Abstract
Bacteria switch the direction their flagella rotate to control movement. FliM, along with FliN and FliG, compose a complex in the motor that, upon binding phosphorylated CheY, reverses the sense of flagellar rotation. The 2.0-Å resolution structure of the FliM middle domain (FliMM) from Thermotoga maritima reveals a pseudo-2-fold symmetric topology similar to the CheY phosphatases CheC and CheX. A variable structural element, which, in CheC, mediates binding to CheD (α2′) and, in CheX, mediates dimerization (β′x), has a truncated structure unique to FliM (α2′). An exposed helix of FliMM (α1) does not contain the catalytic residues of CheC and CheX but does include positions conserved in FliM sequences. Cross-linking experiments with site-directed cysteine mutants show that FliM self-associates through residues on α1 and α2′. CheY activated by BeF3− binds to FliM with ≈40-fold higher affinity than CheY (Kd = 0.04 μM vs. 2 μM). Mapping residue conservation, suppressor mutation sites, binding data, and deletion analysis onto the FliMM surface defines regions important for contacts with the stator-interacting protein FliG and for either counterclockwise or clockwise rotation. Association of 33–35 FliM subunits would generate a 44- to 45-nm-diameter disk, consistent with the known dimensions of the C-ring. The localization of counterclockwise- and clockwise-biasing mutations to distinct surfaces suggests that the binding of phosphorylated CheY cooperatively realigns FliM around the ring.
Many bacteria use flagella operated by rotary motors to swim. Switching the sense of flagellar rotation between clockwise (CW) and counterclockwise (CCW) determines whether the cell tumbles or swims smoothly (for reviews, see refs. 1–4). Binding of the phosphorylated form of the response-regulator CheY (CheY-P) to the motor triggers the change in rotation. The histidine kinase CheA controls generation of CheY-P in response to chemoreceptor occupancy. Depending on the organism, different types of phosphatases terminate the CheY-P signal (e.g., CheC/CheX/FliY in Thermotogae, Bacilli, and Spirochetes or CheZ in β-, δ-, and γ-proteobacteria, which include Escherichia coli) (5).
Bacterial flagella are composed of multiple copies of >20 different proteins (1). EM has generated detailed images of the ≈45-nm Salmonella typhimurium flagellar basal body that embeds in the inner membrane and extends into the cytoplasm (6–8). The MS-ring, formed from ≈26 copies of the protein FliF, is located in the cytoplasmic membrane (8). The C-ring, which extends from the MS-ring into the cytoplasm, is formed mainly from the two proteins FliM (≈35 copies per motor) and FliN (≈100 copies per motor). FliG (≈25 copies per motor), a multidomain protein, associates with FliF in the MS-ring and FliM/FliN in the C-ring and also interacts with the ion-conducting stator protein MotA (1). In vivo genetic experiments, in vitro affinity blotting, coprecipitation, and yeast two-hybrid systems show that FliM, FliN, and FliG together form the “switch complex” (1, 6, 7, 9–16). In Bacillus subtilis and Thermotoga maritima, the switch complex also probably includes the CheY-phosphatase FliY. The switch complex is essential for (i) flagellar assembly, (ii) torque generation, (iii) binding CheY-P, and (iv) changing the sense of the motor rotation (switching).
Structural information is now available for most of the proteins that compose the switch complex. The crystal structure for a FliN fragment (residues 68–154 of 154 residues) reveals a tightly intertwined dimer of largely β-sheet-containing subunits (17). Cross-linking studies (16) indicate that FliN forms a donut-shaped tetramer that could fit into a ring-like feature at the base of the C-ring observed in EM reconstructions (6). The structure for the FliG region that binds to FliM (residues 115–327 of 334 residues) shows two distinct domains linked by a 20-residue-long, possibly flexible linker (18, 19). Mutations in both domains of FliG reduce binding to FliM; mutations in the helical linker affect CCW/CW switching and can suppress motility defects in the stator proteins (18, 20).
To switch rotation direction, CheY-P binds directly to the well conserved FliM N-terminal peptide (LSQXEIDALL) contained on the N-terminal domain of FliM (FliMN). This peptide recognizes the face of CheY-P opposite the phosphorylation site more tightly than unphosphorylated CheY (9–11, 21) because of structural changes that propagate within CheY upon phosphorylation (22).
The middle domain of FliM (FliMM, residues 45–242) has low, but detectable, sequence similarity with the CheC/CheX/FliY family of CheY phosphatases (23, 24). The CheC and CheX phosphatases have pseudo-2-fold symmetry that likely arose from gene duplication (24). Well conserved segments of sequence on two long projecting helices (α1 and α1′) are essential for the dephosphorylation of CheY-P (24). FliM neither conserves these residues nor has phosphatase activity (10, 25, 26). Lastly, FliM also contains a C-terminal domain (FliMC, residues 250–328) that resembles FliN and binds FliN in the overall flagellar assembly (17). T. maritima FliM and FliN form a stable FliM1FliN4 solution complex (17).
Based on the structure of T. maritima CheC (24), we generated a soluble fragment of T. maritima FliM (FliMNM) that does not contain the FliN homology domain. Herein, we report the 2.0-Å resolution crystal structure of FliMM and describe structural relationships among CheC, CheX, and FliM that may have implications for how CheY-P interacts with the flagellar switch. Cross-linking studies based on the FliM structure indicate that FliM self-associates in a side-to-side arrangement, which would allow cooperative switching within the flagella motor. Surprisingly, an α-helix that mediates FliM self-assembly corresponds to regions of CheC and CheX essential for dephosphorylation of CheY.
Results
FliMM Resembles the Chemotaxis Phosphatases CheC and CheX.
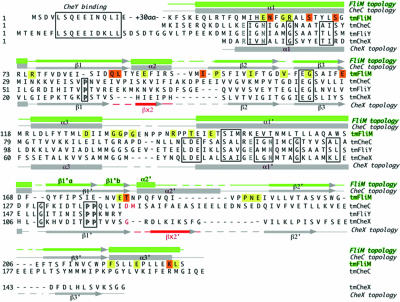
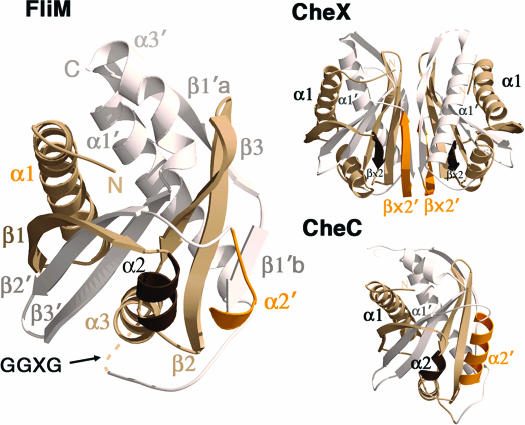
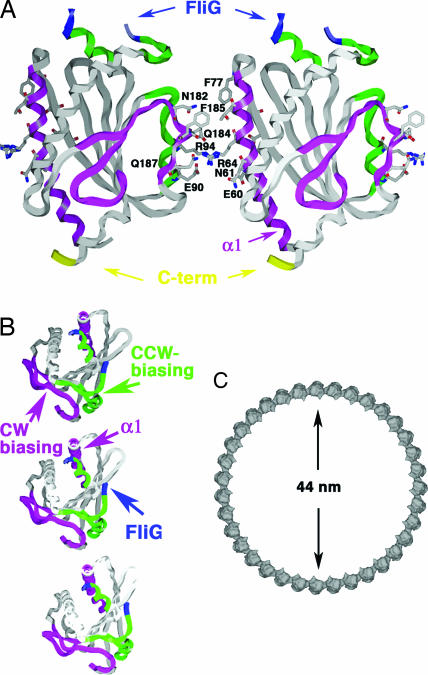
The structure of T. maritima FliMM (residues 44–226) was determined at 2.0-Å resolution by multiwavelength anomalous diffraction of a single-site mercury derivative (Table 1). Among the 1–249 residues of the expressed FliMNM protein, the N-terminal 43 residues and C-terminal ≈20 residues are absent because of a tryptic digestion that was required for crystal growth. The structure of FliMM shares the same topology with the phosphatases CheC and CheX (24) (Figs. 1 and 2). As in CheC, three β-strands (β1–β3) and three α-helices (α1–α3) duplicate to form a pseudosymmetric α/β/α three-layered sandwich. Two long, symmetry-related β-strands wrap around the edges of a central β-sheet platform, and three symmetry-related helices (α1/α1′, α2/α2′, and α3/α3′) surround each face of the β-sheet. As in CheC and CheX, the six β-strands are all antiparallel (β1-β2′-β3′-β3-β2-β1′). Sequence markers for gene duplication are not as pronounced in FliMM as in CheC (Fig. 1), but an internal 2-fold symmetry axis perpendicular to the central β-sheet does relate the two halves of the protein (Fig. 2).
Table 1.
Data collection, phasing, and refinement statistics
| Native | Mercury |
|||
|---|---|---|---|---|
| Peak | Inflection | Remote | ||
| Data collection | ||||
| Space group | P43212 | |||
| Cell dimensions | ||||
| a, b, c, Å | 53.4, 53.4, 130.0 | |||
| Wavelength | 0.9795 | 1.0062 | 1.00936 | 0.9686 |
| Resolution, Å | 30–2.0 (2.07–2.0) | 30–2.7 (2.8–2.7) | 30–2.7 (2.8–2.7) | 30–2.7 (2.8–2.7) |
| Rmerge* | 0.077 (0.436)† | 0.105 (0.453) | 0.115 (0.536) | 0.114 (0.466) |
| I/σI | 31 (11) | 23 (8) | 31 (7) | 25 (9) |
| Completeness, % | 97.7 (96.3) | 99.9 (99.9) | 99.9 (100.0) | 99.9 (100.0) |
| Phasing figure of merit | 0.43 (30.0–2.8 Å) | |||
| Refinement | ||||
| No. of reflections | 12,473 (1,283) | |||
| Rwork/Rfree‡ | 0.224/0.249 | |||
| No. of atoms | 2,913 | |||
| Residues | 179 | |||
| Water | 197 | |||
| B-factors | ||||
| Protein | 28 | |||
| Water | 42 | |||
| Wilson | 25 | |||
| rmsds | ||||
| Bond lengths, Å | 0.007 | |||
| Bond angles, ° | 1.4 | |||
*Rmerge = ΣΣj|Ij − 〈I〉/ΣΣjIj.
†Highest resolution shell for compiling statistics.
‡Rwork = Σ(|Fobs| − |Fcalc|)/Σ|Fobs|. Rfree = Rwork for 10% of the reflections selected at random and removed from the refinement.
Fig. 1.
Sequence alignment of T. maritima FliM, CheC, CheX, and FliY. Secondary structural elements of FliM are more similar to CheC than to CheX, especially in the regions of α2/βx and α2′/βx′ (red). Conserved solvent-exposed FliM residues (yellow) occur at different positions than those of the CheC/CheX/FliY phosphatase family (boxed), with the exception of α1 residues. Solvent-exposed residues that are conserved differently by two general families of chemotactic bacteria (32) indicate potential sites for protein–protein interactions (highlighted in orange).
Fig. 2.
Structure of FliM reveals homology to the CheC/CheX phosphatase family. Ribbon diagrams show topologies and secondary structural elements for FliM (Left), CheX (Upper Right), and CheC (Lower Right). Pseudo-2-fold axes relate one-half of the monomer units (white) to the other (tan). The α2′/βx′ regions (orange), which differ in structure among the three proteins, dimerize CheX, associate CheC with CheD, and mediate FliM self-interactions. The conserved, but disordered, GGXG motif links the two halves of FliM.
Although sequence similarity is very low between FliMM and CheC, both proteins have approximately the same length in most of their α-helices and β-strands (Fig. 1). The most notable differences between FliMM and CheC are a truncated α2–β2 loop and a much shorter α2̀ helix in FliM (Figs. 1, 2). This same secondary structural element distinguishes CheC from CheX: in CheC, this region corresponds to an α-helix (α2′) that binds the activator CheD (27), whereas, in CheX, this region corresponds to a β-strand (βx′) that mediates dimerization (Fig. 1).
The sequence motif “EIGN” contained within α1 and α1′ of CheC is required for phosphatase activity (24). These residues are also conserved in other CheC-like phosphatases CheX and FliY but not in FliM (Fig. 1). However, different residues within the same α1 region (but not α1′) are conserved by FliM proteins [E(D)N(R,K)F(Y)G(A)R; see Fig. 6, which is published as supporting information on the PNAS web site]. Despite the altered consensus motifs, superposition of α1 and α1′ from FliM and CheC shows some similarities in the chemical composition and spatial disposition of exposed side chains (Fig. 7, which is published as supporting information on the PNAS web site). A highly conserved GGXG motif implicated in FliG interactions resides in a disordered loop (α3–α1′) that connects the two pseudosymmetric halves of the molecule.
Binding Constants for T. maritima FliM and CheY.
Isothermal titration calorimetry was used to determine the stoichiometry and binding constants for the interaction between unphosphorylated CheY and T. maritima FliMNM (residues 1–249) or FliM′M (residues 46–242). CheY bound FliMNM with a dissociation constant (Kd) of 1.7 ± 0.2 μM but showed no detectable interaction with FliM′M. Thus, the N-terminal 45 residues in FliM provide the primary contact with unphosphorylated CheY. The affinity of CheY for FliMNM increased 40-fold (Kd = 39 ± 5 nM), when the same titration was performed in the presence of BeF3−, a species known to bind the active-site aspartate of CheY and mimic the phosphorylated state (22, 28–31). No interaction was observed between CheY and FliM′M in the presence of BeF3−. The Kd between unphosphorylated CheY and the CheA P2 domain (Kd = 0.2 μM), which docks CheY for phosphorylation by CheA, lies between the values for FliMNM–CheY and the FliMNM–CheY–BeF3− (32).
In CheC, important residues for CheY dephosphorylation activity reside on α1; hence, we probed whether α1 of FliM participates in direct binding to CheY. We mutated a conserved exposed residue (Glu-60) in FliM α1 to a cysteine residue and modified it with a bulky nitroxide spin-label (MTSSL; methanethiosulfonate spin label). This modification had no appreciable effect on the FliMNM-binding affinity for CheY (Kd = 1.4 ± 0.2 μM) or CheY–BeF3− (Kd = 29 ± 4 nM). Thus, interactions between CheY–BeF3− and α1 of FliMM do not make a major contribution to overall affinity.
The N Terminus of FliM Orders upon Binding to CheY.
FliMNM (29 kDa) elutes on a size-exclusion column at a volume corresponding to a higher molecular mass (≈44 kDa) than expected for a globular protein of its mass. In contrast, FliMM (23 kDa) elutes at a volume appropriate for its size and very similar to that of CheC (also 23 kDa). Thus, the N-terminal segment of FliM (FliMN) increases the hydrodynamic radius of the protein and can be considered structurally disordered. In contrast, both the FliMNM–CheY and the FliMNM–CheY-BeF3− complexes (42 kDa) elute at molecular masses expected for globular complexes of this size. This finding indicates that binding either unphosphorylated or phosphorylated CheY to FliMN generates a more compact structure, at least in the absence of other flagellar components. An ≈35-residue linker of weak sequence conservation connects the CheY-binding N-terminal peptide to the FliMM domain. Near invariance of residues Tyr-39-Asp/Asn-40 in the middle of this linker (Fig. 6) suggests that this region is important for FliM function.
Cross-Linking Studies of FliM Subunit Associations.
End-on views of the C-ring show that it is composed of ≈34 subunits spaced at ≈4-nm intervals (6). On the basis of previous cross-linking results, it has been proposed that FliM is positioned in the middle of the C-ring wall, between FliG and FliN (16). The dimensions of FliMM (3 × 3.5 × 5 nm) are a good fit for this location, with the intermediate dimension corresponding most closely to the observed intersubunit spacing. To test this model for FliM subunit arrangement, we performed targeted cross-linking experiments on E. coli FliM with single or double Cys replacements at positions on the hypothesized FliM–FliM subunit interface. Disulfide cross-linking of the protein in cells was induced by iodine treatment, and products were examined on anti-FliM immunoblots (Fig. 3).
Fig. 3.
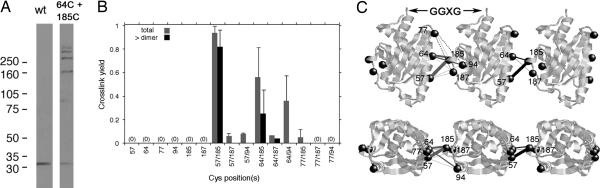
Targeted cross-linking to delineate FliM self-interactions. (A) Anti-FliM immunoblot showing a ladder of cross-linked products formed in cells by the 64/185 double-Cys E. coli FliM mutant. Wild-type (wt) FliM shows no cross-linking. (B) The total cross-link yield (dimer species or larger, gray) and the fraction larger than dimer (black) for single and paired Cys mutants. Error bars represent 1 SD. (C) Three FliM subunits arranged to explain the cross-linking data. Line thickness indicates relative yields, and line colors key to the data panel of B, with gray lines (between Left and Center subunits) representing total cross-linking, black lines (between Center and Right subunits) representing greater-than-dimer yields, and dashed lines indicating pairs of positions that failed to cross-link.
Three Cys pairs (57/185, 64/185, and 64/94 T. maritima numbering) allowed efficient cross-linking into dimers and larger multimers (Fig. 3). Cross-linking yield was greatest for the pairs 57/185 and 64/185 (residues on α1 and α2′); these pairs showed some cross-linking even before the addition of iodine and a ladder of products extending to heptamer after oxidation (Fig. 3A). Other Cys pairs tested showed either a much lower level of cross-linking (57/187, 57/94, 64, 187, and 77/185) or no cross-linking (77/94, 77/187, 77/185, 77/187, 77/94, 57/187, and 57/94) upon treatment with iodine. No cross-linking was observed with any of the single-Cys controls (Fig. 3B). A similar pattern was observed in both flagellate and nonflagellate strains. Thus, the cross-linking reflects a specific interaction between subunits, which appears to occur whether or not the protein is assembled into the motor.
Discussion
The N-Terminal CheY-P-Binding Peptide.
An ≈35-residue linker connects the conserved CheY-binding peptide (LSQXEIDALL) to the rest of the FliM protein. A large increase in hydrodynamic radius for FliMNM compared with FliMM indicates that FliMN has considerable disorder in the absence of CheY and other motor components, which likely explains why crystallization required its removal. In agreement with studies of flagellar proteins from other organisms (9, 11, 33), the N-terminal peptide is the primary interaction site of CheY with T. maritima FliM, and the 1:1 binding increases in affinity ≈10- to 40-fold when CheY becomes activated. However, the absolute affinities of CheY for FliM can differ as much as 100-fold, depending on the parent organisms and context of the measurement (9, 11, 33). Although T. maritima proteins are designed to operate at higher temperatures (≈80°C) than the E. coli proteins (≈37°C), we have found that the T. maritima CheA- and CheY-binding affinities are equivalent to those of the E. coli proteins and largely invariant over a wide temperature range (18–80°C) (32). The different absolute affinities of FliM for CheY most likely reflect either the use of BeF3− as the phosphate mimic or whether FliM is represented by just the N-terminal peptide, FliMNM, the full-length protein, or the intact switch complex.
Linker Region.
An ≈35-aa linker region between the ≈10-residue CheY-binding peptide (LSQXEIDALL) and the central FliM domain is poorly conserved overall, with the exception of two highly conserved residues: Tyr-Asx (Fig. 6). The same CheY-binding peptide is found in FliY, a CheY phosphatase presumed to be part of the switch complex in nonenteric bacteria (26, 34). In FliY, the linker connecting the CheY-binding peptide to the central domain is shorter (10–26 residues) and lacks the invariant Tyr-Asx motif found in all FliM proteins. Mutant studies in S. typhimurium (35) show that single-point mutations in the N-terminal peptide (residues 6–12) and linker (residues 38–48) show phenotypes corresponding to decreased binding of CheY-P. Also, in 10-residue deletion studies of FliM (12, 13), all five deletion mutants in the N-terminal 50 residues support motility but produce a phenotype consistent with a failure to bind CheY-P. Additional residues can be added between the N-terminal CheY-binding peptide and the Y-D/N motif without affecting function in E. coli; however, inserting residues between the equivalents of Tyr-39 and Asp-40 causes a nonchemotactic, CCW-biased phenotype (14). Because calorimetric measurements with BeF3− suggest no additional CheY-P interaction sites in FliM′M, the Tyr/Asx motif of the linker may act as a CheY-P secondary interaction site transmitting the switching signal to other regions of FliM or possibly to FliG.
Helix α1.
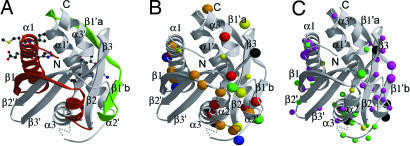
The catalytic EIGN motif of the CheY-P phosphatase CheC resides on α1 and α1′ of the CheC structure. A different ENFGR motif is conserved on α1 in FliM (Figs. 1 and 6). Modification of highly conserved Glu-60 to a bulky substituent did not affect the binding of CheY-BeF3− to FliMM, suggesting that, at least in solution, CheY-P does not strongly interact with α1. However, our disulfide cross-linking studies demonstrate the importance of α1 in FliM oligomerization. Consistent with this finding, dominance studies of 10-residue deletion mutants implicate α1, β1, and α2 of the FliM structure in self-association (Fig. 4A) (12).
Fig. 4.
Mapping mutations that affect function onto the FliMM structure. (A) Ten-residue deletion mutants in the green region produced paralyzed but flagellated phenotypes (12), which suggests that these regions are not important for flagellar assembly per se but necessary for rotation. Deletions in the red region produced nonflagellated cells and were not dominant when expressed with wild-type FliM. Thus, these proteins could not provide contacts for assembly (12). (B) Solvent-exposed positions of conserved hydrophobic (yellow), negatively charged (red), positively charged (blue), polar (green), and glycine (black) residues in FliMM. Larger sphere size indicates invariant residues. Orange indicates residues that are conserved by proteins within each of two families of chemotactic bacteria but differ in residue type between the two families (Fig. 6). (C) Positions of single-point mutations on exposed residues of FliMM that suppress cheY or cheZ mutants and cause either CW motor bias (magenta spheres) or CCW motor bias (green spheres), respectively. The two classes of mutants occur in distinct regions, divided by an area where mutations can cause either phenotype (yellow spheres) (35). Positions of mutations that result in paralyzed phenotypes are shown as black spheres (35). Larger-size spheres indicate mutations that cause greater shifts in the switch bias.
Loop α3–α1′.
A cluster of conserved residues, including the GGXG motif, are contained within a linker between α3 and α1′ that connects the pseudosymmetric halves of FliMM (Figs. 2 and 3C) (14). Weak electron density in this region indicates mobility of the polypeptide and discerns only the first two glycine residues. This mobility probably diminishes upon binding to other switch-complex components. The introduction of one proline into the GGXG motif (GPGDG) reduced binding to FliG and abolished flagellation in E. coli (14). Also, five of six positions where mutations give rise to “paralyzed” (Mot−) phenotypes in S. typhimurium FliM (35) are solvent-exposed and conserved and reside in or near the loop connecting α3 to α1′ (M131, G132, G133, T144, and I146; Fig. 4 B and C). Affinity blotting with the 10-residue deletion mutants (12) also indicates that this region of FliM participates in FliG binding (Figs. 4A and 5A). Taken together, these results suggest that the GGXG motif mediates interactions with another switch protein, most likely the stator-interacting protein FliG.
Fig. 5.
Assembly of FliM in the C-ring. (A) FliM self-association model based on cross-linking data, functional analyses, and intersubunit spacing within the C-ring (see Figs. 3 and 4). FliM self-associates through interactions mediated by largely hydrophilic side chains of α1, β1′, and the α2′–α2 region on the opposing subunit. The GGXG motif implicated in binding FliG is partially disordered (blue). The C terminus of the molecule projects from the bottom to interact with FliN. (B) Top view of three FliM subunits in the C-ring. Switching may involve rotation of the subunits to place the CCW-biasing patch (green) within the subunit interface. (C) An assembly of 35 subunits would generate a C-ring of diameter 44 nm.
Helix α2′.
The most notable structural differences in FliMM compared with CheC are the truncation of helix α2′ and the shortening of loop α2–β2 (Fig. 1). The α2′ region in CheC is important for binding to CheD, which, in turn, activates the phosphatase activity of CheC (27). In CheX, α2′ is replaced with a β-strand (βx′) that mediates dimerization through an extended central β-sheet (24). Conservation of solvent-exposed residues in and around α2′ in FliM (Phe-185 in α2′, Glu-180 in β1′b, and Pro-191-Asn-Glu in loop α2′–β2′) suggests that α2′ is important for protein–protein interactions. All S. typhimurium 10-residue FliM deletions within our model were nonflagellate, except for regions in β1′ and α2′ (Fig. 4B), which gave paralyzed phenotypes. All of these paralyzed mutants could be rescued to some extent by overexpression of FliM or FliN, and so they might affect some aspect of assembly rather than rotation per se (12). The disulfide cross-links between residues on α1 and α2′ demonstrate that these regions contribute to the FliM–FliM interface.
CW- and CCW-Biased FliM Mutants Segregate on the FliMM Structure.
Single-point mutations of S. typhimurim FliM can rescue chemotaxis defects caused by cheY or cheZ mutations, which generate CCW-biased or CW-biased phenotypes, respectively (35). These FliM suppressors are not allele-specific, and many appear to act by stabilizing one state of the switch relative to the other rather than by directly affecting the interaction with CheY. It is more likely for a mutation to destabilize some state than to stabilize a competing state; thus, the CW-biasing mutants probably disfavor the CCW state, whereas the CCW-biasing mutants disfavor the CW state. Mapping the CW-biased and CCW-biased mutation sites onto the structure of FliMM reveals a clear clustering according to phenotype (Fig. 4C). Most of the mutations are in solvent-exposed residues and, hence, suggest a defect in interactions with other subunits/proteins (Fig. 4C). CW-biasing mutations localize on α1 and to a region on the opposite side of FliMM composed of the β2–β3 hairpin, β1b′, and α2′ (Fig. 4C, magenta spheres). CCW-biasing mutants localize to an adjacent area that involves α2, α3, and the α3–α1′ loop (Fig. 4C, green spheres). Thus, sites important for the stability of the CCW or CW states cluster on distinct surfaces of FliM. These patches also coincide with positions of exposed conserved residues (Fig. 4B). Interestingly, mutations at a few sites result in either CW or CCW biases depending on the type of residue introduced; these positions lie at the border of the CW- and CCW-biasing regions (Fig. 4C, yellow spheres).
Cross-Linking Studies Support FliM Self-Association Mediated by α1 and α2′.
Efficient cross-linking of the 57/185 and 64/185 Cys pairs (Fig. 3) indicates that adjacent FliM subunits in the C-ring are in close contact through these positions. The structure shows that, within a FliM subunit, these residues are separated by 33 Å (β-carbon to β-carbon), thus making intramolecular cross-links very unlikely. Given a typical separation of 4 Å between β-carbons of disulfide-bonded Cys residues, adjacent FliM subunits are predicted to be spaced at ≈37 Å, in close agreement with the spacing of 39 Å deduced from EM images (6, 7). Other positions that cross-linked fall on the same edges of FliM as residues 65 and 185 and are also expected to be within cross-linking distance if adjacent FliM subunits orient similarly (i.e., with the same end up) and associate by interactions between α1 and α2′.
FliM Self-Assembly in the C-Ring.
Data from cross-linking, mutant studies, and residue conservation when taken with the FliMM structure and overall dimensions of the C-ring (6, 7) indicate that 33–35 copies of FliM can associate in a ring of the appropriate size (Fig. 5). Orientations of FliM that maintained <10-Å separations between the major cross-linking sites (Cβ-positions) and <16 Å between the minor sites were computationally arranged into a ring of radius 220 Å that contained 33–35 copies of FliM, with the convex site of FliMM facing the convex side of the ring. Side-chain orientations and subunit positions were optimized within the interfacial regions with the program MULTIDOCK (36) (Fig. 5A). The resulting subunit interactions (Fig. 5B) associate α1 and β1 with α2′, α2, and the β1′ C-terminal end in the adjacent subunit, thereby matching α1 with the CW-biasing region (magenta spheres in Fig. 4C). The GGXG motif (Fig. 5A) and α3–α1′ loop project from the top of the ring, permitting interaction with FliG. The position of β1′ on the exterior of the ring and not within the subunit interface is consistent with deletions in this region permitting flagellation but not motility (Fig. 4A). The latter suggests that β1′ is important for directly or indirectly mediating functional interactions with the stator. The C terminus of the FliMM domain extends on the bottom face for attachment to the FliN-interacting domain. The surface area buried between FliM subunits in this association is only ≈170 Å2 per subunit and involves mainly long hydrophilic side chains (57% hydrophilic surface, Fig. 5A). In CW mode, minor rotation of the FliM subunits may disrupt this interface and allow the CCW-biasing region (green regions on Figs. 4C and 5) to mediate contacts between subunits. Although this model assumes a symmetric assembly of FliM in the C-ring, this may not necessarily be the case. The symmetry of FliF–FliG rings allows only ≈26 copies of FliG to engage the ≈34-fold symmetric C-ring (6–8). Thus, FliM could be found in at least two different states, with some copies lacking interactions with FliG or perhaps sharing interactions with the same FliG.
How Does CheY-P Mediate Switching?
Could CheC and FliM have similar structures because they conserve a mode of interaction with CheY-P mediated through α1 or α1′? The current data weigh against this possibility. No CCW-biasing mutants localize to these helices, and we have not been able to detect binding of activated CheY to these regions. Instead, cross-linking shows that α1 participates in an interface with β1′–α2′ on an adjacent subunit. Studies of mutants suggest that CCW rotation requires this mode of assembly (because CW-biasing mutation sites localize here). On the contrary, CCW-biasing mutants localize to a neighboring patch involving α2 and the α3–α1′ loop. Thus, CheY-P may cause switching by favoring an alternative mode of FliM association accessed by a concerted rotation of the subunits. Changes in the assembly of FliN tetramers below FliM could also be coupled to FliM reorganization (16). Reorientation of FliMM and FliN likely alters how FliG engages the ion-conducting stator protein MotA. Importantly, a side-to-side association of FliM around the C-ring allows for a cooperative transition between two alignment states; consistent with the remarkable sensitivity of the switch to CheY-P concentration (33, 37).
Experimental Procedures
Protein Preparation.
The genes encoding T. maritima FliM residues 1–249 (FliMNM), which includes the CheY-binding peptide and the CheC-like domain, FliM residues 46–242 (FliM′M), which includes the CheC-like domain only, and CheY (full-length, residues 1–120) were PCR cloned into the vector pET28a (Novagen) and expressed with a 6-histidine (His) tag in E. coli strain BL21-DE3 (Novagen). The proteins were purified on Nickel-NTA columns, and their His-tags were removed by thrombin digestion (38). Further purification on a Superdex75 sizing column (Amersham Pharmacia) was followed by concentration (Centriprep; Amicon) in GF buffer (50 mM Tris, pH 7.5, and 150 mM NaCl). The complex of FliMNM and CheY was coeluted on the Superdex75 column.
Crystallization and Data Collection.
Initial FliMNM (≈40 mg/ml) crystals appeared after 3 months in an ≈2-μl drop (1:1 mixture of protein in GF buffer and reservoir) from a sealed well under vapor diffusion against a reservoir of 30–40% PEG 4000, 0.1 M Tris·HCl, pH 8.5, and 0.2 M sodium acetate. Optimized crystals grew overnight by adding ≈1 μg/ml trypsin, which cleaves after residue 43 and removes FliMN. Mercury-derivatized crystals were grown in the presence of 1 mM ethyl mercury chloride. Diffraction data for both native (2.5 Å) and mercury-derivatized (2.7 Å) FliMM crystals (space group P43212, one molecule per asymmetric unit, 30% solvent) were collected under a 100 K nitrogen stream at the National Synchrotron Light Source (X25) on a CCD detector (Quantum 315; Area Detector Systems). Diffraction data were collected at three wavelengths chosen to optimize the mercury anomalous signal and processed by HKL2000 (39) (Table 1).
A higher resolution (2.0 Å) data set for FliMM was obtained at the Cornell High-Energy Synchrotron Source (F2) under a 100 K nitrogen stream on a CCD detector (Q4; Area Detector Systems). These latter crystals grew in 1 month from a FliMNM–CheY complex (≈100 mg/ml) in an ≈2-μl drop equilibrated by vapor diffusion against a reservoir of 0.8 M Na K tartrate and 0.1 M Hepes, pH 7.5 (Hampton Research). These crystals also contained protein that underwent proteolysis of FliMN at residue 43 and were isomorphous to those grown from the PEG conditions.
Structure Determination and Refinement.
Patterson analysis revealed one mercury atom bound per asymmetric unit. The program SOLVE/RESOLVE (40, 41) was used to generate experimental phases from the multiwavelength anomalous diffraction data collected at three wavelengths (Table 1). The initial FliMM model was built manually with XFIT (42) to 2.8-Å resolution and then improved with the program ARP/wARP (43) against 2.0-Å resolution data. The final model (residues 44–228) was refined with the program CNS (44) after water molecule placement (final R-factor = 0.224, Rfree = 0.249; Table 1). Although the latter crystal was grown from preformed FliMNM–CheY complex, no additional electron density corresponding to FliMN or CheY was observed.
Site-Directed Mutagenesis and Cysteine Blockage.
A cysteine point mutation in FliM (E60C) was introduced by QuickChange mutagenesis (Stratagene) and verified by DNA sequencing. The mutant protein was expressed and purified as described above and bound to a nickel-NTA-affinity column. The column bed was exchanged with GF buffer containing 5–10 mM MTSSL (1-oxyl-2,2,5,5-tetramethylpyrolinyl-3-methyl)-methanethiosulfonate; Toronto Research). After 4 h at room temperature and overnight at 4°C, unreacted MTSSL was washed off with GF buffer. After a 6- to 12-h incubation of the column with thrombin, the protein was eluted with GF buffer. Incorporation of the label was confirmed by ESR spectroscopy.
Isothermal Titration Calorimetry (ITC).
Protein concentrations were determined by the RC/DC assay (Bio-Rad, Hercules, CA) with cytochrome c as standards for FliMNM (29.1 kDa), FliMNM E60C-MTTSL (29.1 kDa), FliM′M (22.8 kDa), and CheY (13.2 kDa). Before titration, samples of FliMNM, FliM′M, and CheY were dialyzed against GF buffer. To mimic the phosphorylated state of CheY, BeF3− was added to the dialysis GF buffer (GF buffer plus 0.5 mM BeCl2, 3 mM NaF, and 1 mM MgCl2) before the titration experiments. [Higher concentrations of BeF3− and Mg2+ (5 mM BeCl2, 27 mM NaF, and 8 mM MgCl2) give a similar binding affinity.] Calorimetric measurements with a VP-ITC titration calorimeter (MicroCal, Northampton, MA) were carried out by titrating CheY (0.5–1 mM) into FliM (50–100 μM) at 26°C. The thermodynamic parameters were determined by fitting to a single site-binding model with the Origin software package (MicroCal).
Disulfide Cross-Linking Studies.
Cross-linking studies of FliM were carried out in E. coli, as described (45). Briefly, cells were cultured, pelleted, and resuspended in a buffer containing 20 mM sodium phosphate, pH 7.4, and 150 mM NaCl, to an OD600 of 10. Cells were incubated for at least 10 min on ice, and then controls were treated with N-ethylmaleimide (NEM) (final concentration 20 mM); experimental samples were treated with 0.2 mM I2 added from a 20 mM stock in 95% ethanol. Samples were left on ice for 10 min, then NEM was added to experimental samples to block unreacted sulfhydryls. After 5 min, cells were pelleted, resuspended in nonreducing gel-loading buffer containing 7% SDS, boiled, loaded on gels, and analyzed by SDS/PAGE and immunoblotting (16, 45).
C-Ring Modeling.
Orientations of FliM were computationally placed in a C-ring of radius 43–46 nm that contained 33–35 subunits. Knowledge of the efficient cross-linking sites put strong constraints on possible orientations of subunits relative to the C-ring axis. Orientations and side-chain positions were optimized for an interacting dimer with MULTIDOCK (36) and the resulting subunits placed back into a C-ring of appropriate size and checked for maintenance of the cross-linking distances.
Supplementary Material
Acknowledgments
We thank the National Synchrotron Light Source and Cornell High-Energy Synchrotron Source for access to data collection facilities and Peter Borbat for assistance with ESR spectroscopy. This work was supported by National Institutes of Health Grants GM64664 (to D.F.B.) and GM066775 (to B.R.C.).
Abbreviations
- CCW
counterclockwise
- CheY-P
phosphorylated CheY
- CW
clockwise.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2HP72HP7).
References
- 1.Berg H. C. Annu. Rev. Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S., Blair D. F. Int. Rev. Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 3.Wadhams G. H., Armitage J. P. Nat. Rev. Mol. Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 4.Sourjik V. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Szurmant H., Ordal G. W. Microbiol. Mol. Biol. Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas D., Morgan D. G., deRosier D. J. J. Bacteriol. 2001;183:6404–6412. doi: 10.1128/JB.183.21.6404-6412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young H. S., Dang H., Lai Y., DeRosier D. J., Khan S. Biophys. J. 2003;84:571–577. doi: 10.1016/S0006-3495(03)74877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki H., Yonekura K., Namba K. J. Mol. Biol. 2004;337:105–113. doi: 10.1016/j.jmb.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 9.Sagi Y., Khan S., Eisenbach M. J. Biol. Chem. 2003;278:25867–25871. doi: 10.1074/jbc.M303201200. [DOI] [PubMed] [Google Scholar]
- 10.Bren A., Eisenbach M. J. Mol. Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 11.McEvoy M. M., Bren A., Eisenbach M., Dahlquist F. W. J. Mol. Biol. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 12.Toker A. S., Kihara M., Macnab R. M. J. Bacteriol. 1996;178:7069–7079. doi: 10.1128/jb.178.24.7069-7079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toker A. S., Macnab R. M. J. Mol. Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 14.Mathews M. A., Tang H. L., Blair D. F. J. Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang H., Braun T. F., Blair D. F. J. Mol. Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 16.Paul K., Blair D. F. J. Bacteriol. 2006;188:2502–2511. doi: 10.1128/JB.188.7.2502-2511.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown P. N., Mathews M. A., Joss L. A., Hill C. P., Blair D. F. J. Bacteriol. 2005;187:2890–2902. doi: 10.1128/JB.187.8.2890-2902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown P. N., Hill C. P., Blair D. F. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd S. A., Whitby F. G., Blair D. F., Hill C. P. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 20.Marykwas D. L., Berg H. C. J. Bacteriol. 1996;178:1289–1294. doi: 10.1128/jb.178.5.1289-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch M., Oosawa K., Aizawa S. I., Eisenbach M. Biochemistry. 1994;33:10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- 22.Lee S. Y., Cho H. S., Pelton J. G., Yan D. L., Henderson R. K., King D. S., Huang L. S., Kustu S., Berry E. A., Wemmer D. E. Nat. Struct. Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- 23.Kirby J. R., Kristich C. J., Saulmon M. M., Zimmer M. A., Garrity L. F., Zhulin I. B., Ordal G. W. Mol. Microbiol. 2001;42:573–585. doi: 10.1046/j.1365-2958.2001.02581.x. [DOI] [PubMed] [Google Scholar]
- 24.Park S. Y., Chao J., Gonzalez-Bonet G., Beel B. D., Bilwes A. M., Crane B. R. Mol. Cell. 2004;16:563–574. doi: 10.1016/j.molcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Bischoff D. S., Ordal G. W. Mol. Microbiol. 1992;6:23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 26.Szurmant H., Bunn M. W., Cannistraro V. J., Ordal G. W. J. Biol. Chem. 2003;278:48611–48616. doi: 10.1074/jbc.M306180200. [DOI] [PubMed] [Google Scholar]
- 27.Chao X., Muff T. J., Park S. Y., Zhang S., Pollard A. M., Ordal G. W., Bilwes A. M., Crane B. R. Cell. 2006;124:561–571. doi: 10.1016/j.cell.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 28.Lee S. Y., Cho H. S., Pelton J. G., Yan D. L., Berry E. A., Wemmer D. E. J. Biol. Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- 29.Cho H., Wang W., Kim R., Yokota H., Damo S., Kim S. H., Wemmer D., Kustu S., Yan D. Proc. Natl. Acad. Sci. USA. 2001;98:8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho H. S., Lee S. Y., Yan D. L., Pan X. Y., Parkinson J. S., Kustu S., Wemmer D. E., Pelton J. G. J. Mol. Biol. 2000;297:543–551. doi: 10.1006/jmbi.2000.3595. [DOI] [PubMed] [Google Scholar]
- 31.Yan D., Cho H. S., Hastings C. A., Igo M. M., Lee S. Y., Pelton J. G., Stewart V., Wemmer D. E., Kustu S. Proc. Natl. Acad. Sci. USA. 1999;96:14789–14794. doi: 10.1073/pnas.96.26.14789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S. Y., Beel B. D., Simon M. I., Bilwes A. M., Crane B. R. Proc. Natl. Acad. Sci. USA. 2004;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sourjik V., Berg H. C. Proc. Natl. Acad. Sci. USA. 2002;99:12669–12674. doi: 10.1073/pnas.192463199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szurmant H., Muff T. J., Ordal G. W. J. Biol. Chem. 2004;279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- 35.Sockett H., Yamaguchi S., Kihara M., Irikura V. M., Macnab R. M. J. Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson R. M., Gabb H. A., Sternberf M. J. E. J. Mol. Biol. 1998;276:265–285. doi: 10.1006/jmbi.1997.1519. [DOI] [PubMed] [Google Scholar]
- 37.Cluzel P., Surette M., Leibler S. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 38.Bilwes A. M., Alex L. A., Crane B. R., Simon M. I. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski A., Minor W. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger T. C. Acta Crystallogr. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McRee D. E. J. Mol. Graphics. 1992;10:44–47. [Google Scholar]
- 43.Perrakis A., Morris R. M., Lamzin V. S. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 44.Brunger A. T., Adams P. D., Clore G. M., Delano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Lowder B. J., Duyvesteyn M. D., Blair D. F. J. Bacteriol. 2005;187:5640–5647. doi: 10.1128/JB.187.16.5640-5647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.