Abstract
Biomolecules often undergo large-amplitude motions when they bind or release other molecules. Unlike macroscopic machines, these biomolecular machines can partially disassemble (unfold) and then reassemble (fold) during such transitions. Here we put forward a minimal structure-based model, the “multiple-basin model,” that can directly be used for molecular dynamics simulation of even very large biomolecular systems so long as the endpoints of the conformational change are known. We investigate the model by simulating large-scale motions of four proteins: glutamine-binding protein, S100A6, dihydrofolate reductase, and HIV-1 protease. The mechanisms of conformational transition depend on the protein basin topologies and change with temperature near the folding transition. The conformational transition rate varies linearly with driving force over a fairly large range. This linearity appears to be a consequence of partial unfolding during the conformational transition.
Keywords: conformational transition, cracking, partial unfolding, funnel
To function, biomolecules often undergo large-amplitude structural changes upon binding or releasing ligands. These structural changes organize the workings of biomolecular machines such as the ribosome, molecular chaperones, and molecular motors. Structural information on the conformational ensembles before and after the conformation changes is often available through x-ray crystallography or NMR. These experiments, however, provide primarily quasistatic information. They reveal directly less about the transition dynamics between two end structures. The overall dynamics of basin-hopping can be studied by pump-probe experiments or from NMR relaxation. These experiments, however, usually monitor directly only a few local structure changes in what is typically a huge system. Thus, we see that global time-dependent structural information at high resolution is rarely obtained directly by experiments. Simulations can potentially provide full time-dependent structural information on biomolecular machines. Yet conventional atomistic simulations currently only reach times up to microseconds (1). This time scale falls orders of magnitude short of the typical physiologically important time scales of milliseconds to seconds. To overcome this limitation, one approach is to coarse-grain the molecular representation (2). Reduction in complexity allows one to simulate much longer times. This so-called “minimalist approach” has been quite successful for studying protein folding (3–8). The purpose of this article is to investigate a minimal structure-based model, which we call the “multiple-basin model,” to simulate large-scale conformational changes when structures for the endpoints of the transition are available. This approach can be used for simulations of even very large biomolecular complexes.
To motivate the present model, we first note that two qualitatively different kinds of protein motions occur depending on the amplitude of motion. For small deviations from the fiducial native structure motions are well approximated by quasi-harmonic dynamics. Indeed, very simple quasi-harmonic topology-based models reproduce the size of fluctuations at single-residue resolution as evidenced by experimental B-factors (9, 10). Structure changes upon ligand binding usually occur predominantly in directions that correspond to combinations of a few low-frequency modes (11–16). Ikeguchi et al. (17) have shown using their linear response theory that the direction of structural change can be predicted from the dominant principal components of the structural fluctuations in the unbound states multiplied vectorially by the force exerted by the ligand. Conformational transitions, however, must involve rearrangement of nonlocal contacts of amino acid pairs (see Fig. 1 Lower). Such notions clearly require going beyond the quasi-harmonic picture (18–21): The protein breaks some contacts specific to the initial conformation and forms new contacts that are specific to the final conformation. This second, large-amplitude regime of protein dynamics has been termed a “proteinquake” (22) and may involve “cracking” (18, 21) or local unfolding. As the protein softens and its fluctuations increase with temperature, a third regime where the protein may globally unfold begins to play a further role in function especially for the so-called “natively unfolded proteins” (23, 24).
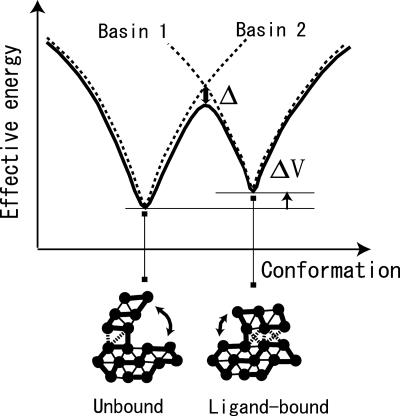
Fig. 1.
Schematic view of multiple-basin energy landscape of proteins. Two single funnels used for model construction are depicted by dashed lines. Conformational change is associated with the rearrangement of some contacts. Contacts specific to the unbound conformation are broken, and new contacts are formed in bound conformation. Thick solid bonds correspond to covalent linkages.
We see that a minimal requirement for describing conformational transitions is to be able to model both the quasi-harmonic fluctuations around each basin and the transient and partial unfolding near the transition region. A natural framework to formalize such a model is provided by energy landscape theory. Energy landscape theory has established that proteins have evolved to have funnel-like energy landscapes (25–27). The bottom of the funnel corresponds to a fiducial native structure having the lowest effective solvent averaged free energy (as an individual conformation). As the protein unfolds, the number of structures multiplies while the effective energy increases. An ideal funnel-like energy landscape can be realized by the so-called Gō model (6, 7, 28–30), first introduced for lattice models (where of course there is no harmonic motion) in which a protein is represented as a chain with attractive interactions between pairs of residues that interact in the native structure and repulsive interactions for all other residue pairs. These models have been very effective for studying folding mechanisms. Moreover, the off-lattice version of perfect funnel models (6, 7) already goes far to meet our present needs because it models quite well the quasi-harmonic motion in the limit of the weak fluctuations (31).
In the standard energy landscape for folding proteins, however, only a single dominant minimum, which corresponds to the native structure, is assumed. Studying conformational changes of functional proteins requires more than one basin to be taken into account. Each basin should correspond to the structure with or without ligands. The standard structure-based models are not directly applicable to this case of multiple basins. Here we explicitly build up an energy landscape encoding multiple near-degenerate basins. Given two reference structures supplied by x-ray crystallography, we first created two independent structure-based potentials (32), which were then smoothly connected to make a double-well energy landscape. Very recently, a closely related approach having the same spirit was put forward by Best et al. (20). We will survey several systems with the model and highlight the cracking phenomenon.
Specifically, we simulated conformational transitions for four proteins, glutamine-binding protein (GBP), S100A6 (which is a structural analog of calmodulin), dihydrofolate reductase, and HIV-1 protease. The global character of the conformational motions, the residue-specific involvement in transition pathways, and the temperature dependence of rates were then investigated and related to changes in protein topology. We also show that the linear dependence of rate on driving force arises as a consequence of local unfolding near the transition region.
Results
Multiple-Basin Model.
We explicitly describe the model for the case with two major structurally characterized basins, basin 1 and 2 as in Fig. 1. (For a generalization to n basins see Materials and Methods.) The model's energy landscape within each individual basin is a perfect funnel. We start with two virtual funnels (dashed curves in Fig. 1). The target energy landscape (solid curve) coincides with one of the dashed curve near basin 1 and merges with the other dashed curve near basin 2. Around the transition region, the energy landscape should be smoothly connected. From a mathematical viewpoint, the way to connect smoothly two basins is not unique. Here we choose a smooth connection that is computationally convenient.
Mathematically we construct first two single-basin potentials as V(R | R1) and V(R | R2) + ΔV (dashed lines in Fig. 1), where R collectively represents the coordinates of the protein structure and R1 and R2 correspond to the coordinates of the fiducial structures. For convenience, the input potential V(R | Rν) is defined so that the energy at the bottom of basin ν is (nearly) zero (see Materials and Methods for details). Then ΔV is introduced to modulate the relative stability of the two basins to correspond to empirically determined energetics; a larger ΔV makes basin 2 less stable than basin 1. We then introduce a coupling between the two potentials to make a smoothed double basin potential VMB. We use (merely for construction) an analogy with the quantum mechanics of electron transfer to define such a smooth potential, VMB, as the eigenvalue of the characteristic equation:
where Δ is a coupling constant and (c1, c2) is the eigenvector. The condition that a nontrivial solution exists leads to the secular equation:
We use the lower-energy solution as the multiple-basin potential:
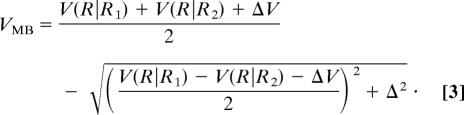 |
VMB being continuous and differentiable can directly be used for molecular dynamics (MD) simulations. The corresponding eigenvector (c1, c2) indicates whether the system resides in basin 1 or 2; thus, we also can use χ = 1n (c2/c1) as a reaction coordinate for the transition. This present interpolation is particularly useful because it allows us to freely tune one of the two fundamental energy scales: The coupling constant Δ modifies directly the energy barrier, and ΔV modulates the relative stability.
Simulation of Conformational Changes.
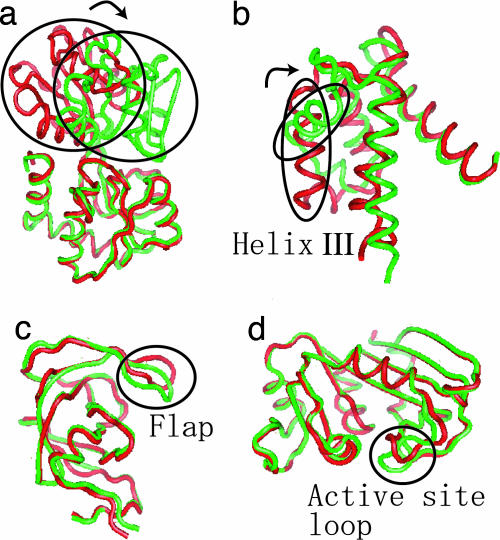
We first illustrate the results using GBP. GBP is composed of two domains connected by a hinge (see Fig. 2a). Without substrates, GBP is found in an open form (red in Fig. 2). Upon binding to glutamine, the hinge swings to make the closed form (green). Using the open form for basin 1 and basin 2 as the closed form as fiducial structures, we tuned ΔV so that the protein spends equal time in each basin. At this tuning, ligand/protein concentration coincides with the dissociation constant. The simulation temperature T was set to 0.8 times the folding transition temperature (see Materials and Methods). One finds that ΔVeq = −4.4 kBT. We obtained a reversible transition between two basins as in Fig. 3b. (Movie 1, which is published as supporting information on the PNAS web site, shows a transition trajectory.) Here we see that the protein resides in each basin for reasonably long times. The transition occurs infrequently, but very rapidly, without any detectable intermediate state.
Fig. 2.
Conformational change of four proteins studied. (a) GBP. (b) S100A6. (c) HIV-1 protease. (d) DHFR. The red structures are those of unbound states (occluded in the case of DHFR), and the green structures are those of bound states (41).
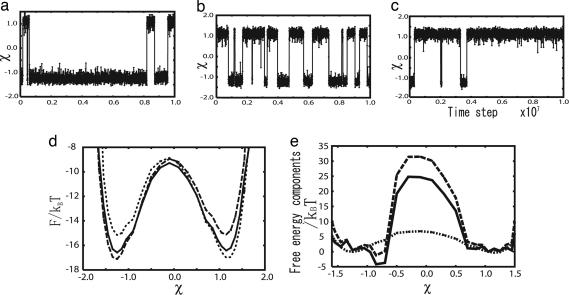
Fig. 3.
Trajectories and free-energy profiles of conformational changes of GBP plotted for the reaction coordinate χ. (a) A trajectory with ΔV = 0 kBT. (b) A trajectory with ΔV = −4.4 kBT. (c) A trajectory with ΔV = −8.9 kBT. (d) The free-energy profiles for three different values of ΔV. The dotted curve corresponds to ΔV = −8.9 kBT, the solid curve corresponds to ΔV = −4.4 kBT, and the dashed curve corresponds to ΔV = 0 kBT. (e) Energetic 〈E〉 (dashed line) and entropic TS (solid line) contributions to the free-energy profile (dotted line) for the case of ΔV = −4.4 kBT.
We plot the free-energy profile F(χ) in Fig. 3d. The open and closed conformations χ ∼ ±1.3 have roughly the equal free energies separated by a single free-energy barrier of modest height ∼ 7 kBT. In contrast, the average energy profile 〈E(χ)〉 shown in Fig. 3e suddenly increases at χ ∼ ±0.7, and the purely energetic contribution to the barrier becomes ∼32 kBT. This large increase in energy is compensated by an entropic contribution −TS = F − 〈E〉 ∼ −25 kBT (Fig. 3e). The sudden increase in conformational entropy at χ ∼ ±0.7 is the hallmark of cracking (18). Importantly, the ability to crack drastically lowers the free-energy barrier. We also simulated the conformational change with ΔV = 0 kBT, where the open conformation is more stable (Fig. 3 a and d), and with ΔV = −8.9 kBT, where the closed conformation is more stable (Fig. 3 c and d).
Dominant Pathways of the Transition.
How are contacts specific to the initial basin broken, and how are the new contacts specific to the final basin formed? To quantitatively answer this question, we used three types of Q scores (i.e., the fraction of formed contacts). The contacts of the two reference structures 1 and 2 are classified into three types: (i) those that are unique to structure 1, (ii) those that are unique to structure 2, and (iii) those contacts that are common to both structures 1 and 2. For each, we define the fraction of those contacts actually formed for any given structure, namely, Q(struct 1) for the type 1 contact set, Q(struct 2) for the type 2, and Q(common) for the type 3. A contact is defined as “formed” when its pair distance falls within a distance 1.15 times that of the reference structure.
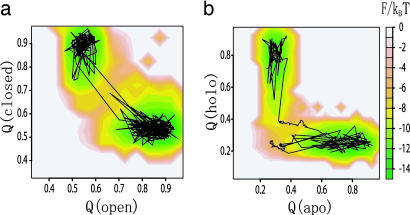
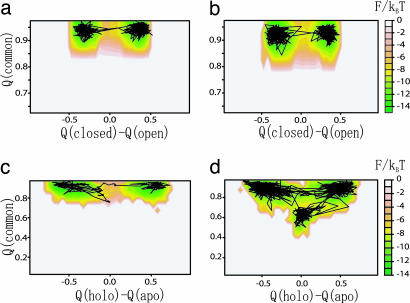
The free-energy surface for GBP is drawn on the Q(closed)–Q(open) plane in Fig. 4a. A representative trajectory superimposed on this surface illustrates the typical transition dynamics. There are two free-energy minima corresponding to the closed (top left basin) and open (right basin) states. These two minima are connected by a straight valley, indicating that breaking of contacts specific to the initial basin and formation of contacts specific to the final basin occur simultaneously. The simultaneity of the transitions is characteristic of the GBP topology change.
Fig. 4.
Free-energy surfaces of conformational change of two proteins. (a) Conformational change of GBP. The y and x axes are the fraction of formed native contacts that are specific to the closed and open conformations, respectively. (b) Conformational change of S100A6. The y and x axes are a fraction of formed native contacts that are specific to the holo and apo conformations, respectively. A representative trajectory is superimposed.
The corresponding free-energy surface for S100A6 is shown in Fig. 4b. S100A6 is the calcium binding domain, a structural analog of a domain of calmodulin (structures depicted in Fig. 2b). The conformational change from the apo (unbound) to holo (bound) states involves an 86° reorientation of helix III leading to a relatively large-scale shear motion. In contrast to the GBP case, the free-energy surface for S100A6 suggests that, upon changing from apo to holo, the contacts specific to apo first are broken, and then contacts specific to holo are formed. The transition is sequential.
The different characteristics of the two free-energy surfaces reflecting distinct mechanisms of conformational change may in part be attributed to the difference in the type of motion; a hinge-type motion for GBP and a shear-type motion for S100A6. In the interdomain hinge motion of GBP the residues that lose contacts upon conformational change are different from those that gain new contacts. Thus, disrupting some contacts and forming new contacts can proceed concomitantly (see Figs. 7 and 8, which are published as supporting information on the PNAS web site). In contrast, the shear motion in S100A6 requires the same residues to exchange contact partners and thus has to be inherently sequential.
For dihydrofolate reductase (DHFR) and HIV-1 protease, in which the conformational changes are rather smaller than these systems, no remarkable features are apparent in the free-energy surface in Q (data not shown).
Temperature Dependence of the Rate.
At low temperature, proteins do not have enough thermal energy to break contacts, and transitions will be very slow. When the temperature reaches nearly the folding temperature TF, the conformational change becomes coupled with transient but global unfolding.
We compare the free-energy surfaces of GBP at T = 0.8 TF (Fig. 5a) with that at T = 0.88 TF (Fig. 5b) (here TF is that of the open form). Using Q(closed)–Q(open) and Q(common), the former functions as the conformational reaction coordinate, and the latter monitors local and global unfolding of the core. There was no qualitative change of the surface between two temperatures, but the population shifts to the open form at the elevated temperature.
Fig. 5.
Temperature dependence of conformational change dynamics of GBP and S100A6. The y axis is a fraction of formed native contacts that are common to two reference structures. The x axis is the difference between the fraction of native contacts specific to holo conformations and native contacts specific to apo conformations. (a) GBP T = 0.8 TF(open). (b) GBP T = 0.88 TF(open). (c) S100A6 T = 0.8 TF(apo). (d) S100A6 T = 0.88 TF(apo). Here TF(apo) is the folding transition temperature of the single Gō model funneled to the apo state. A representative trajectory is superimposed.
In contrast, the same analysis for S100A6 reveals a change of mechanism. At 0.80 TF the protein proceeds directly from one basin to the other (Fig. 5c), but when the temperature is increased to 0.88 TF an additional free-energy minimum emerges (Fig. 5d). Conformations in the minimum are somewhat extended because of loose packing between helices. At this higher temperature there are two possible paths: one direct, and the other via an extended intermediate conformation.
The Transition Rate Coefficient vs. the Driving Force: Tafel Plot.
How does the transition rate depend on the driving force of the conformation change, i.e., ΔV ? This driving force depends on the ligand concentration if binding/unbinding is sufficiently fast. Although the transition rate may be estimated from the barrier in free-energy profile (like Fig. 3d), this estimate of the rate may depend on the specific choice of the reaction coordinate (χ in Fig. 3d) reflecting recrossing effects (33). By using this estimate the dependence of the transition rate on the driving force can be determined. In the electron-transfer processes, a quadratic dependence of the barrier on stability is predicted (34). This finding has been experimentally proven to be fairly accurate (35). For the conformational changes of proteins, Miyashita et al. (18) also argued that a fully elastic model would also give such a curved dependence. On the other hand, they suggested that local unfolding, or cracking, will lead to a linear dependence over a large range of the driving force. We now examine this notion using multibasin model MD simulations.
The conformational transition rate coefficient kchange was estimated as the inverse of the first passage time as in ref. 36. The transition is considered complete when χ first reaches the value at the minimum of the final basin. For very large driving forces, the conformational transition becomes barrier-less and limited only by diffusion, as expected. In this regime the rate becomes saturated. Here we limit ourselves to thermodynamic conditions having significant barriers: The trajectories reside in the initial basin at least 100 MD steps (on average) before making the first transition.
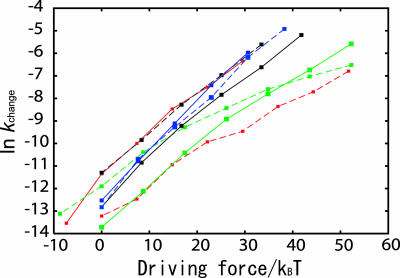
For the four proteins studied, we calculated both the rates going from open to closed and in the other direction over as large a range of the driving force without reaching the barrier-less regime. The resulting Tafel plots (Fig. 6) are surprisingly linear (with the exception of the binding reaction for S100A6, which is curved). The linear dependence is the consequence of the local unfolding as described by Miyashita et al. (18, 21).
Fig. 6.
The transition rate constant as a function of the driving force of the conformational transition (Tafel plot). For each of four proteins, both transitions from unbound to bound (dashed lines) and those from bound to unbound (solid lines), were investigated. The vertical axis is the logarithm of the transition rate coefficient, and the horizontal axis is the driving force. The driving force is ±ΔV, where the sign is chosen so that the increase in the driving force corresponds to stabilize the final state. GBP is in red, S100A6 is in green, DHFR is in black, and HIV-1 protease is in blue.
Discussion
Building on the elastic picture of Miyashita and collaborators, the multiple-basin models somewhat similar to the present one have recently been proposed. Maragakis and Karplus (19) put forward a plastic network model in which individual basins are approximated by the Tirion harmonic model and are then smoothly connected by the secular equation formulation, as we do in Eq. 3. Being locally purely harmonic local unfolding is not taken into account in this model. This plastic network model should work best for small-amplitude conformational changes. The model developed by Best et al. (20) is close in spirit to the present multibasin Hamiltonian because it also connects single-basin potentials in a smooth function. Their interpolation was achieved by the analogy to Boltzmann averaging instead of the secular equation formula. Their model should give similar results to ours when there are two basins. When there are more than two basins, the present model allows the possibility to modulate the barrier heights of each basin-hopping motion individually and thus is somewhat flexible.
Directly modeling the multiple-basin energy landscapes is at an early stage. There is considerable room for creativity and improvement at this low-resolution scale. An ingredient that is missing in the present model as well as in the others (18–21) is precisely how to correctly account for the interaction with ligands. In the current model the effects of ligand are implicit in the value of ΔV, which modulates the overall stability. But such a term does not account for the local nature of the interactions with the ligand. For example, S100A6 binds to calcium ions at the EF-hand loops, and the interactions with calcium ions make the EF-hand loop more rigid. This locality may play an important role in allostery. Further work in this direction is necessary.
The present multiple-basin model can directly be used for MD simulation of very large biomolecular systems, such as molecular motors. Recently, the molecular mechanisms of the rotary motor F1-ATPase were studied by using the switching Gō model (37). In that work, the change in the nucleotide state was modeled as a “vertical excitation,” resulting in switching between single-basin models. The multiple-basin model proposed here provides a natural framework for realizing thermally activated conformational motions coupling ligand binding and release.
Materials and Methods
Proteins Studied.
We studied four proteins: GBP, S100A6, DHFR, and HIV-1 protease. For each, two Protein Data Bank (PDB) structures were used to construct the multiple-basin model. GBP is composed of two domains. Without substrates, GBP is found in an open form (PDB ID code 1GGG; red in Fig. 2a), and upon glutamine binding the hinge between domains swings, forming the closed structure (PDB ID code 1WDN; green). S100A6 is a structural analog of calmodulin. Its conformational change, a shear motion (38), occurs between the apo state (PDB ID code 1K9P; red in Fig. 2b) and the calcium-bound holo state (PDB ID code 1K9K; green). DHFR changes its active-site loops via a shear motion (38) between the occluded state (PDB ID code 1RX6; red in Fig. 2c) and the closed state (PDB ID code 1RX2; green). HIV-1 protease has a β-hairpin loop (the flap) that adopts an open conformation (PDB ID code 3HVP; red in Fig. 2d) without an inhibitor but acquires a closed conformation (PDB ID code 4HVP; green) with an inhibitor. The conformational change is relatively small and is of the hinge type (38).
Multiple-Basin Model: Mathematical Expressions.
The multiple-basin model VMB is defined in Eq. 1 in terms of the single-basin model potentials V(R | Rν) where Rν stands for the reference structure, the structure at the bottom of the basin ν. For the single-basin model, we used Clementi et al.'s version of the off-lattice Gō model (6, 7). In this model, each amino acid is represented as a bead located at the Cα position, two consecutive amino acids are connected by harmonic springs, and local and nonlocal interactions are designed to bias the surface toward the reference structure Rν. (In case of folding, the reference structure is the native.)
The local interactions in the original version of Clementi et al. (6, 7) are:
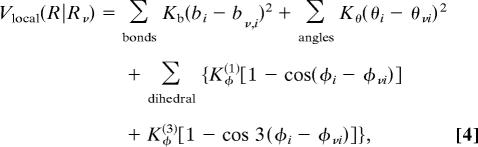 |
where bi is the bond length between ith and (i + 1)th Cα atoms, θi is the ith bond angle between ith and (i + 1)th bonds, and φi is the dihedral angle around the (i + 1)th bond. Parameters with the subscript ν indicate the values of the corresponding variables at the reference structure Rν. Constants K are independent of residue number i, Kb = 100.0, Kθ = 20.0, Kφ(1) = 1.0, and Kφ(3) = 0.5. When a protein's landscape possesses two basins, 1 and 2, we define the local strain energies as the local interaction energies at R1 for the single-basin model defined with the reference structure R2. For example, the local strain energy for θi is Kθ (θ1i − θ2i)2, which monitors the degree of structural change in this portion. Often, a large-amplitude conformational change between states 1 and 2 induces a strong strain in limited and specific portions of the protein, such as the hinge region. Physically, the strain is relieved by breaking fragile local interactions involving this portion. Taking this into consideration, we have reduced the coefficient Kθ of this portion when the strain energy is larger than the cutoff value εθ = 1.0. Explicitly, Kθ in Eq. 4 is replaced with the site-specific constant Kθi, which is defined as Kθi = min[Kθ, εθ/(θ1i − θ2i)2]. In the same way, for the potential on φ, we have reduced the values of Kφi(1) = 2Kφi(3) when the strain energy in the ith dihedral angle exceeds the cutoff value εϕ = 0.5.
The nonlocal interactions in the single-basin model have specific attractive and repulsive interactions for the amino acid pairs that make contacts in the fiducial structure and generic repulsive interactions for the rest of the pairs. Here, an ij pair is considered to be in “contact” when at least one nonhydrogen atom of the ith amino acid is within 6.5 Å of any nonhydrogen atom of the jth amino acid. For the multiple-basin model, the sets of fiducial contact pairs of the reference structures are not equivalent. We need to classify the amino acid pairs into three types: (i) those pairs that make contact in all reference structures, (ii) pairs that make contact in some of, but not all of the reference structures, and (iii) pairs that do not make contact in any reference structures.
For the type 1 and 2 pairs, we modify the functional form for the following reasons. In general, the repulsive part of the pair interaction is quite sharp, and thus the pair energy increases very rapidly when they come closer than the critical distance. For example, in the two-basin case, for type 1 and 2 pairs the critical distances of two reference structures are not the same. This difference induces a large energy gap between the two single-basin potential values, leading to a transition barrier from one basin to another that is unphysically large. To avoid this, we changed the energy function so that the repulsive interactions are identical for the two basins; of the two critical distances, the smaller one is used. Conversely, the attractive part, if any, depends on the basin.
Mathematically, we can express this change in the energy for type 1 and 2 pairs as follows. We divide the nonlocal interactions into Vnative–attr and Vrepul. The former is the attraction interaction between the native contact pairs and is given by:
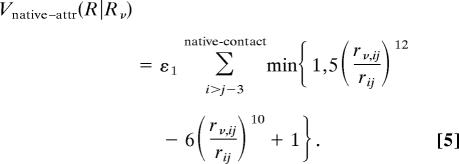 |
Here ε1 = 0.18 was used. We note that, for convenience, we shifted the zero energy at the bottom of the curve instead of the energy at infinite distance, as in the original Clementi et al. version (6, 7). For the type 3 pairs, we can just use the same functional form as the original one because the repulsive force has a generic form. The repulsive part for all types can be written as:
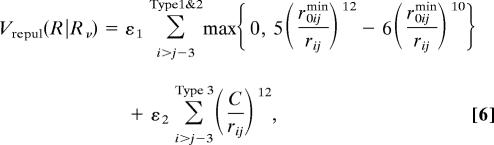 |
where
which is the smallest characteristic distance, ε2 = 0.18, and C = 4.0 Å.
Combining these terms, the total potential of the single-basin model that constitutes the input to the multiple-basin model is V(R | Rν) = Vlocal + Vnative–attr + Vrepul. We note that, with this definition, the single-basin potential is always positive and its value at the bottom of the basin V(Rν | Rν) vanishes.
Although the multiple-basin model was explicitly described for the two-basin case in this article, its generalization to the n-basin case is straightforward. The multiple-basin model potential energy VMB is defined as the solution of the n × n secular equation:
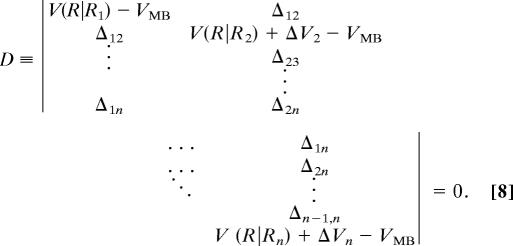 |
Practically, unless more than three V(R | Rν) are near degenerate, we can solve 2 × 2 equations with two smallest V values. In case of the degeneracy, calculating the force on each residue needs a slightly involved procedure. From ∂D/∂ri = 0, we can calculate the force Fi acting on ith residue as:
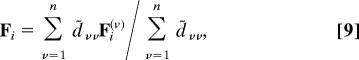 |
where d̃νμ is the minor determinant and Fi(ν) = −∂V(R | Rν)/∂ri.
Simulation Protocol.
In this work, MD simulation was carried out by using the constant-temperature Newtonian dynamics. The velocity Verlet algorithm (39) was used for time propagation, and the temperature was controlled by the simple Berendesen thermostat (40). The mass for all amino acids was set to be the same.
The simulation temperatures were determined by using a crude estimate of the folding transition temperature TF for each reference structure of studied proteins. For S100A6 and DHFR, TF was calculated by the protocol of ref. 36. After this, we assumed that TF scales with the number of contacts per residue. Thus, using the data on S100A6 and DHFR, we estimated the TF values of other proteins. Default simulation temperatures of conformational change were set as 0.8 TF(min), where TF(min) is the smaller of the two TF values associated with two reference structures. (If TF ∼ 80°C, 0.8 TF becomes 9°C) Preliminary tests indicated that at higher temperatures proteins would globally unfold, whereas at lower temperatures the conformational transitions were too slow to sample in reasonable simulation times.
It was necessary to fix the values of the two parameters introduced for the multiple-basin model, the coupling term Δ and the relative stability ΔV. The former controls the energy barrier between two states; the larger is Δ, and the smaller is the barrier. ΔV modulates the relative stability of two states. Both parameters can be determined by using experimental input. For convenience, we adjusted these parameters so that reversible transitions between two conformations were realized for each of the proteins studied. Starting with a fairly small value, Δ is gradually increased until the transitions take place within acceptable computation times. Second, ΔV was tuned so that transitions from each conformation take place with equal frequency. The resulting value is ΔVeq. The parameters so obtained are as follows (in units of kBT): Δ = 59 and ΔVeq = −4.4 for GBP, Δ = 66 and ΔVeq = 11 for S100A6, Δ = 6 and ΔVeq = −2 for DHFR, and Δ = 20 and ΔVeq = 2 for HIV-1 protease.
Supplementary Material
Acknowledgments
N.K. is supported by a Japan Society for the Promotion of Science Research Fellowship for Young Scientists. This work was partly supported by the Water and Biomolecules program of the Ministry of Education, Culture, Sports, Science, and Technology, Japan. Work in University of California at San Diego was supported by National Science Foundation Grants PHY0216576 and PHY0225630.
Abbreviations
- GBP
glutamine-binding protein
- DHFR
dihydrofolate reductase
- MD
molecular dynamics
- PDB
Protein Data Bank
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Daggett V. Protein Simulations. San Diego: Elsevier Academic; 2003. [Google Scholar]
- 2.Tozzini V. Curr. Opin. Struct. Biol. 2005;15:144–150. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Ueda Y., Taketomi H., Go N. Int. J. Pept. Res. 1975;7:445–459. [PubMed] [Google Scholar]
- 4.Socci N. D., Onuchic J. N., Wolynes P. G. Proc. Natl. Acad. Sci. USA. 1999;96:2031–2035. doi: 10.1073/pnas.96.5.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takada S., Luthy-Schulten Z., Wolynes P. J. Chem. Phys. 1999;110:11616–11629. [Google Scholar]
- 6.Clementi C., Jennings P. A., Onuchic J. N. Proc. Natl. Acad. Sci. USA. 2000;97:5871–5876. doi: 10.1073/pnas.100547897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementi C., Nymeyer H., Onuchic J. N. J. Mol. Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 8.Honycutt J. D., Thirumalai D. Biopolymers. 1992;32:695–709. doi: 10.1002/bip.360320610. [DOI] [PubMed] [Google Scholar]
- 9.Tirion M. M. Phys. Rev. Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 10.Bahar I., Jernigan R. L. J. Mol. Biol. 1998;281:871–884. doi: 10.1006/jmbi.1998.1978. [DOI] [PubMed] [Google Scholar]
- 11.McCammon J. A., Gelin B. R., Karplus M., Wolynes P. G. Nature. 1976;262:325–326. doi: 10.1038/262325a0. [DOI] [PubMed] [Google Scholar]
- 12.Cui Q., Li G., Ma J., Karplus M. J. Mol. Biol. 2004;340:345–372. doi: 10.1016/j.jmb.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 13.Krebs W. G., Alexandrov V., Wilson C. A., Echols N., Yu H., Gerstein M. Proteins. 2002;48:682–695. doi: 10.1002/prot.10168. [DOI] [PubMed] [Google Scholar]
- 14.Hayward S., Kitao A., Berendsen H. J. Proteins. 1997;27:425–437. doi: 10.1002/(sici)1097-0134(199703)27:3<425::aid-prot10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 15.Tama F., Sanejouand Y. H. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Tama F., Valle M., Frank J., Brooks C. L., III Proc. Natl. Acad. Sci. USA. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeguchi M., Ueno J., Sato M., Kidera A. Phys. Rev. Lett. 2005;94:078102. doi: 10.1103/PhysRevLett.94.078102. [DOI] [PubMed] [Google Scholar]
- 18.Miyashita O., Onuchic J. N., Wolynes P. G. Proc. Natl. Acad. Sci. USA. 2003;100:12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maragakis P., Karplus M. J. Mol. Biol. 2005;352:807–822. doi: 10.1016/j.jmb.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Best R. B., Chen Y. G., Hummer G. Structure (London) 2005;13:1755–1763. doi: 10.1016/j.str.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Miyashita O., Wolynes P. G., Onichic J. N. J. Phys. Chem. 2005;109:1959–1969. doi: 10.1021/jp046736q. [DOI] [PubMed] [Google Scholar]
- 22.Ansari A., Berendzen J., Bowne S. F., Frauenfelder H., Iben I. E., Sauke T. B., Shyamsunder E., Young R. D. Proc. Natl. Acad. Sci. USA. 1985;82:5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bracken C., Iakoucheva L. M., Romero P. R., Dunker A. K. Curr. Opin. Struct. Biol. 2004;14:570–576. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Shoemaker B. A., Portman J. J., Wolynes P. G. Proc. Natl. Acad. Sci. USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 26.Bryngelson J. D., Wolynes P. G. Proc. Natl. Acad. Sci. USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leopold P. E., Montal M., Onuchic J. N. Proc. Natl. Acad. Sci. USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Go N. Annu. Rev. Biophys. Bioeng. 1983;12:183–210. doi: 10.1146/annurev.bb.12.060183.001151. [DOI] [PubMed] [Google Scholar]
- 29.Shoemaker B. A., Wang J., Wolynes P. G. Proc. Natl. Acad. Sci. USA. 1997;94:777–782. doi: 10.1073/pnas.94.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takada S. Proc. Natl. Acad. Sci. USA. 1999;96:11698–11700. doi: 10.1073/pnas.96.21.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takano M., Higo J., Nakamura H. K., Sasai M. Natural Comput. 2004;3:377–393. [Google Scholar]
- 32.Zuckerman D. M. J. Phys. Chem. B. 2004;108:5127–5137. [Google Scholar]
- 33.Frauenfelder H., Wolynes P. G. Science. 1985;229:337–345. doi: 10.1126/science.4012322. [DOI] [PubMed] [Google Scholar]
- 34.Marcus R. A., Sutin N. Biochim. Biophys. Acta. 1985;811:265–332. [Google Scholar]
- 35.Miller L. T., Calcaterra L. T., Gloss G. L. J. Am. Chem. Soc. 1984;106:3047. [Google Scholar]
- 36.Koga N., Takada S. J. Mol. Biol. 2001;313:171–180. doi: 10.1006/jmbi.2001.5037. [DOI] [PubMed] [Google Scholar]
- 37.Koga N., Takada S. Proc. Natl. Acad. Sci. USA. 2006;103:5367–5372. doi: 10.1073/pnas.0509642103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerstein M., Krebs W. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frenkel D., Smit B. Understanding Molecular Simulations: From Algorithms to Applications. San Diego: Academic; 1996. [Google Scholar]
- 40.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 41.DeLano W. L. pymol User's Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.