI) was used for DNA staining (red) without fixation. Bars, 10 µm.
To determine the localization of Ppe1, GFP was fused to the C-terminus of Ppe1, and the resulting Ppe1–GFP gene was chromosomally integrated with the native promoter. The Ppe1–GFP signal was indistinguishable from Ekc1–YFP localization (Figure 2B); the signal was enriched at the nuclear chromatin and seen as diffused in cytoplasm except for vacuole-like regions. However, the integrant strain showed cs growth and a cell shape defect. As we could not determine Ppe1 localization by immunofluorescence using polyclonal anti-Ppe1 antibodies, it remains to be determined whether the localization of functional Ppe1 was identical to that of Ppe1–GFP shown here. The ectopically expressed N-terminus of Ppe1 (amino acids 1–94) tagged with GFP showed a chromatin-enriched localization, similar to full-length Ppe1–GFP (data not shown). It is hence likely that native Ppe1 is targeted to chromatin by its N-terminal domain.
Microtubule inhibitor causes unequal segregation in ekc mutants
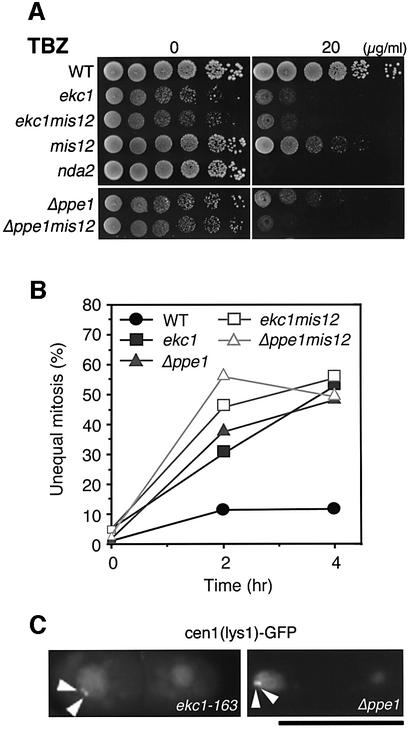
In ekc1 and ekc2 (and also Δppe1) mutants, chromosomes appeared to be equally segregated. As these mutants were found to be hypersensitive to the tubulin poison, thiabendazole (TBZ), at the permissive temperature, 33°C (Figure 3A), we examined whether chromosome segregation was still equal if TBZ was added to the culture medium. When treated with 20 µg/ml TBZ, ekc1-163 and Δppe1mutant cells produced a high frequency (52 and 48% of mitoses, respectively) of unequal segregation of chromosomes at 33°C (Figure 3B). Wild-type controls at 33°C exhibited only 11% unequal segregation under this condition. The occurrence of mis-segregation was verified by light microscopy using CEN1–GFP (Nabeshima et al., 1998) as shown in Figure 3C. Two sister centromere signals (arrowheads) were often found in one daughter nucleus in both mutants. The role of Ppe1–Ekc1 phosphatase in equal sister chromatid separation thus became evident when microtubules were inhibited in semi-permissive concentrations of TBZ. The TBZ sensitivities of the double mutants ekc1 mis12 and Δppe1 mis12 were similar to those of the single ekc mutant (Figure 3A and B).
Fig. 3. Chromosome missegregation in ekc1 and Δppe1 mutants in the presence of TBZ. (A) Moderate hypersensitivity of ekc1-163, Δppe1, mis12, ekc1-163 mis12 and Δppe1 mis12 mutants to TBZ, a microtubule-destabilizing drug. Exponentially growing cells were diluted and spotted onto YPD plates that contained 20 µg/ml TBZ, and incubated at 33°C. nda2-KM52, an α-tubulin mutant, was used as a control hypersensitive mutant. (B) Unequal chromosome segregation frequently occurred in ekc1-163, Δppe1, ekc1 mis12 and Δppe1 mis12 mutant cells in the presence of TBZ. Wild-type and mutant cells were cultured at 33°C, and TBZ (20 µg/ml) was added (time 0). The frequency of dividing cells did not change after TBZ addition, whereas the frequency of mutant cells displaying unequal nuclear division increased to 48–55% (of binucleate cells) after 4 h. (C) Mis-segregation of sister chromatids in ekc1-163 and Δppe1 was confirmed through the use of cen1[lys1]–GFP, a cen1-proximal DNA marker (arrowheads) (33°C, 4 h after TBZ addition). Bar, 10 µm.
Kinetochore localization of Mis12 is restored in the double mutant ekc1 mis12
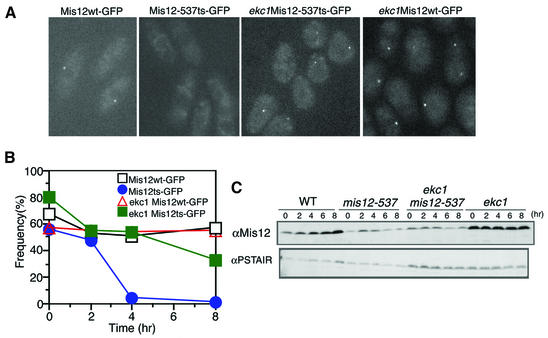
To investigate how the phenotype of mis12-537 was rescued by ekc mutations, we determined the localization of mutant Mis12 protein in an ekc1 mutant background (Figure 4A). For this purpose, we constructed a strain (designated Mis12ts–GFP) in which the wild-type mis12+ gene was replaced with the GFP-tagged mutant mis12-537ts (G52E) gene. This strain exhibited the phenotypes of ts growth and unequal chromosome segregation at 36°C, just like the original mis12-537 strain. Mis12ts–GFP cells were cultured at the permissive temperature (26°C) and then shifted to the restrictive temperature, 36°C (time 0). Cells were observed by microscopy without fixation after 2, 4 and 8 h (Figure 4A and B). The dot-like localization of Mis12-537ts–GFP mutant protein that was clearly seen in 60% of the cells before the temperature shift up disappeared after 4 h at 36°C (<5%). Control cells expressing Mis12wt–GFP protein showed a high frequency of dot-like localization (∼60%) at both 26 and 36°C. Note that GFP observation was carried out at a single focal plane, so that the signals of GFP could not be seen in a fraction of cells in which the centromeres were out of focus. To test the effect of the ekc1 mutation, Mis12ts–GFP was observed in the ekc1-163 background. The ekc1 Mis12ts–GFP double mutant was cultured at 36°C, and the GFP signal was observed. The dot-like localization of mutant Mis12 protein was restored, strongly suggesting that localization of Mis12 is under the control of Ekc1.
Fig. 4. Restoration of Mis12 mutant protein localization in the ekc1-163 background. (A) The dot-like localization of Mis12 mutant protein was restored in the double mutant mis12 ekc1. Images represent Mis12–GFP and Mis12-537–GFP expressed in the wild-type and ekc1 background cells (36°C, 8 h). (B) The frequencies of the centromeric dot-like appearance of Mis12ts–GFP were restored in ekc1 mutant cells. Mis12ts–GFP signals in the wild-type background became diffused after 4 h. (C) Schizosaccharomyces pombe cell extracts of wild-type and mutant cells cultured at 36°C were prepared at 2 h intervals. Immunoblotting of extracts was performed using anti-Mis12 and control anti-PSTAIR antibodies.
Immunoblotting of mis12-537 extracts prepared from the culture at the restrictive temperature, 36°C, showed that Mis12-537 mutant protein was less stable than that of the wild-type control, and that the instability was not restored to the wild-type level in the double mutant ekc1 mis12-537 (Figure 4C). This result strongly suggested that mutant Mis12 protein gained the ability to be incorporated into the centromere in the background of ekc1 mutation at 36°C.
Mis12, Ppe1 and Ekc1 are bound to chromatin
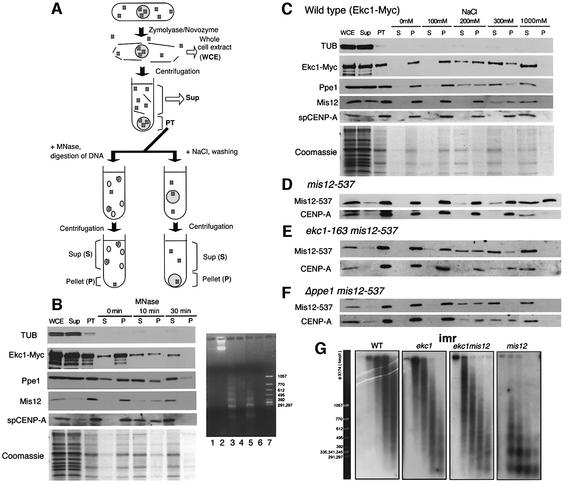
The above results suggested that Ppe1 and Ekc1 might directly or indirectly regulate Mis12. To examine whether wild-type and mutant Mis12 proteins behave as chromatin proteins and to study how ekc mutations affect the behaviour of Mis12, a chromatin fractionation assay was introduced for spheroplasted cell lysates of S.pombe (schematized in Figure 5A). Cell lysates (designated whole-cell extract; WCE) were centrifuged and separated into soluble (Sup) and pellet (PT) fractions. Whole chromatin DNA and chromatin-interacting proteins remained in the PT fraction at this stage. This pellet was treated by either MNase (Figure 5A, left) or NaCl extraction (right). After MNase treatment for 10–30 min, the average length of chromatin DNA was fragmented to <10 kb (Figure 5B, right panel lanes 3 and 5), resulting in the release of chromatin-associated proteins to the Sup (S). Hence, in this assay, those proteins that were first in the PT and then released into the S fraction after MNase treatment were defined as chromatin-associated proteins.
Fig. 5. Chromatin fractionation assay of Mis12, CENP-A, Ekc1 and Ppe1 proteins. (A) Schematic representation of the chromatin fractionation assay employed in the present study. See text for explanations. (B) Left: immunoblotting of tubulin (TUB), Ekc1-Myc, Ppe1, Mis12 and spCENP-A in various fractions: WCE, whole-cell extracts; PT, the pellet fraction after spheroplasting; S, supernatant; and P, pellet after MNase treatment for 0, 10 and 30 min. Four-fold concentrated proteins were loaded in the PT, S and P lanes. Mis12, CENP-A, Ekc1-Myc and Ppe1 were found to be solubilized after MNase digestion. Coomassie Blue staining is shown at the bottom. Right: DNA samples extracted before or after MNase digestion (0, 10 and 30 min) were electrophoresed in an agarose gel, followed by ethidium bromide staining. Lane 1, S at 0 min; lane 2, P at 0 min; lane 3, S at 10 min; lane 4, P at 10 min; lane 5, S at 30 min; lane 6, P at 30 min; lane 7, size marker. (C) The PT fraction was washed with the buffer containing 0–1000 mM NaCl followed by centrifugation. Four-fold concentrated proteins were loaded in the PT, S and P lanes. Approximately 30% of Mis12 was solubilized by treatment with 300 mM NaCl. Most Ppe1 and Ekc1-Myc was solubilized in 300 mM NaCl. Coomassie Blue staining is shown at the bottom. (D–F) The same NaCl treatment experiment as (C) was performed for mis12-537 (D), ekc1 mis12-537 (E) and Δppe1 mis12-537 (F) mutants. (G) Recovery of centromere chromatin structure by ekc1 mutation. Nuclear chromatin was prepared from spheroplasts of wild type and mis12, ekc1 and ekc1 mis12 mutants cultured at 36°C for 8 h. Chromatin was digested with MNase for 0, 1, 2, 4 and 8 min (left to right lanes). Digested DNAs were electrophoresed in an agarose gel. Southern hybridization using the central centromere DNA probe imr1 was performed. The size markers are shown on the left.
Supernatants and pellets were analysed by SDS–PAGE, followed by immunoblot and also Coomassie Blue staining (Figure 5B, left panel). Simultaneously, DNAs before and after the MNase treatment were electrophoresed in an agarose gel (right panel). Coomassie Blue staining indicated that most proteins were in the Sup at the first stage, and the PT proteins almost completely went into the S fraction after MNase treatment. Digested DNA was detected in the solubilized fraction (lanes 3 and 5). Immunoblot using anti-Mis12 antibody revealed that wild-type Mis12 was first found in PT but was completely released to S after MNase digestion. spCENP-A behaved similarly to Mis12. As a control, we found tubulin to be exclusively in the Sup after cell lysis. These results established that most Mis12 and spCENP-A were chromatin bound. Ppe1 and Ekc1-Myc were present initially in both Sup and PT, and their subpopulation in the PT moved to S after MNase treatment, indicating that the subfractions of Ppe1 and Ekc1-Myc were chromatin bound.
To estimate the concentration of NaCl required for extracting Mis12 from chromatin, PT was washed with buffer containing various concentrations of NaCl (Figure 5C). After 10 min on ice, samples were centrifuged and separated into soluble (S) and pellet (P). Proteins, such as histones, tightly bound to DNA and chromatin were not dissociated from DNA by the low salt treatment. The PT fraction was washed with 100–1000 mM NaCl-containing buffer. Coomassie Blue staining indicated that most proteins in the PT were solubilized by 300 mM NaCl. Thirty to forty percent of Mis12 was solubilized after extraction with 300 mM NaCl, and almost all of the protein was solubilized by 1000 mM NaCl. spCENP-A was more resistant than Mis12 to the salt treatment, and it remained in P after 300 mM NaCl treatment. Ppe1 and Ekc1 were solubilized after 200–300 mM NaCl, similar to Mis12. These properties indicated that Mis12, Ppe1 and Ekc1 behaved in a manner similar to non-histone chromatin proteins, and associated fairly strongly with chromatin but less tightly than histone proteins.
ekc mutations restore the Mis12-containing inner centromere chromatin
We performed the chromatin assay using mis12-537 (Figure 5D), ekc1 mis12-537 (Figure 5E) and Δppe1 mis12-537 (Figure 5F) strains (36°C, 6 h), and found that Mis12-537 protein was more resistant to NaCl extraction in the presence of functional Ppe1 and Ekc1 (Figure 5D). Unlike wild-type Mis12, mutant Mis12-537 protein detected in the PT was not solubilized by 300 mM NaCl treatment. Further, more than half of the Mis12-537 protein remained in the pellet fraction even after treatment with 1000 mM NaCl, conditions that solubilized CENP-A. In sharp contrast, most of the Mis12-537 protein became solubilized by 200–300 mM NaCl treatment in the background of ekc1-163 or Δppe1 (Figure 5E and F). Down-regulated Ppe1–Ekc1 thus dramatically recovered the salt solubility property of Mis12-537 to that of wild-type Mis12.
We then examined whether ekc1 mutation restores the specialized centromere chromatin in the central regions. Southern hybridization was performed after MNase digestion of chromatin DNAs using the inner centromere sequence (imr) as a probe. The smeared centromere chromatin pattern present in wild-type and single ekc1 mutant cells after MNase treatment was abolished in mis12-537 at 36°C, but restored in the ekc1 mis12 double mutant cells cultured at 36°C (Figure 5G). The outer repetitive heterochromatin was not affected by either ekc1 or mis12, and displayed regular nucleosomal ladders at 36°C (data not shown). The functional defect of Ekc1 could thus result in restoration of the central centromere chromatin structure in mis12-537 mutant.
The dosage increase of Gsk3 kinase suppresses the phenotype of mis12-537
Extragenic suppressor analysis identified Ppe1 as a phosphatase that interacts with Mis12. We next screened for high gene dosage suppressors for the ts phenotype of mis12-537 using two S.pombe genomic DNA libraries based on the multicopy plasmid vector. A number of high gene dosage suppressors were found, including Gsk3, Ssp1, Spi1, Sds23 and Mts2 (Matsumoto and Beach, 1991; Gordon et al., 1993; Matsusaka et al., 1995; Ishii et al., 1996; Plyte et al., 1996). Gsk3 and Ssp1 are described below, and other suppressors will be described elsewhere.
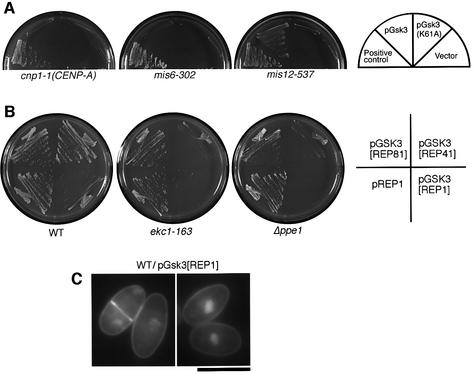
A strong suppressor gene obtained from both libraries resembled the mammalian GSK-3β kinase. Gsk3 kinase in fission yeast is non-essential for cell viability, but does suppress cytokinesis-defective cdc14 when overexpressed [it was also called Skp1 (shaggy kinase in S.pombe); Plyte et al., 1996]. Mitotic defects have not been shown in the Δgsk3 disruptant. Multicopy plasmid carrying the gsk3+ gene fully suppressed the phenotype of mis12-537 at 36°C, but the kinase-dead mutant (K61A) did not (Figure 6A), strongly suggesting that the elevated Gsk3 kinase activity was required for the suppression of mis12. Other central kinetochore mutants cnp1-1 (spCENP-A) and mis6-302 were not suppressed by Gsk3, suggesting that suppression was specific to the mis12 mutation.
Fig. 6. Overproduced Gsk3 kinase suppresses mis12 and antagonizes Ppe1. (A) Multicopy plasmid pGsk3 suppressed the ts phenotype of mis12-537 at 36°C, but did not suppress cnp1-1 and mis6-302 mutants even at 33°C. (B) Gsk3 is antagonistic to Ppe1 and Ekc1, as its moderate overexpression by REP41 plasmid in the absence of thiamine prevented colony formation of ekc1-163 and Δppe1, but not of wild-type. Even the milder expression of Gsk3 by REP81 plasmid still caused retarded growth of ekc1 and Δppe1 mutants. REP1 caused the strongest overexpression. (C) Gsk3 overexpression in the wild-type cells led to the accumulation of pear-shaped cells. Wild-type cells in which Gsk3 was expressed by REP1 plasmid in the absence of thiamine for 24 h at 26°C. Cells were stained by DAPI. Bar, 10 µm.
Moderate overproduction of Gsk3 under the inducible promoter inhibited colony formation of ekc1 or ppe1 mutant cells, whereas intense overexpression was required for wild-type inhibition (Figure 6B). This suggests that Ppe1–Ekc1 phosphatase and Gsk3 kinase are functionally counteracting. Consistently, cells in which Gsk3 was intensely expressed were round in shape, similar to ekc1- and ppe1-defective mutant cells (Figure 6C).
We then searched for other suppressor kinases by transforming multicopy plasmids that expressed Ark1 (aurora kinase), Plo1 (polo-like kinase), Ssp1 (a cell shape-controlling kinase), Pka1 (PKA), Spc1, Wis1, Spk1 or Pek1 (MAPK pathway kinases). Among them, Ssp1 was obtained as another high dosage suppressor kinase. Ssp1 was closely related to Ppe1 as ssp1 was identified originally as an extragenic suppressor for ppe1, and was essential for correct actin organization in the cortex (Matsusaka et al., 1995; Rupes et al., 1999). Mitotic phenotypes have not been found in Δssp1. Overexpression of Ssp1 induced round cells like the case of Gsk3 (Matsusaka et al., 1995). Gsk3 is a high copy suppressor for ssp1 disruptant (T.Toda and T.Matsusaka, personal communication). Ssp1 may also counteract against Ppe1.
The C-terminus of Mis12 is essential for chromatin localization and is phosphorylated
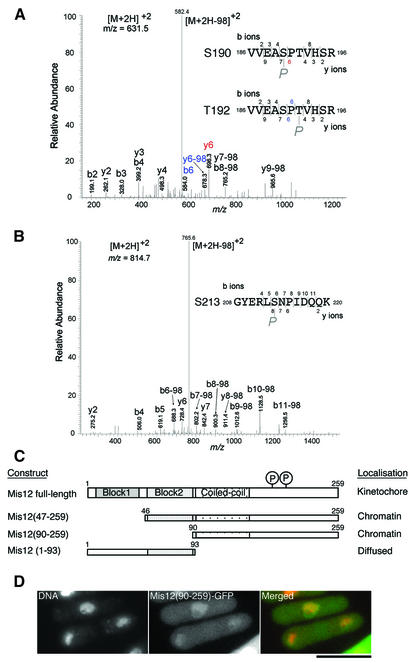
To test whether Mis12 was actually phosphorylated in vivo, we first tried to purify Mis12 produced by the single copy gene in S.pombe, but the amount obtained so far was insufficient for analysis of phosphopeptides. Mis12 overproduced in S.pombe was then purified using the haemagglutinin (HA)- and histidine-tagged Mis12 gene under the nmt1 promoter. Mis12-HAHis6 protein was purified under denaturing condition using TALON beads, and was detected as a single band at the expected molecular weight. Liquid chromatography and tandem mass spectrometry (MS) analyses (Ohta et al., 2002) showed that Ser190 or Thr192, and Ser213 were indeed phosphorylated (Figure 7A and B). Attempts to compare the levels of Mis12 phosphorylation in the presence or absence of Ekc1 and Ppe1, however, have not been successful.
Fig. 7. Residues in the C-terminus of Mis12 are phosphorylated. (A and B) Ser190 or Thr192 and Ser213 are phosphorylated in vivo. HA- and histidine-tagged Mis12 overproduced in wild type was purified using TALON beads under denaturing conditions, and subjected to SDS–PAGE. The band of Mis12-HAHis6 was cut out and trypsinized in-gel, and subjected to survey of the phosphorylation site using MS (Ohta et al., 2002). The MS/MS spectra of the tryptic phosphopeptide amino acids 186–196 (A) and 208–220 (B) of Mis12 obtained by collision-induced dissociation of the [M + 2H]2+precursor ions, m/z 631.5 and 814.7, are shown. In both cases, intense [M + 2H –98]2+ ions, m/z 582.4 in (A) and 765.8 in (B), due to neutral loss (H2O + phosphate, 97.8) of precursor ions were observed, indicating that the potential phosphorylated residues are serine or threonine. Three potential sites of phosphorylation (two serine and one threonine) are found in the peptide 186–196 (A). The fragment ions enable localization of the phosphorylated residue to one of the two central residues, Ser190 and Thr192. An ion fragment at m/z 696.3 is assignable to y6 (red letters) when the peptide is phosphorylated at Ser190, and ion fragments at m/z 584.0 and 678.3 are assignable to b6 and y6-98, respectively, when the peptide is phosphorylated at Thr192 (blue letters). There is one possible residue in the peptide 208–220 (B), and the fragment ions were matched to the peptide phosphorylated at Ser213. (C) Localization of three Mis12-truncated proteins. Each construct is tagged with GFP at the C-terminus and expressed from REP41 plasmid that allows moderate overexpression. (D) Chromatin localization of truncated Mis12 (amino acids 90–259)–GFP protein. DNA was counterstained by DAPI. Bar, 10 µm.
Phosphorylated residues were located at the hydrophilic C-terminal region. Construction and expression of three truncated Mis12 showed that the C-terminal half was sufficient for chromatin localization (Figure 7C). Truncated Mis12(90–259) lacking the conserved two-block sequences was enriched in chromatin (Figure 7D), while another truncation, Mis12(1–93) containing only the conserved domain, was localized diffusely throughout the cell (data not shown). Kinetochore-specific localization thus requires both the N- and C-terminal regions.
Discussion
Previous studies (Goshima et al., 1999, 2003; Takahashi et al., 2000) established that Mis12 is independent of CENP-A, a kinetochore-specific histone H3 variant. The aim of the present investigation was to understand what kind of specific gene products directly or indirectly interact with Mis12. By introducing the chromatin fractionation assay and MNase release experiment for fission yeast kinetochore proteins, we were able to demonstrate that both Mis12 and spCENP-A were chromatin-associating proteins, but their behaviours were different. Salt-washing experiment indicated that Mis12 was released before nucleosomal histones. Although Mis12 is essential for the formation of specialized chromatin in the central centromeres, the behaviour of Mis12 is like that of a non-histone chromatin protein. It may be added to a pre-formed centromere protein–DNA complex or may bind directly to specific centromere DNA sequences, and be easily extracted by salt.
Extragenic suppressor analysis resulted in the finding that Mis12 interacts with Ppe1 and its partner Ekc1, an evolutionarily highly conserved protein phosphatase complex. All the 24 cs suppressors turned out to cause downregulation of Ppe1, either ekc1 or ekc2 (identical to ppe1) mutants. Suppression of mis12 mutants by deletion of Ppe1 is quite specific, as mis6-302 was not suppressed by ppe1 mutation. Ppe1–Ekc1 showed no interaction with CENP-A. Ppe1 is the sole phosphatase so far that is functionally related to Mis12. A chromatin-associated pool of Ppe1 and Ekc1 behaved similarly to Mis12 and is released in a soluble form after MNase treatment. Although an interaction was not detected with our immunoprecipitation conditions, it is entirely possible that Mis12 has a transient but direct interaction with Ppe1 or its positive regulator, Ekc1, in the centromere chromatin. Mis12 may be the direct target of phosphorylation and dephosphorylation, since two phosphorylation sites were identified by MS analysis. The sites of phosphorylation are in the region required for chromatin localization of Mis12. In order to determine whether phosphorylation is cell cycle regulated, phosphoprotein produced by the native gene must be identified, and phosphopeptide antibodies are essential for such analyses.
The Ppe1–Ekc1 phosphatase might ensure the dynamic connection between kinetochore microtubules and kinetochore chromatin, as both mutants were sensitive to TBZ. The phenotypes of the ekc1 and Δppe1 mutants in the presence of semi-permissive TBZ concentrations were strikingly similar to that of mis12 at 36°C. This provides evidence that the Ppe1/PP6 family has an important role in equal chromosome segregation in mitosis even in the presence of functional Mis12. The microtubule– kinetochore linkage itself should exist in cells displaying the unequal segregation phenotype, because no lagging chromosomes were observed in mis12 and ekc mutants. Ppe1/PP6 family proteins are likely to have multiple roles, as is often the case for phosphatases (Cohen, 1997). Budding yeast Sit4, reported to have a cytoplasmic distribution, is essential for cell growth and cell cycle control (e.g. Sutton et al., 1991; Fernandez-Sarabia et al., 1992; Beck and Hall, 1999). Drosophila PPV is enriched in the cytoplasm in cellular blastoderm embryos (Mann et al., 1993), although functional analysis has not been carried out for animal orthologues. In fission yeast, Ppe1 has roles in cell morphogenesis and cell cycle control (Shimanuki et al., 1993). Ekc1 and Ppe1 appear to be present in the nuclear chromatin and cytoplasm. These observations support the idea that Ppe1 phosphatase plays roles in both the cytoplasm and nucleus.
We showed that the high gene dosage of two distinct protein kinases, Gsk3 and Ssp1, rescued the ts phenotype of mis12-537. They may counteract Ppe1 so that mis12 could be suppressed. Overproduction of these functionally related kinases caused a pear-like cell shape like ppe1 and ekc1 mutations. Suppression of mis12 by these kinases and phosphatase may become possible at the expense of normal cell shape. These kinases might compete at the level of the substrates with Ppe1. One of the direct substrates may be Mis12. Alternatively, Mis12 may be downstream of the direct targets of these kinases or phosphatase. Mis12-interacting protein(s) are also candidate substrates. These hypotheses are not inconsistent with the fact that the biochemical properties of Mis12 are altered in the chromatin fractionation assay of ekc mutant extracts (see Figure 5D–F). It remains to be determined which kinase is responsible for phosphorylation of Ser190/Thr192 or Ser213 of Mis12. Gsk3 is a highly conserved kinase, and it is known that Mck1, a budding yeast GSK3-like kinase, can suppress mutations in the centromere DNA or the genes encoding kinetochore proteins (Shero and Hieter, 1991; Jiang et al., 1995).
It recently was reported that mammalian GSK3 is present at the mitotic spindle and is implicated in chromosome movements during mitosis (Wakefield et al., 2003). Aurora kinase B and type I phosphatase (PP1) antagonistically regulate kinetochore function in budding yeast and mammalian cells (e.g. Francisco et al., 1994; Biggins et al., 1999; Zeitlin et al., 2001). However, in fission yeast, Ark1 (aurora homologue) is localized at the outer region of the centromere in early mitosis, and ark1 mutant phenotypes are different from those of inner centromere protein mutants such as mis12 (Morishita et al., 2001; Petersen et al., 2001). Further, fission yeast PP1 (Dis2 and Sds21) mutants exhibit defects in sister chromatid separation, and do not cause unequal chromosome segregation like inner centromere protein mutants (Ohkura et al., 1989). We hence suppose that the active regions at kinetochores are distinct for GSK3/Ppe1 and Aurora/PP1.
In summary, the present study showed that Mis12 is a non-histone-type kinetochore chromatin protein that behaves in a fashion quite distinct from CENP-A. The mechanism of Mis12 binding to the centromeres is unknown, but is likely to be regulated by Gsk3-dependent phosphorylation and Ppe1/PP6-dependent dephosphorylation.
Materials and methods
Strains, media and antibodies
Schizosaccharomyces pombe mutants used in this study were described previously (Shimanuki et al., 1993; Goshima et al., 1999). The deletion of Ppe2 phosphatase was made by one-step gene replacement and verified by the lack of a detectable Ppe2 protein band by polyclonal anti-Ppe2 antibodies. Δppe2 was found to be viable and produced no appreciable defective phenotypes. Ekc1-Myc, Ekc1–YFP, Ppe1–GFP and Mis12ts–GFP strains were made by integration of the plasmid containing tagged genes into the endogenous locus. Complete YPD and minimal EMM2 media were used for the culture of S.pombe. Thiamine (2 µM) was added when the nmt1 promoter was used for the repressive condition. Polyclonal antibodies (rabbit) against full-length Mis12 were obtained by a method similar to that described in Goshima et al. (2003).
Isolation of the ekc1+ gene
A genetic complementation test was performed for 24 cs revertants by random spore analysis. In this test, if parental cells have mutations at the same genetic loci, none of the spores can grow at 22°C. By this analysis, 21 strains were mapped at the ekc1 locus, while the remaining three strains were at ekc2. Plasmid pGG800 that rescued the cs phenotype of ekc1-163 was obtained from the S.pombe genomic library. By subcloning pGG800, plasmid pGG806 containing the minimal functional fragment was found to carry one open reading frame (ORF; SPCC663.01). This ORF was found to complement ekc1. Tetrad analysis established the tight linkage (PD:TT:NPD = 13:1:0) between the ekc1 and arg1 mutation (located on cos976, which is adjacent to cos663), so that this ORF was verified to be the product of the ekc1+ gene.
Cell extracts
Extracts were prepared using HB buffer (for anti-myc immunoprecipitation; Moreno et al., 1989) or TEG buffer (50 mM Tris pH 7.5, 1 mM EDTA, 10% glycerol, 150 mM NaCl, 0.1% NP-40) (for anti-Ppe1 immunoprecipitation). Monoclonal anti-myc and polyclonal anti-Ppe1 antibodies were used. To purify Mis12-HAHis6, 2 × 109 cells, which overexpressed Mis12-HAHis6 by the REP1 plasmid in EMM2 medium in the absence of thiamine, were collected and suspended in 1 ml of NG8 buffer (10 mM Tris–HCl at pH 8.0, containing 100 mM Na-phosphate, 50 mM NaCl, 6 M guanidine hydrochloride and 0.1% NP-40). Glass beads were added to the cell suspension, for preparation of extracts. The extracts were centrifuged (14 000 r.p.m.) and 100 µl of TALON beads (Clontech) were added to the supernatant. After incubation for 40 min at room temperature, the beads were washed twice with NG8 buffer, twice with NG7 (pH 7.0) and then twice with buffer N8 (as NG8 but without guanidine hydrochloride). The beads were boiled in SDS sample buffer after the addition of 100 mM EDTA.
Chromatin fractionation and Southern hybridization
The chromatin fractionation assay was performed with modifications of the method previously described for MNase digestion (Takahashi et al., 1992). Cells (1 × 109) were harvested, washed once with 5 ml of 10 mM Tris–HCl pH 7.5 buffer, and then incubated in 5 ml of PEMS (100 mM PIPES pH 6.9, 1 mM EGTA, 1 mM MgSO4, 1.2 M sorbitol) containing 0.6 mg/ml zymolyase 100T (Seikagaku) and 1.2 mg/ml lysing enzyme (Sigma) for 20 min at 36°C. The cell suspension was centrifuged, and the resulting pelleted cells were suspended in 200 µl of SCPT solution [1 M sorbitol, 0.1 mM CaCl2, 20 mM PIPES pH 6.3, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF)]. Cells were lysed for 5 min on ice by adding 8 ml of lysis buffer (20 mM PIPES pH 6.3, 0.1 mM CaCl2, 9% Ficol-400, 1 mM PMSF, 0.5% Triton X-100). The resulting lysate was the WCE. Supernatant (Sup) and pellet (PT) fractions were obtained by centrifugation of the WCE (8000 r.p.m., 5 min, 4°C). In the first assay, PT was resuspended in 800 µl of SCP solution containing 200 U of MNase and incubated at 36°C for 0–30 min. The reaction was terminated by transferring the samples on ice. The digested solution was centrifuged (10 000 r.p.m. for 5 min at 4°C) to separate the PT into supernatant (S) and pellet (P), from each of which DNA was extracted after treatment with proteinase K. The resulting samples were subjected to SDS–PAGE. In the second assay, the PT fraction was suspended in the SCPT solution containing 0–1000 mM NaCl, and kept on ice for 10 min, followed by centrifugation (10 000 r.p.m., 5 min, 4°C). Immunoblotting was performed using the following antibodies; anti-myc [9E10 (Calbiochem), 1:100], anti-Mis12 (rabbit, 1:30), anti-CENP-A (rabbit, 1:50), anti-Ppe1 (rabbit, 1:1000) and anti-tubulin (TAT1, mouse, 1:1000). For Southern hybridization, plasmid pKT108 was used as a hybridization probe (Takahashi et al., 1992).
Acknowledgments
Acknowledgements
We are grateful to Mizuki Shimanuki, Shigeaki Saitoh and Koji Hizume who assisted in the early stages of this study. We thank Takashi Toda and Takahiro Matsusaka for communicating unpublished results, Takashi Toda for helpful comments on the manuscript, and Ken-ichi Yoshino for valuable comments on the interpretation of MS/MS spectra. The work was supported by a CREST grant of the Japan Science and Technology Corporation and a COE Scientific Research Grant from the Ministry of Education, Culture, Sports, Science and Technology. G.G. was the recipient of pre- and post-doctoral fellowships of the Japan Society for Promotion of Science (JSPS).
References
- Bastians H. and Ponstingl,H. (1996) The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J. Cell Sci., 109, 2865–2874. [DOI] [PubMed] [Google Scholar]
- Beck T. and Hall,M.N. (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature, 402, 689–692. [DOI] [PubMed] [Google Scholar]
- Biggins S., Severin,F.F., Bhalla,N., Sassoon,I., Hyman,A.A. and Murray,A.W. (1999) The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev., 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D. and Karpen,G.H. (2001) The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol., 3, 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P.T. (1997) Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem. Sci., 22, 245–251. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sarabia M.J., Sutton,A., Zhong,T. and Arndt,K.T. (1992) SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2 and HCS26 RNAs during late G1. Genes Dev., 6, 2417–2428. [DOI] [PubMed] [Google Scholar]
- Francisco L., Wang,W. and Chan,C.S. (1994) Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol., 14, 4731–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Vardy,L., Koonrugsa,N. and Toda,T. (2001) Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J., 20, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C., McGurk,G., Dillon,P., Rosen,C. and Hastie,N.D. (1993) Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature, 366, 355–357. [DOI] [PubMed] [Google Scholar]
- Goshima G., Saitoh,S. and Yanagida,M. (1999) Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev., 13, 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu,T., Yoda,K. and Yanagida,M. (2003) Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol., 160, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps N.R., Brewis,N.D., Lineruth,K., Davis,T., Kaiser,K. and Cohen,P.T. (1998) Protein phosphatase 4 is an essential enzyme required for organisation of microtubules at centrosomes in Drosophila embryos. J. Cell Sci., 111, 1331–1340. [DOI] [PubMed] [Google Scholar]
- Howman E.V., Fowler,K.J., Newson,A.J., Redward,S., MacDonald,A.C., Kalitsis,P. and Choo,K.H. (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA, 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Kumada,K., Toda,T. and Yanagida,M. (1996) Requirement for PP1 phosphatase and 20S cyclosome/APC for the onset of anaphase is lessened by the dosage increase of a novel gene sds23+. EMBO J., 15, 6629–6640. [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Lim,M.Y., Yoon,H.J., Thorner,J., Martin,G.S. and Carbon,J. (1995) Overexpression of the yeast MCK1 protein kinase suppresses conditional mutations in centromere-binding protein genes CBF2 and CBF5. Mol. Gen. Genet., 246, 360–366. [DOI] [PubMed] [Google Scholar]
- Kinoshita N., Ohkura,H. and Yanagida,M. (1990) Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell, 63, 405–415. [DOI] [PubMed] [Google Scholar]
- Luke M.M., Della Seta,F., Di Como,C.J., Sugimoto,H., Kobayashi,R. and Arndt,K.T. (1996) The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol., 16, 2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D.J., Dombradi,V. and Cohen,P.T. (1993) Drosophila protein phosphatase V functionally complements a SIT4 mutant in Saccharomyces cerevisiae and its amino-terminal region can confer this complementation to a heterologous phosphatase catalytic domain. EMBO J., 12, 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T. and Beach,D. (1991) Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell, 66, 347–360. [DOI] [PubMed] [Google Scholar]
- Matsumoto T. and Beach,D. (1993) Interaction of the pim1/spi1 mitotic checkpoint with a protein phosphatase. Mol. Biol. Cell, 4, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusaka T., Hirata,D., Yanagida,M. and Toda,T. (1995) A novel protein kinase gene ssp1+ is required for alteration of growth polarity and actin localization in fission yeast. EMBO J., 14, 3325–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Hailey,D.W., Pot,I., Givan,S.A., Hyland,K.M., Cagney,G., Fields,S., Davis,T.N. and Hieter,P. (2002) Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev., 16, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Hayles,J. and Nurse,P. (1989) Regulation of p34cdc2 protein kinase during mitosis. Cell, 58, 361–372. [DOI] [PubMed] [Google Scholar]
- Morishita J., Matsusaka,T., Goshima,G., Nakamura,T., Tatebe,H. and Yanagida,M. (2001) Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells, 6, 743–763. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa,T., Straight,A.F., Murray,A., Chikashige,Y., Yamashita,Y.M., Hiraoka,Y. and Yanagida,M. (1998) Dynamics of centromeres during metaphase–anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell, 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Goshima,G., Morishita,J. and Yanagida,M. (2001) M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol., 11, 537–549. [DOI] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi,T., Hiraoka,Y., Ikemura,T., Regnier,V., Dodson,H., Earnshaw,W.C. and Fukagawa,T. (2002) CENP-I is essential for centromere function in vertebrate cells. Dev. Cell, 2, 463–476. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai,A., Rybina,S., Kirkham,M. and Hyman,A.A. (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol., 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Kinoshita,N., Miyatani,S., Toda,T. and Yanagida,M. (1989) The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell, 57, 997–1007. [DOI] [PubMed] [Google Scholar]
- Ohta S., Shiomi,Y., Sugimoto,K., Obuse,C. and Tsurimoto,T. (2002) A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2-5 complex as a novel PCNA-binding protein. J. Biol. Chem., 277, 40362–40367. [DOI] [PubMed] [Google Scholar]
- Petersen J., Paris,J., Willer,M., Philippe,M. and Hagan,I.M. (2001) The S.pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci., 114, 4371–4384. [DOI] [PubMed] [Google Scholar]
- Plyte S.E., Feoktistova,A., Burke,J.D., Woodgett,J.R. and Gould,K.L. (1996) Schizosaccharomyces pombe skp1+ encodes a protein kinase related to mammalian glycogen synthase kinase 3 and complements a cdc14 cytokinesis mutant. Mol. Cell. Biol., 16, 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi C. and Clarke,L. (1991) The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J. Cell Biol., 112, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronne H., Carlberg,M., Hu,G.Z. and Nehlin,J.O. (1991) Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol., 11, 4876–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupes I., Jia,Z. and Young,P.G. (1999) Ssp1 promotes actin depolymerization and is involved in stress response and new end take-off control in fission yeast. Mol. Biol. Cell, 10, 1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Takahashi,K. and Yanagida,M. (1997) Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell, 90, 131–143. [DOI] [PubMed] [Google Scholar]
- Shero J.H. and Hieter,P. (1991) A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1). Genes Dev., 5, 549–560. [DOI] [PubMed] [Google Scholar]
- Shimanuki M., Kinoshita,N., Ohkura,H., Yoshida,T., Toda,T. and Yanagida,M. (1993) Isolation and characterization of the fission yeast protein phosphatase gene ppe1+ involved in cell shape control and mitosis. Mol. Biol. Cell, 4, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.M. (2002) Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol., 14, 279–285. [DOI] [PubMed] [Google Scholar]
- Stoler S., Keith,K.C., Curnick,K.E. and Fitzgerald-Hayes,M. (1995) A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev., 9, 573–586. [DOI] [PubMed] [Google Scholar]
- Sutton A., Immanuel,D. and Arndt,K.T. (1991) The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol., 11, 2133–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Murakami,S., Chikashige,Y., Funabiki,H., Niwa,O. and Yanagida,M. (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell, 3, 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen,E.S. and Yanagida,M. (2000) Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science, 288, 2215–2219. [DOI] [PubMed] [Google Scholar]
- Toda T., Shimanuki,M. and Yanagida,M. (1991) Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev., 5, 60–73. [DOI] [PubMed] [Google Scholar]
- Toyoda Y., Furuya,K., Goshima,G., Nagao,K., Takahashi,K. and Yanagida,M. (2002) Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle–kinetochore interaction in fission yeast. Curr. Biol., 12, 347–358. [DOI] [PubMed] [Google Scholar]
- Wakefield J.G., Stephens,D.J. and Tavare,J.M. (2003) A role for glycogen synthase kinase-3 in mitotic spindle dynamics and chromosome alignment. J. Cell Sci., 116, 637–646. [DOI] [PubMed] [Google Scholar]
- Zeitlin S.G., Shelby,R.D. and Sullivan,K.F. (2001) CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol., 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]