Abstract
The Rtf1 subunit of the Paf1 complex is required for proper monoubiquitination of histone H2B and methylation of histone H3 on lysines 4 (H3K4) and 79 in yeast Saccharomyces cerevisiae. Using RNAi, we examined the role of Rtf1 in histone methylation and gene expression in Drosophila melanogaster. We show that Drosophila Rtf1 (dRtf1) is required for proper gene expression and development. Furthermore, we show that RNAi-mediated reduction of dRtf1 results in a reduction in histone H3K4 trimethylation levels on bulk histones and chromosomes in vivo, indicating that the histone modification pathway via Rtf1 is conserved among yeast, Drosophila, and human. Recently, it was demonstrated that histone H3K4 methylation mediated via the E3 ligase Bre1 is critical for transcription of Notch target genes in Drosophila. Here we demonstrate that the dRtf1 component of the Paf1 complex functions in Notch signaling.
Keywords: chromatin, elongation, RNA polymerase II, transcription, monoubiquitination
Interplay between transcriptional activators and repressors regulates gene expression by RNA polymerase II (RNA Pol II). In several cases, chromatin structure is implicated in transcriptional activation and repression. Posttranslational methylation of lysines on the N-terminal tails of histones is thought to modulate higher-order chromatin folding and can activate or repress transcription, depending on the residue being methylated, the regulatory protein recruited by the methyl mark, and whether the lysine is mono-, di-, or trimethylated (1).
Histone modifications can be interdependent, such that one modification requires another preexisting modification (1). Histone H3 methylation at lysines 4 and 79 is catalyzed by the complex of proteins associated with Set1 (COMPASS) and Dot1p, respectively in Saccharomyces cerevisiae (1–8). Methylation at these residues requires monoubiquitination of histone H2B at lysine 123 by Rad6/Bre1 (9–11). The Paf1 complex indirectly regulates histone methylation through its regulation of H2B monoubiquitination and interaction of COMPASS with RNA Pol II (12–14).
The Paf1 complex in yeast is composed of five subunits, Paf1, Rtf1, Cdc73, Ctr9, and Leo1, and is associated with the elongating form of RNA Pol II (1, 15–18). The Rtf1 component of Paf1 is required for H2B ubiquitination by Rad6 (12–14) and for the recruitment of Set1/COMPASS to elongating RNA Pol II (12, 13). Because Rtf1 is essential for histone monoubiquitination, methylation, and transcriptional control in yeast, we sought the Drosophila homologue dRtf1 to characterize its role in higher eukaryotes. Here we use RNAi to reduce dRtf1 expression levels and examine the in vivo effect of dRtf1 reduction on transcription and development in the fly. We show that RNAi knockdown of dRtf1 causes pupal lethality. To demonstrate a role for dRtf1 in gene expression, we tested the effect of dRtf1 RNAi knockdown on heat shock gene expression and found that Rtf1 knockdown results in a reduction in heat shock (Hsp70) gene expression. Recently, Bray et al. (19) demonstrated that the Drosophila homologue of Bre1 is required for proper histone H3K4 methylation and is critical for transcription of Notch target genes. In this study, we show that the dRtf1 component of the Paf1 complex participates in Notch signaling in the wing margins. Our studies indicate that transcriptional regulation via the Paf1 complex is highly conserved among eukaryotes.
Results
Paf1 Complex Regulation of Histone Methylation.
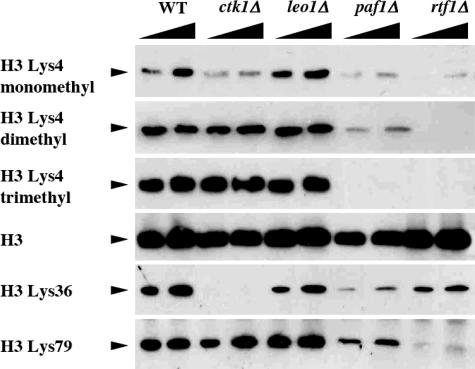
Many of the subunits of COMPASS (the yeast homologue of the mammalian MLL complex and the Drosophila trithorax complex) are required for the proper mono-, di-, and/or trimethylation of H3K4 (1). In addition to COMPASS, the E2 conjugating enzyme Rad6 and its E3 ligase Bre1 are required for proper H3K4 methylation via the regulation of H2B monoubiquitination (9–11). Also, it has been shown that deletion of components of the Paf1 complex and the Bur1/Bur2 kinase can greatly reduce histone H2B monoubiquitination and, thereby, H3K4 methylation (12–14, 20, 21). However, deletion of RTF1, which is required for the activation of Rad6, seems to be required for mono-, di-, and trimethylation mediated by COMPASS (Fig. 1). This observation mirrors that of effects observed when either RAD6 or BRE1 is deleted. Although loss of H2B monoubiquitination is not fully required for H3K4 monomethylation by COMPASS, this observation can be explained by the fact that Rtf1 is not only required for the regulation of H2B monoubiquitination but also plays a role in the recruitment of COMPASS to the transcribing RNA Pol II (13). In addition to regulation of H3K4 methylation, different components of the Paf1 complex have varying effects on other types of H3 tail methylation (Fig. 1).
Fig. 1.
Rtf1 regulation of histone H3 methylation. Histone H3 methylation patterns in the indicated yeast strains were tested by using Western blot analyses with specific antibodies generated toward each modified histone H3.
Because Paf1 is required for histone H3K4 and K79 methylation, we tested the effect of Paf1 loss on histone methylation stability in vivo (data not shown). We used a tetracycline-regulated PAF1 gene strain grown under normal conditions, turning off the expression of Paf1 in the presence of tetracycline. Cell extracts were prepared at different time points, and histone H3 modification stability was tested in the absence of Paf1. After approximately 4 hours in the presence of tetracycline, Paf1 levels were reduced by >95%. However, even 12 hours after Paf1 loss, histone H3K4 and K79 methylation levels appear to be unaffected. Our observation substantiates a report by Ng et al. (14), who found that H3K4 methylation seems to be a stable mark once established on the Gal gene. We verify that histone H3K4 methylation seems to be stable, further supporting a role for the stability of this type of histone modification.
Developmental Expression of the Drosophila Rtf1, dRtf1.
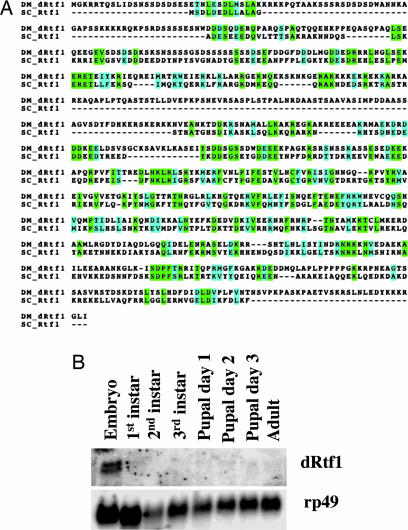
Loss of the Rtf1 subunit of the yeast Paf1 complex has the greatest effect on histone H2B monoubiquitination and H3K4 methylation (Fig. 1). To investigate the conservation of Rtf1's role in histone methylation and regulation of gene expression in a higher eukaryote, we characterized the role of subunit Rtf1 in histone methylation and transcriptional regulation in Drosophila. CG10955 is the Drosophila structural homologue of yeast Rtf1, dRtf1 (Fig. 2A). Fig. 2B shows dRtf1 mRNA levels visualized by Northern blot during eight developmental stages. A transcript of ≈3 kb was detected by using dRtf1-radiolabeled antisense RNA probe. Much higher levels of transcript were observed in embryos than in any other developmental stage (Fig. 2B). In this respect, dRtf1 expression was similar to elongation factor dELL (22), but it contrasted with elongation factor dEloA expression, which is most abundant in late larvae and during pupariation (23).
Fig. 2.
Drosophila Rtf1 expression patterns. (A) The gene CG10955 was identified as the Drosophila homologue of Rtf1. (B) Total RNA was isolated from each developmental stage of Drosophila. Fifteen micrograms of total RNA from each stage was loaded and analyzed by Northern blot analysis using a dRtf1-specific probe.
dRtf1 Is Essential for Development in Drosophila.
We used RNAi to lower Rtf1 mRNA and protein levels, to determine whether dRtf1 is required for Drosophila viability. RNAi knockdown of dRTF1 was accomplished by using P element-mediated transformation of the SympUAST vector with a 600-bp insertion of dRtf1 cDNA sequence. The SympUAST vector contains two convergent Gal4-inducible UAS promoters that can be used to transcribe double-stranded RNA from the dRtf1 fragment, activating the Drosophila RNAi machinery (24). SympUAST–dRtf1 flies were crossed to the Actin5C–Gal4/CyO, y+ driver line to activate RNAi. Rtf1 RNAi is expressed only in Actin5C–Gal4/SympUAST–dRtf1 progeny, and not their CyO, y+/SympUAST–dRtf1 siblings. When we used one line, 10A, 235 CyO, y+/SympUAST–dRtf1 adults were recovered with no Actin5C–Gal4/SympUAST–dRtf1 progeny observed. When we used a second independent line, 17C, 310 CyO, y+/SympUAST–dRtf1 adults were recovered with no Actin5C–Gal4/SympUAST–dRtf1 progeny observed. A large number of dead pupae were observed in both crosses, suggesting that the Rtf1 RNAi flies were dying during the pupal stage. Because lethality was observed by using two independent SympUAST–dRtf1 insertion lines, lethality was unlikely to be due to ectopic gene activation at the SympUAST–dRtf1 insertion site.
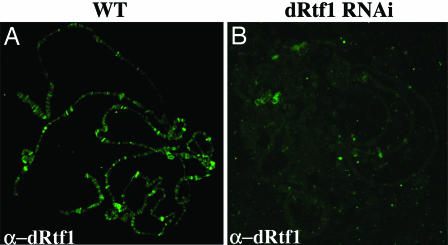
In Actin5C–Gal4/SympUAST–dRtf1 third instar larvae, Rtf1 protein levels on polytene chromosomes (Fig. 3B) were reduced compared with those in their CyO, y+/SympUAST–dRtf1 siblings (Fig. 3A). Although Northern blot analysis showed that dRtf1 transcripts decreased by ≈20% (data not shown), this level of reduction seems to be sufficient to cause 100% lethality. Our data indicate that, upon expression of Rtf1 RNAi in Actin5C–Gal4/SympUAST–dRtf1 progeny, the overall level of chromosomal Rtf1 was substantially reduced, suggesting that the small RNAs generated by Dicer from the long double-stranded Rtf1 RNA precursors may act as microRNAs (miRNAs) and may inhibit Rtf1 translation as well.
Fig. 3.
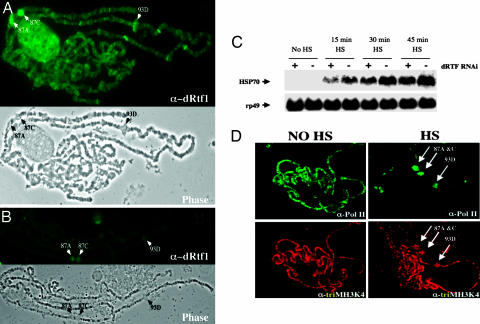
Loss of Rtf1 levels on chromatin. SympUAST–dRtf1 flies were crossed to the Actin5C–Gal4/CyO, y+ driver line to activate RNAi. Fixed polytene chromosome squashes were prepared from CyO (WT) (A) and Actin 5C (dRtf1 RNAi) (B) larvae and were immunostained with an antibody specific for dRtf1.
dRtf1 Is Required for Histone H3K4 Trimethylation on Polytene Chromosomes.
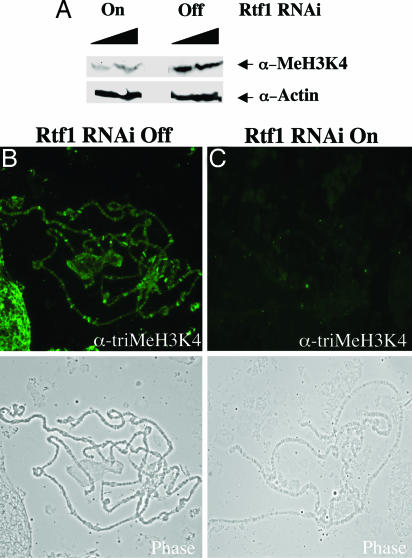
Because Rtf1 loss in yeast results in the abrogation of H3K4 methylation, we tested whether dRtf1 in Drosophila functions in the same pathway. Extracts prepared from both Actin5C–Gal4/SympUAST–dRtf1 progeny (Rtf1 RNAi on) and CyO, y+/SympUAST–dRtf1 progeny (Rtf1 RNAi off) were tested for methylated H3K4 levels. As shown in Fig. 4A, dRtf1 RNAi knockdown resulted in a significant reduction in total cellular trimethylated H3K4. To test whether this reduction occurs throughout the genome, fixed polytene chromosomes squashes were prepared from Actin5C–Gal4/SympUAST–dRtf1 and CyO, y+/SympUAST–dRtf1 larvae and were immunostained with an antibody specific for trimethylated H3K4. In WT polytene chromosomes, trimethylated H3K4 is widely distributed throughout the euchromatic chromosome arms and is highly enriched at developmental puffs, sites of active transcription (Fig. 4B). In contrast, the polytene chromosomes from dRtf1 knockdown larvae consistently show reduced staining with the same antibody (Fig. 4C). Together, our data suggest that the loss of Rtf1 results in the loss of H3K4 methylation in Drosophila.
Fig. 4.
dRtf1 is required for proper histone H3K4 trimethylation. (A) Extracts from Actin5C–Gal4/SympUAST–dRtf1 progeny (Rtf1 RNAi On) and CyO, y+/SympUAST–dRtf1 progeny (Rtf1 RNAi Off) were applied to SDS/PAGE and Western blot analysis by using antibodies specific to methylated H3K4. (B and C) Trimethylated H3K4 levels are reduced on dRtf1 knockdown chromosomes. (B) CyO, y+/SympUAST–dRtf1 progeny chromosomes (Rtf1 RNAi Off) immunostained with anti-trimethyl H3K4 serum (Upper) and corresponding phase-contrast image of the same chromosome (Lower). Trimethylated H3K4 is widely distributed and enriched at sites of active transcription in WT chromosomes. (C) Actin5C–Gal4/SympUAST–dRtf1 progeny (Rtf1 RNAi On) chromosomes immunostained with anti-trimethyl H3K4 serum (Upper) and corresponding phase-contrast image of the same chromosome (Lower). H3K4 trimethylation is dramatically reduced in dRtf1 knockdown chromosomes.
dRtf1 Affects Transcription of Heat Shock Genes.
Rtf1 in S. cerevisiae is required for COMPASS recruitment to elongating RNA Pol II (12–14), implicating it in RNA Pol II transcription. Accordingly, we tested the effect of dRtf1 reduction on transcription in flies. Because RNAi knockdown of dRtf1 in cultured S2 cells reduces Hsp70 expression by ≈25% (25), we used the Hsp70 heat shock genes as reporters for the effect of dRtf1 knockdown in whole animals. We found that, in Actin5C–Gal4/SympUAST–dRtf1 third instar larvae dRtf1 levels at the 87A and 87C loci (Hsp70 puff sites) was reduced (Fig. 5A and B) and Hsp70 transcripts were reduced at various time points throughout a 60-min heat shock (Fig. 5C). This observation indicates that dRtf1 is required for proper regulation of heat shock gene expression in whole flies and suggests that histone methylation is required for full heat shock gene expression.
Fig. 5.
dRtf1 is required for proper heat shock gene expression. (A and B) CyO, y+/SympUAST–dRtf1 chromosomes (Rtf1 RNAi off) (A) and Actin5C–Gal4/SympUAST–dRtf1 chromosomes (Rtf1 RNAi on) (B) were immunostained with anti-Rtf1 after heat shock to determine the levels of Rtf1 at heat shock puff sites upon Rtf1 RNAi induction. (C) Larvae with RNAi-mediated depletion of dRtf1 express less Hsp70 transcript than larvae that do not carry the RNAi driver construct. Male dRtf1 RNAi-carrying flies were crossed to the female (Actin5C–Gal4/CyO driver) flies. The progeny were heat shocked (HS) for various times as indicated. Total RNA was prepared from both nonheat shocked and heat shocked Actin 5C–Gal4 and CyO larvae. A Northern blot probed for Hsp70 mRNA shows that transcript levels are reduced in RNAi+ animals. rp49 mRNA was probed as a load control. (D) WT (Oregon R) chromosomes were immunostained with anti-RNA Pol II (Upper) and anti-trimethyl H3K4 serum (Lower) before and after heat shock (HS). Sites of heat shock puffs are indicated by arrows.
A previous report showed that dRtf1 is recruited to heat shock loci in polytene chromosomes upon heat shock (25). To test whether dRtf1 recruitment is associated with H3K4 methylation at heat shock loci, we immunostained polytene chromosomes from heat shocked larvae with an antibody to trimethylated H3K4. Trimethylated H3K4 accumulates at heat shock puff sites (87A, 87C, and 93D) in response to heat shock (Fig. 5D). Note that extensive H3K4 methylation remains throughout the rest of the genome (Fig. 5D), consistent with the observation that a significant fraction of dRtf1 remains at non-heat shock loci during heat shock (25).
Knockdown of dRtf1 by RNAi Enhances Nnd-1.
The Notch signaling pathway is required for regulation of cell fate decisions throughout metazoan development. Although Notch signaling is known to activate transcription of numerous target genes, little is known regarding the molecular details of this process. Recently, Bray et al. (19) reported that the Drosophila homologue of yeast Bre1 is required for Notch target gene expression in Drosophila. Because Rtf1 is also required for regulation of Rad6/Bre1 in yeast, we tested whether dRtf1 is required for proper Notch signaling.
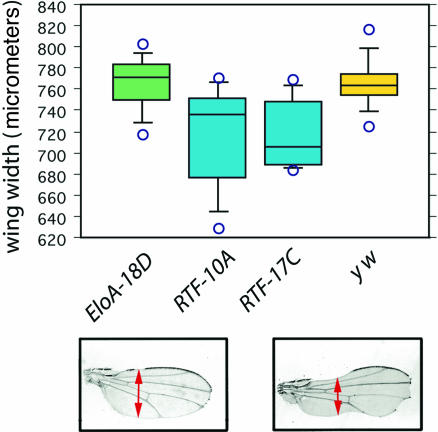
The hypomorphic Notch allele Nnd-1 causes limited nicking in the margin of the adult wing. Mutations in factors required for Notch signaling enhance this wing nicking; strong enhancement results in a reduced wing width. Mild, nonlethal activation of two independent dRtf1 RNAi knockdown transgenes, dRtf1–10A and dRtf1–17C, by an Hsp70–Gal4 driver at 27°C decreased the width of Nnd-1 wings relative to a no-RNAi control (y w) and relative to RNAi directed against dEloA (dEloA–18D) (Fig. 6). Both dRtf1 RNAi insertions gave significant reductions in wing width relative to the dEloA and y w controls (P < 0.003 in all cases by the Bonferroni/Dunn test). There was no significant difference in wing width between y w and dEloA RNAi, demonstrating that the effect on Nnd-1 was not due to ectopic Gal4 expression.
Fig. 6.
Partial knockdown of Rtf1 by RNAi increases Nnd-1 hypomorphic mutant wing phenotype severity. Two independent RNAi P element insertions against Rtf1 (Rtf1–10A and Rtf1–17C) decreased the width of Nnd-1 wings relative to no RNAi (y w) or RNAi directed against EloA (EloA–18D). The wing width distributions are shown as box plots, where the horizontal lines indicate the 10th, 25th, 50th, 75th, and 90th percentiles. Wing width was measured from the anterior margin to the posterior margin between the cross-veins, as diagrammed below the box plots. Decreased wing width indicates reduced Notch function. Both Rtf1 RNAi insertions gave significant reductions in wing width relative to the EloA and y w controls (P < 0.003 in all cases by the Bonferroni/Dunn test). There was no significant difference in wing width between y w and EloA RNAi.
Discussion
The yeast Paf1 complex interacts with RNA Pol II and regulates the pattern of histone modifications; deletion of the Rtf1 subunit in yeast has the strongest phenotype on histone H3 modification loss (Fig. 1). Analysis of the functional homologue of the Paf1 complex from human cells indicated that this complex includes hCtr9, hPaf1, hLeo1, and hCdc73, and a higher eukaryotic-specific subunit, hSki8 (26). The Rtf1 subunit does not appear to have a stable interaction with Paf1 complex from either human or Drosophila once purified. However, Rtf1 colocalizes broadly with actively transcribing, phosphorylated RNA Pol II in a pattern very similar to that of Paf1 and CDC73 (25). These data suggest that Rft1 functions with the Paf1 complex in vivo. Our findings regarding the role of Rtf1 in gene expression, Notch signaling, and fly development extend the essential role of this complex in regulating gene expression during development.
Much of our understanding of gene regulation and its role in development and differentiation comes from studies initially performed in yeast and Drosophila. The MLL gene, a frequent partner in chromosome translocations leading to leukemia, exists in a large macromolecular complex, similar to its yeast homologue SET1 in the COMPASS complex (1, 2, 27, 28). The methylation of histones by yeast COMPASS requires ubiquitination of histone H2B by the Rad6 and Bre1 proteins, and the Paf1 complex is required to activate Rad6 and Bre1 histone ubiquitination activity (9, 12, 13). Here we report that the Drosophila Rtf1 subunit of the Paf1 complex facilitates Notch signaling, linking histone ubiquitination and methylation to gene activation by the Notch pathway. Previously, it was reported that the Drosophila homologue of Bre1 is also required for expression of Notch target genes and histone H3K4 methylation (19). Work here indicates that Rtf1 contributes to Notch signaling and histone methylation. Therefore, the functional connection among the Paf1 complex, Rad6/Bre1, complex, and the histone H3K4 methylation seen in yeast is conserved in Drosophila. In addition to revealing some of the molecular mechanisms underlying MLL action, these observations predict that the human Paf1 complex will also play a role in Notch signaling and suggest that the Notch pathway may be linked to the pathogenesis of leukemia via the MLL translocations.
Materials and Methods
Plasmid Construction.
The published S. cerevisiae Rtf1 sequence (GenBank accession no. NP_011270) was used to search the National Center for Biotechnology Information (NCBI) database for the Drosophila melanogaster homologue, dRtf1 (GenBank accession no. NM_137821, gene CG10955). To create the SympUAST–dRtf1 construct, oligonucleotides were designed to PCR-amplify 600 bp (nucleotides 841–1,440) from the dRtf1 cDNA with nested EcoRI and BglII restriction sites (5′-TACTACAGATCTGATGCGGCTTCCACTTCAGCGGCAGTGG-3′ and 5′-TACTACGAATTCAATTTCGGCCACTCGATACACGGGTTTC-3′). The amplification product was ligated to the EcoRI–BglII fragment (≈10.5 kb) of SympUAST–w (24). The resulting SympUAST–dRtf1 vector was used for P element-mediated transformation as described below. An NCBI BLAST search of the dRtf1 600-bp fragment for short, nearly exact matches indicated that there were no unintentional RNAi targets.
Northern Blot Analysis.
For the developmental Northern blot data, 15 μg of total RNA was isolated from each developmental stage. To compare RNA in dRtf1 RNAi and control flies, 15 μg of total RNA was isolated from SympUAST–dRtf1/Actin5C–Gal4 or SympUAST–dRtf1/CyO, y+ third instar larvae, respectively. For heat shock Northern blot data, SympUAST–dRtf1/Actin5C–Gal4 or SympUAST–dRtf1/CyO, y+ third instar larvae were heat shocked at 37°C for 0, 15, 30, and 45 min.
Immunostaining of Polytene Chromosomes.
Chromosomes were dissected from third instar larvae of the indicated genotypes and fixed first in Buffer A (15 mM Tris·HCl, pH 7.4/60 mM KCl/15 mM NaCl/0.5 mM spermidine/0.1% Triton X-100) plus 2% formaldehyde for 30 sec and then in 45% acetic acid plus 2% formaldehyde before squashing. RNA Pol II-specific antisera (Covance, Princeton, NJ) were used at 1:100 dilution, trimethyl H3K4 antisera (Upstate Biotechnology, Lake Placid, NY) were used at 1:50 dilution, dRtf1 antisera [a gift from John Lis (Cornell University, Ithaca, NY)] were used at 1:200 dilution, and an FITC-coupled anti-rabbit secondary antibody was used at 1:100 dilution.
Assay for Notch Signaling.
y w Males or y w; P{Sym–pUAST–RNAi} males with RNAi insertions were crossed to y w Nnd-1; P{w[+mC] = GAL4–Hsp70.PB}2 virgin females at 27°C. P{w[+mC] = GAL4–Hsp70.PB}2 provides heat shock-inducible Gal4 and was obtained from the Bloomington Drosophila Stock Center at Indiana University [stock 2077, donated by N. Perrimon (Harvard Medical School, Boston, MA)]. At 27°C with this driver, the RNAi for Rtf1 is nonlethal. Ten randomly chosen wings from male progeny were mounted on microscope slides in Permount (Sigma) and digitally photographed and measured by using Northern Eclipse software (Empix Imaging, Mississauga, ON, Canada). Statistical analysis was performed by using Statview software (SAS Institute, Cary, NC).
Acknowledgments
We thank Kristy Wendt for editorial assistance. Work in D.D.'s laboratory is supported by National Institute of General Medical Sciences Grant R01 GM055683. Work in J.C.E.'s laboratory was supported by National Science Foundation Grant MCB 0131414. Work in A.S.'s laboratory is supported by National Institutes of Health Grants 2R01CA089455 and 1R01GM069905 and by the American Cancer Society. A.S. is a Scholar of the Leukemia and Lymphoma Society.
Abbreviation
- RNA Pol II
RNA polymerase II.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Shilatifard A. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 2.Miller T., Krogan N. J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J. F., Shilatifard A. Proc. Natl. Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krogan N. J., Dover J., Khorrami S., Greenblatt J. F., Schneider J., Johnston M., Shilatifard A. J. Biol. Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 4.Roguev A., Schaft D., Shevchenko A., Pijnappel W. W., Wilm M., Aasland R., Stewart A. F. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng H. H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 7.Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., Dent S. Y., Winston F., Allis C. D. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Leeuwen F., Gafken P. R., Gottschling D. E. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 9.Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., Shilatifard A. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 10.Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., et al. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 11.Sun Z. W., Allis C. D. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 12.Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., et al. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 13.Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 14.Ng H. H., Sudhanshu D., Struhl K. J. Biol. Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 15.Mueller C. L., Jaehning J. A. Mol. Cell. Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber M., Shilatifard A. J. Biol. Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- 17.Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., Jennings J. L., Link A. J., Arndt K. M., Hartzog G. A. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J. F. Mol. Cell. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray S., Musisi H., Bienz M. Dev. Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. Mol. Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Laribee R. N., Krogan N. J., Xiao T., Shibata Y., Hughes T. R., Greenblatt J. F., Strahl B. D. Curr. Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 22.Gerber M., Ma J., Dean K., Eissenberg J. C., Shilatifard A. EMBO J. 2001;20:6104–6114. doi: 10.1093/emboj/20.21.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerber M., Eissenberg J. C., Kong S., Tenney K., Conaway J. W., Conaway R. C., Shilatifard A. Mol. Cell. Biol. 2004;24:9911–9919. doi: 10.1128/MCB.24.22.9911-9919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordano E., Rendina R., Peluso I., Furia M. Genetics. 2002;160:637–648. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelman K., Wei W., Ardehali M. B., Werner J., Zhu B., Reinberg D., Lis J. T. Mol. Cell. Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu B., Mandal S. S., Pham A. D., Zheng Y., Erdjument-Bromage H., Batra S. K., Tempst P., Reinberg D. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes C. M., Rozenblatt-Rosen O., Milne T. A., Copeland T. D., Levine S. S., Lee J. C., Hayes D. N., Shanmugam K. S., Bhattacharjee A., Biondi C. A., et al. Mol. Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 28.Tenney K., Shilatifard A. J. Cell. Biochem. 2005;95:429–436. doi: 10.1002/jcb.20421. [DOI] [PubMed] [Google Scholar]