Abstract
Little is known about the assembly and turnover of cellulose synthase complexes commonly called rosettes. Recent work indicates that rosette assembly could involve the dimerization of CesA (cellulose synthase catalytic subunit) proteins regulated by the redox state of the CesA zinc-binding domain (ZnBD). Several studies in the 1980s led to the suggestion that synthase complexes may have very short half-lives in vivo, but no recent work has directly addressed this issue. In the present work, we show that the half-life of cotton fiber GhCesA1 protein is <30 min in vivo, far less than the average membrane protein. We also show that the reduced monomer of GhCesA1 ZnBD is rapidly degraded when exposed to cotton fiber extracts, whereas the oxidized dimer is resistant to degradation. Low rates of degradation activity were detected in vitro by using extracts from fibers harvested during primary cell-wall formation, but activity increased markedly during transition to secondary cell-wall synthesis. In vitro degradation of reduced GhCesA1 ZnBD is inhibited by proteosome inhibitor MG132 and also by E64 and EGTA, suggesting that proteolysis is initiated by cysteine protease activity rather than the proteosome. We used a yeast two-hybrid system to identify a putative cotton fiber metallothionein and to confirm it as a protein that could interact with the GhCesA1 ZnBD. A model is proposed wherein active cellulose synthase complexes contain CesA proteins in dimerized form, and turnover and degradation of the complexes are mediated through reductive zinc insertion by metallothionein and subsequent proteolysis involving a cysteine protease.
Keywords: cell wall, protein turnover, rosette, zinc finger
Cotton (Gossypium hirsutum) fibers represent a very useful model for studying cellulose biosynthesis in plants. Fibers are differentiated epidermal single cells that arise from the developing seed. The development of the fiber can be divided into four phases: initiation, elongation (primary cell-wall synthesis and deposition), secondary cell-wall synthesis, and maturation that is suggested to involve programmed cell death (PCD) (1, 2). Fibers initiate on the day of anthesis by forming expanded epidermal cells. Although subject to varietal differences, elongation of fiber cells generally continues for 15–21 days postanthesis (DPA) followed by the deposition of the cellulose-rich secondary cell wall, peaking at 24 DPA, and the maturation process that begins at ≈40 DPA (3).
The transition from primary to secondary cell-wall synthesis is also characterized by an oxidative burst and induction of small GTPases of the Rho subfamily (4). In a number of biological systems, generation and accumulation of reactive oxygen species induce enzymes from the ubiquitin/proteosome pathway and other proteolytic enzymes, such as cysteine proteases (5, 6). By determining protein turnover and controlling protein half-life, cysteine proteases can serve important functions in cellular proliferation, differentiation, and PCD. For example, ectopic expression of cystatin, a cysteine protease inhibitor, reduced protease activity and blocked PCD in soybean cells (6).
Fiber secondary cell-wall formation is characterized by massive synthesis of cellulose comprising multiple 1,4-β-glucan chains that assemble into microfibrils (7). These chains are synthesized at the plasma membrane by multisubunit synthase complexes known as rosettes (8, 9). Substantial evidence supports the notion that the cellulose synthase catalytic subunit proteins (CesA proteins) responsible for glucan-chain elongation within rosettes are encoded by genes referred to as CesA (7). CesA genes were discovered in cotton fibers (8, 10) and comprise a family of 10–15 genes in all plants investigated to date (11, 12) (http://cellwall.stanford.edu). In addition to conserved regions surrounding the active site, all CesA proteins contain plant-specific regions not found in bacterial CesAs, including two zinc fingers hereafter referred to as the zinc-binding domain (ZnBD), located at the cytoplasmic N-terminal region of the proteins (13, 14). This conserved region is cysteine-rich, shows significant sequence similarity to several soybean putative transcription factors, and resembles the LIM or RING domains found in a number of proteins involved in protein–protein interactions (13–15). The prediction that the CesA ZnBD lies on the cytoplasmic side of the plasma membrane suggests that this domain is likely to be involved in protein–protein interactions. We have recently reported that CesA proteins are able to homo- and heterodimerize and that this dimerization occurs through redox-regulated disulfide bond formation between at least some of the cysteine residues in their ZnBD and might represent one step in rosette assembly (14).
Beyond the study of Kurek et al. (14), little is known of the processes of assembly and turnover of cellulose synthase complexes in plants. A number of observations have suggested that rosettes may have relatively short half-lives: (i) generally, rosettes are quite labile and difficult to preserve in freeze–fracture (8); (ii) Rudolph et al. (16) found that rosette disassembly occurs within minutes within the plasma membrane of Funaria hygrometrica in the presence of vesicle-transport inhibitors; (iii) Robinson and Quader (17) concluded that cellulose synthesis in Oocystis depended on continued protein synthesis, with an estimated half-life of complexes being <4 h; (iv) the recent visualization in vivo of moving cell-surface complexes containing CesA proteins (18) is suggestive of relative short half-lives that, from more recent studies, are estimated to be ≈20 min (C. R. Somerville, personal communication). The availability of a specific antibody to the ZnBD of G. hirsutum CesA1 (GhCesA1) has allowed us to address the question of CesA protein half-life in vivo by using cotton ovules cultured with their associated fibers. Our results do indeed indicate a high rate of turnover of GhCesA1 in vivo; we also identify a cysteine protease and a metallothionein (MET) as two candidates that might be involved in regulation of zinc availability and turnover of CesA proteins.
Results
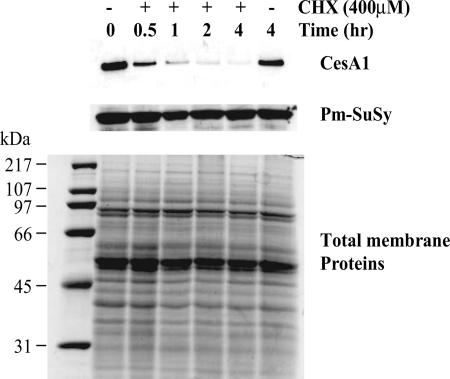
To estimate the half-life of the GhCesA1 protein in vivo, 24-DPA cultured cotton fibers were incubated in the presence or absence of 400 μM cycloheximide. By metabolically labeling the cellular proteins with [35S]methionine, we determined that 400 μM cycloheximide caused essentially complete inhibition of new protein synthesis and that after a 4-h incubation in the presence of the inhibitor, the steady-state level of soluble and membrane proteins was reduced by only 17 ± 4% and 19 ± 6%, respectively (not shown). In addition, a qualitatively similar protein profile was observed by silver staining SDS/PAGE-separated proteins throughout the cycloheximide treatment (Fig. 1Bottom), and one specific and abundant protein implicated in cellulose synthesis, the plasma membrane sucrose synthase (Pm-SuSy) (19), was completely stable throughout the 4-h treatment period (Fig. 1 Middle). By marked contrast, after a 30-min treatment with cycloheximide, the intensity of the GhCesA1 band detected by antibody against GhCesA1 ZnBD was less than half that of the band detected at t0, and it completely disappeared after the 4-h treatment. Thus, the intact GhCesA1 protein appears to have a half-life of <30 min, a turnover rate far faster than the average membrane protein.
Fig. 1.
Half-life of GhCesA1 in 24-DPA cultured cotton fibers. Fibers, with their associated ovules, were incubated for the indicated times in the presence or absence of 400 μM cycloheximide (CHX). Western blot analysis of total membrane proteins (5 μg per lane) was carried out by using antibody against the GhCesA1 ZnBD (Top) and Pm-SuSy (Middle). Total membrane proteins (20 μg per lane) were separated by SDS/PAGE and detected by silver staining (Bottom).
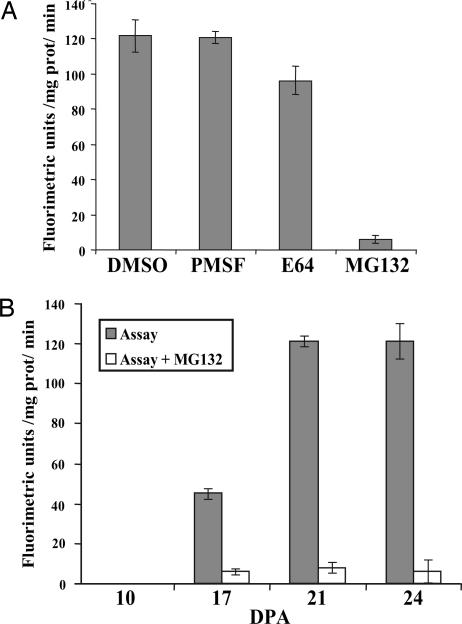
To obtain insight into the possible mechanism of GhCesA1 degradation, a crude soluble fraction from 24-DPA fibers was assayed for proteolytic activity by monitoring the breakdown of the peptide succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (sLLVY-NH-Mec), which is widely used as a substrate to measure proteosome activity. High breakdown rates of the sLLVY-NH-Mec peptide were detected in the control experiment (DMSO) and in the presence of the protease inhibitor PMSF (Fig. 2A). The cysteine protease inhibitor E64 (5) reduced peptide breakdown by only 15 ± 4%, whereas the proteosome inhibitor MG132 resulted in complete inhibition of activity, suggesting that 24-DPA fibers have a high potential for ubiquitin-mediated degradation. Activity was also analyzed during cotton fiber development. During the phase of primary cell-wall synthesis (10 and 17 DPA), low MG132-sensitive activity was detected that rose markedly during the transition from primary to secondary cell-wall formation (21 DPA) and continued to be high at the peak of secondary cell-wall cellulose synthesis (24 DPA) (Fig. 2B).
Fig. 2.
Proteolytic activity in cotton fiber extracts monitored by the breakdown of the peptide sLLVY-NH-Mec. (A) Activity detected in the soluble fraction extracted from 24-DPA fibers in the absence or presence of the indicated inhibitors. (B) Activity at the indicated age in the absence (gray bars) and presence (unshaded bars) of MG132.
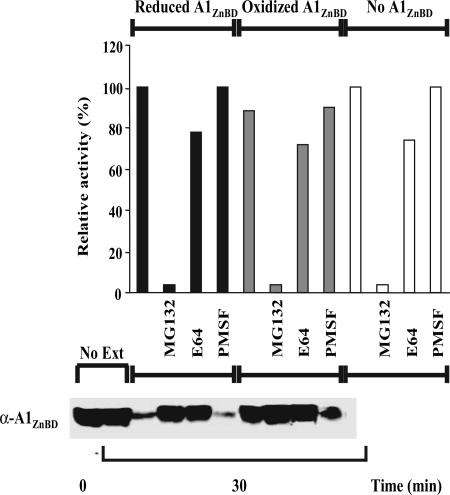
To analyze the effect of the redox state of the GhCesA1 ZnBD on susceptibility to degradation, the reduced and oxidized forms of the recombinant His6-tagged CesA1 ZnBD [referred to as A1ZnBd by Kurek et al. (14)] were incubated in the presence of 24-DPA cotton fiber extract. Consistent with the data of Fig. 2A, total proteolytic activity was extremely sensitive to MG132, and the presence of added recombinant reduced and oxidized ZnBD did not alter the pattern of overall proteolysis (Fig. 3Upper). Western blot analysis with antibodies to recombinant ZnBD revealed similar levels of recombinant ZnBD in the absence of the 24-DPA extract at 0 and 30 min, indicating that the protein itself is stable in the absence of cotton extract (Fig. 3 Lower). Substantial degradation of the reduced ZnBD was detected in the presence of the extract, and this activity was insensitive to PMSF but completely inhibited by either MG132 or E64. MG132 completely inhibits proteosome activity, but it can also affect cysteine protease activity, whereas E64 inhibits specifically cysteine protease activity (5). Therefore, these results suggest that initiation of the degradation of ZnBD by cotton extracts involves a cysteine protease activity and not the ubiquitin pathway. It is quite striking that in the absence of protease inhibitors, the reduced monomeric form of ZnBD is quite susceptible to degradation but that the oxidized dimer is much more resistant.
Fig. 3.
Effect of redox state on the sensitivity of GhCesA1 ZnBD to proteolysis. (Upper) Proteolytic activity of a crude soluble extract from 24-DPA fibers incubated in the absence (unshaded bars) or presence of reduced (black bars) or oxidized (gray bars) GhCesA1 ZnBD as affected by various protease inhibitors. (Lower) Western blot analysis of the samples described above using antibodies to monitor the proteolysis of GhCesA1 ZnBD.
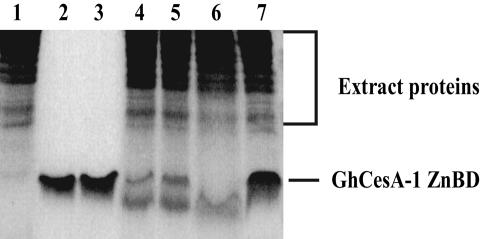
Because cysteine proteases are stimulated by Ca2+, we studied the effect of the Ca2+ chelator EGTA on the proteolytic activity. In the presence of EGTA only, the 8-kDa recombinant, reduced ZnBD remained intact for 30 min (Fig. 4, lanes 2 and 3; t0 and 30 min, respectively). After a 30-min incubation with extract, a faint amount of an ≈5-kDa truncated version of ZnBD remained, a conversion that was completely inhibited by E64 (Fig. 4, lanes 6 and 7). The addition of 5 and 10 mM EGTA to the reduced ZnBD in the presence of cell extract resulted in partial inhibition of the proteolysis with low levels of both the intact and the 5-kDa truncated variant detected (Fig. 4, lanes 4 and 5). The partial inhibition by EGTA further supports the notion that the reduced ZnBD is targeted for proteolysis by a Ca2+-stimulated protease, although the oxidized form is much more resistant to proteolysis.
Fig. 4.
Effect of EGTA on the proteolysis of recombinant GhCesA1 ZnBD by cotton fiber extracts. Cotton fiber extracts (lane 1) were incubated with ZnBD for 30 min in the presence of 5 and 10 mM EGTA (lanes 4 and 5, respectively) and the absence of EGTA (lanes 6 and 7). The stability of ZnBD incubated for 0 and 30 min in the absence (lanes 2 and 3, respectively) and the presence of cotton fiber extracts and E64 for 30 min (lanes 6 and 7, respectively) is shown. After incubation, proteins were separated by SDS/15% PAGE, and the proteins were detected by silver staining.
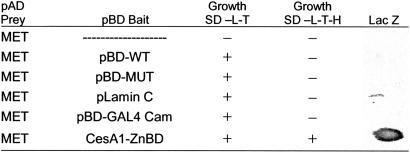
In our search for proteins that might regulate the transition of the GhCesA1 ZnBD between oxidized and reduced forms, we have used the yeast two-hybrid system to search for cotton fiber proteins that might interact with this domain of GhCesA1. An initial screen was performed employing the GhCesA1 ZnBD as bait and a 17-DPA cotton fiber library as prey. Among the yeast transformants that grew on appropriate selective medium and were β-galactosidase-positive, a clone that appeared two or more times in each of at least two independent library screens was determined by sequencing and BLAST search to encode a MET.¶
Interaction of the MET-activating domain (MET-AD) fusion protein with ZnBD was confirmed by performing a secondary yeast two-hybrid assay (Table 1). MET-AD induced the his and lacZ reporter genes, allowing colony growth in the absence of histidine and expression of β-galactosidase. A plasmid encoding MET was then isolated and retransformed back into yeast either alone or in combination with control plasmids. MET alone failed to induce the reporter genes alone or in combination with control plasmids, but it did so when combined with ZnBD.
Table 1.
Specificity of interaction between GhCesA1 ZnBD and MET
Yeast cells transformed with plasmids containing the GAL4-binding domain (pBD bait control sequences or CesA1 ZnBD) and the GAL4-activating domain (pAD prey for a clone encoding the MET sequence) were grown on SD medium lacking lysine, tryptophan, and histidine (−L-T-H) or only lysine and tryptophan (−L-T); β-galactosidase assays (LacZ) were carried out on individual yeast colonies. pBD-WT, wild-type fragment of C λ cl repressor, specific interaction control; pBD-MUT, E233K mutant fragment of λ cl repressor, specific interaction control; pLamin C, human lamin C, negative control. pBD-GAL4 Cam, specific interaction control.
Previous work showed that the ZnBD does not self-activate in the two-hybrid system and that it cannot form heterodimers with the pGAL4-BD (14). This domain is known to contain a C2H2-type zinc finger, which by itself can form dimers and binds DNA (20). These data therefore suggest that the interactions between ZnBD and MET are specific. Although Kurek et al. (14) have shown that ZnBD can interact with itself to dimerize under oxidizing conditions, in the case of the two-hybrid system, interactions in yeast occur in the nucleus, so it is not likely that we would have detected any interaction of ZnBD with itself because the native form of GhCesA1 would have been targeted to the plasma membrane.
Complete sequence analysis of the MET clones found to interact with ZnBD showed 61% identity and 83% similarity to a MET-like protein type 2 from kiwifruit (GenBank accession no. L27813) (21) and to several other plant MET-like genes. The reconstructed cotton fiber gene, homologous to higher-plant MET-like genes, is designated GhMet-1, and it has been submitted to the GenBank database with accession no. AY667624. GhMet-1 cDNA contains the entire coding region for a predicted polypeptide of 82 amino acids with a calculated molecular mass of 8.7 kDa.
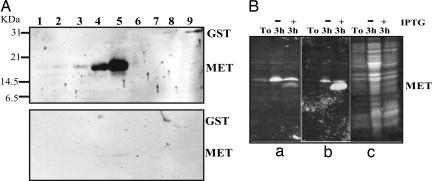
METs and MET-like proteins have been identified in diverse organisms, and they have been shown to associate with several metal ions, most commonly Zn2+ and Cu+ (22); they could therefore play some role in the provision of zinc to ZnBDs during the cycling between oxidized and reduced states. In view of this potential role, we further verified CesA interactions with MET by using an in vitro assay. Recombinant His6-tagged MET fusion protein was expressed in Escherichia coli and purified. Increasing amounts of purified recombinant MET (or recombinant GST as a control) were subjected to SDS/PAGE and electroblotted onto nitrocellulose membranes. The proteins were allowed to renature in PBS overnight, and they were then overlaid with purified recombinant ZnBD. After extensive washing, the membranes were incubated with anti-ZnBD antibodies. Duplicate membranes were subjected to the same treatment and incubated with anti-ZnBD antibodies and secondary antibodies without recombinant ZnBD in the overlay solution as a control for antibody cross-reactivity. The ZnBD was shown to interact with MET in vitro in a concentration-dependent manner but not with the control protein, GST (Fig. 5A Upper). These interactions were shown to be specific and not caused by antibody cross-reactivity (Fig. 5A Lower). Similar specific interactions between ZnBD and MET were obtained by using whole-cell lysates of E. coli after induction, whereas no interaction was detected with uninduced E. coli whole-cell lysates (Fig. 5B). These results are therefore consistent with the results from the two-hybrid screen and further support the notion of a specific interaction between the ZnBD of GhCesA1 and MET.
Fig. 5.
In vitro analysis of the interaction of GhCesA1 ZnBD with MET. (A Upper) His6-tagged MET and GST (control) were expressed in E. coli, purified, subjected to SDS/PAGE, and electroblotted. Membranes were overlaid with purified recombinant ZnBD, washed, and detected by using anti-ZnBD antibodies as described in Materials and Methods. (A Lower) Membranes were treated as previously but without recombinant ZnBD in the overlay solution (control antibodies alone). Lanes 1–5: 10, 50, 100, 200, and 400 ng of MET, respectively. Lanes 6–9: 150, 300, 600, and 1,500 ng of GST, respectively. (B) Whole-cell lysates of MET-expressing E. coli before (T0) and after a 3-h incubation with (+) and without (−) isopropyl β-d-thiogalactoside (IPTG) induction were subjected to SDS/PAGE and electroblotted. Membrane treatments were the same as for A. (Ba) No ZnBD in overlay solution. (Bb) Overlay with recombinant ZnBD. (Bc) Ponceau staining of total protein.
Levels of expression of the GhMet-1 gene, monitored in developing cotton fibers by semiquantitative RT-PCR, indicate that high levels are present at all stages of fiber development (not shown). Because GhCesA1 is highly expressed at the onset and throughout secondary cell-wall formation (10), both GhCesA1 and MET would clearly be present at the same stage of development and available for potential interactions.
Discussion
Our data provide additional support for the suggestions of Rudolph et al. (16) and Robinson and Quader (17) that synthase complexes may undergo rapid turnover in the plasma membrane. By using antibodies directed against the GhCesA1 ZnBD, we showed that in the presence of cycloheximide, the full-length GhCesA1 protein disappeared with a half-life of <30 min, and it was not detected at all after 4 h (Fig. 1). Under these same conditions, general total membrane protein profiles of the most abundant proteins visualized through silver staining after SDS/PAGE showed no obvious differences with or without cycloheximide, total membrane protein levels were reduced by only ≈20%, and Pm-SuSy levels remained unchanged even after 4 h of cycloheximide treatment. These results certainly indicate that the CesA protein, a major component of the synthase complex, is subject to much more rapid turnover than the average membrane protein.
Developmental regulation via the ubiquitin/proteosome pathway has been described for a large number of plant genes/gene products involved in different pathways such as hormone-regulated events, cell cycle, circadian rhythms, pathogen responses, photomorphogenesis, and senescence (23, 24). Although GhCesA1 contains a putative RING motif under reduced conditions similar to others shown to act as E3 ligases and promote their own degradation (25), and despite the high presumed proteosome activity sensitive to MG132 that was detected in 21- to 24-DPA cotton fibers, our data suggest that the initiation of proteolysis of GhCesA1 more likely involves the activity of a cysteine protease(s). We deduce this possibility for several reasons. First, this activity is quite sensitive to E64, a specific cysteine protease inhibitor (5). Second, SDS/PAGE analysis (Fig. 4) indicates that at least the first stage of proteolysis is limited to cleavage at a specific site, something that is typical of cysteine proteases (26). In this regard, it may be that such a cysteine protease only initiates a degradation process that is completed through the action of yet other proteases. We note that we saw no evidence of the truncated product in our in vivo experiments, and it appears to be transient as well in our in vitro experiments. Finally, partial inhibition of the proteolysis by EGTA is indicative of a calcium-stimulated cysteine protease activity such as the maize DEK1, which belongs to the calpain family (27). DEK1 is a membrane-bound protein that contains a calpain catalytic triad, and it displays high structure homology in domains II and III to the animal small opticlobes (SOL), and SOHL (small homolog from human) calpains (28). In contrast to the animal calpains, DEK1 exhibits strong proteolytic activity in the absence of calcium that increases further in the presence of Ca2+, similar to what we observed in this study (Fig. 4). A search for plant calpain sequences and comparison with three different human calpain members revealed a cotton calpain partial cDNA sequence from immature fiber (29) as well as a papain-like sequence expressed in fibers (GenBank accession no. AA018731).
The involvement of cysteine proteases in senescence and PCD is widely documented (5, 30). Inhibition of PCD by protease inhibitors such as YVAD-aldehyde, DEVD-CHO, E64, 4-(2-aminomethyl)benezenesulfonyl fluoride (AEBSF), and tosyl-l-lysine chloromethyl ketone indicates that plant cell death is regulated by caspases and cysteine and serine proteases. Although low doses of reactive oxygen species induce antioxidant enzymes, accumulation of reactive oxygen species above a certain level induces PCD in plants (31). In this respect, Potikha et al. (4) demonstrated high rates of H2O2 production in 24-DPA cotton fibers coinciding with secondary cell-wall cellulose synthesis, and they proposed a model in which the small GTPase Rac regulates the oxidative burst that initiates secondary cell-wall formation and ultimately leads to PCD in fibers. The high proteosome and cysteine protease activity we detected during secondary cell-wall formation is consistent with this hypothesis. Activation of cysteine proteases in plants has been demonstrated by applying H2O2 at a concentration shown to induce PCD in soybean suspension-cultured cells (6), and strong inhibition of PCD was detected after ectopic expression of the cysteine protease inhibitor Sbcys cystatin.
From the results presented along with those from the work of Kurek et al. (14), suggesting that dimerization in the Golgi or plasma membrane and degradation in the plasma membrane of GhCesA1 or GhCesA2 are regulated by the redox state of the ZnBD, one might propose a plausible scenario for the assembly and turnover of cellulose synthase complexes. Because the onset of synthesis of GhCesA1 and GhCesA2 coincides with the onset of H2O2 production and secondary cell-wall deposition, one might propose that GhCesA1 and GhCesA2 could quickly form homo- or heterodimers under such oxidative conditions, that they could be regulated by enzymes such as protein disulfide isomerases, and that this event is one of the early steps in rosette assembly that could occur in the secretory system of the endoplasmic reticulum or Golgi before delivery to the plasma membrane. These dimerized forms are resistant to proteolysis, and such oxidized dimers would represent the active form of GhCesA proteins within fully assembled complexes. The rate of GhCesA/rosette turnover might be controlled by the extent of the interaction the ZnBDs have with MET in the cell wall, and this interaction might promote the formation of CesA monomers and thus initiate degradation by means of a cysteine protease. With MET operating as part of the metallothionein/thionein (MT/T) system (32), an interaction of the ZnBD with MET could well reflect its involvement in either insertion or removal of zinc from this domain, something that must occur when this domain cycles between reduced and oxidized states. In fact, MET has been shown to transfer zinc atoms to zinc-depleted sorbitol dehydrogenase in a redox-dependent manner (33). It has also been proposed that the MT/T system serves as both a cellular reservoir for zinc and a controlled system that can supply different amounts of zinc to apoproteins according to demand (34, 35). The biological specificity of this system seems to be embedded not in the recognition between MET and apoproteins, but rather in signals effecting a change of the cellular redox state (33, 34, 36, 37). Thus, MET is a redox protein, and it could be involved in the regulation of CesA complex formation, activation, or deactivation by release or binding of zinc ions to the ZnBD of GhCesA proteins.
These data do not offer any insights into the reason why CesA proteins might need to have very short half-lives in vivo. Estimates for the time it takes to synthesize one glucan chain are on the order of a few minutes (38), and it is possible that this time is tied to the time for turnover of an individual CesA protein; yet, it must also be noted that it appears that glucan chains are initiated and terminated many times during the synthesis of a microfibril. Clearly, further studies on rates and reasons for rapid turnover in a variety of systems are warranted. It is important also to note that the model suggested may only be relevant for the control of assembly and turnover of complexes involved in secondary cell-wall cellulose synthesis. Because the CesA proteins involved and the redox state are different during primary cell-wall synthesis, additional studies are needed for this different stage of cell-wall synthesis, although the recent work that visualizes complexes moving in Arabidopsis suggests that a similar phenomenon may also apply to expanding hypocotyls (18). It is our hope that the results presented here will stimulate additional research on the role of redox regulation and proteolysis in the assembly and turnover of cellulose synthase complexes during development in a range of plant species.
Materials and Methods
GhCesA Stability in Cultured Cotton Fibers.
Preliminary experiments using [35S]methionine indicated that 400 μM cycloheximide was sufficient to inhibit completely the incorporation of methionine into proteins (data not shown). To measure the effects of cycloheximide on longer-term levels of total protein, 20-DPA cotton cultured fibers (39) were incubated in the absence and presence of 30 μCi/ml [35S]methionine by using EasyTag EXPRESS35S protein-labeling mix (PerkinElmer) in the presence or absence of 400 μM cycloheximide. All samples, including controls, contained 0.01% DMSO. Treated cultured cotton fibers with their associated ovules were washed twice with culture medium, and detached fibers were frozen in liquid N2. Fiber protein was prepared as described in ref. 39 with the following modification: the crude extract was centrifuged at 15,000 × g for 20 min at 4°C, and the supernatant was then centrifuged at 400,000 × g for 90 min at 4°C. The membrane pellet was resuspended and analyzed by Western blotting using anti-GhCesA1 ZnBD, as described in ref. 14. To determine the effects on total protein synthesis, 5 μl of radiolabeled membrane pellet was resuspended in extraction buffer (39), loaded onto a glass fiber GFA filter (Millipore), and washed with 10 ml of ice-cold 10% (vol/vol) trichloroacetic acid. [35S]Methionine incorporation was quantified and normalized to the total membrane protein concentration as determined by the assay of Bradford (40).
Assays for Proteolytic Activities.
Fresh cotton fibers were homogenized in ice-cold extraction buffer (50 mM Hepes/KOH, pH 7.2/2 mM DTT/0.25 M sucrose) and centrifuged at 15,000 × g for 10 min at 4°C. The soluble protein fraction was then diluted to a final concentration of 1 mg/ml in extraction buffer, and 100-μg aliquots were incubated at 30°C for 5 min in proteolysis buffer containing 100 mM Hepes/KOH (pH 7.8), 5 mM MgCl2, 10 mM KCl, and 2 mM ATP. To initiate the reaction, 2 mM peptide sLLVY-NH-Mec (Sigma) in DMSO was added. The breakdown of the peptide was monitored as described in refs. 41 and 42. Twenty micrograms of treated proteins was analyzed by SDS/15% PAGE developed by silver staining or immunodetected with anti-GhCesA1 ZnBD antibodies as described in ref. 14.
Cotton Fiber Two-Hybrid Library Construction.
G. hirsutum cv. Coker 130 was grown in a greenhouse, and bolls were collected at 17 DPA, as described in ref. 19. Total RNA was isolated as described by Wan and Wilkins (43), and poly(A)+ RNA was further purified by using the PolyATract isolation system III (Promega). Approximately 6 μg of poly(A)+ RNA was used for cDNA synthesis according to the protocol of the HybriZAP 2.1 two-hybrid cDNA synthesis kit (Stratagene). cDNAs ranging from 0.5 to 4 kb were directionally cloned into the λ HybriZAP 2.1 vector as EcoRI–XhoI fragments. The primary library contained 1 × 107 pfu; it was amplified (3 × 1010 pfu) and converted to plasmid form by mass in vivo excision according to the manufacturer’s protocol. The GhCesA1 N-terminal region (Met-1 to Ile-88) (ZnBD) was PCR-cloned into the pBD-GAL4 vector (Stratagene) as bait. All clones were sequenced to verify the correct reading frame.
Library Screening and Interaction Assay.
All media, buffers, and methods for the yeast two-hybrid work were adopted from the λ HybriZAP 2.1 system protocol. Saccharomyces cerevisiae YRG2 cells were first transformed with the bait pBDGhCesA1 (ZnBD) and subsequently transformed with cDNA library plasmid DNA by the lithium acetate method. Transformants were plated onto SD medium lacking leucine, tryptophan, and histidine (SD −L-T-H). Colonies that grew on selection media were restreaked onto SD −L-T-H, transferred to nitrocellulose membranes, and assayed for β-galactosidase (LacZ) activity by the X-Gal filter assay. LacZ-positive colonies growing on SD −L-T-H medium were considered putative positive clones. Plasmids were recovered from these yeast colonies, transformed, and amplified in the E. coli XL1-Blue strain. The recovered plasmids were then transformed back into YRG2 cells either alone or in combination with control plasmids. Those able to elicit expression of the two reporter genes only in the presence of pBDGhCesA1 bait, but not alone or in combination with control plasmids, were considered true positives.
In Vitro Protein Overlay Assay.
The ZnBD (Met-1 to Val-66) was PCR-cloned into the His6-tagged expression vector pQE30 (Qiagen, Valencia, CA), expressed in E. coli BL21 cells, affinity-purified according to the protocols supplied by the manufacturer, and dialyzed against 20 mM Hepes (pH 7.4). Similarly, full-length MET (Met-1 to Lys-82) was PCR-cloned into pQE30 and expressed in E. coli BL21 cells. The N-terminal His6-tagged fusions of MET were expressed and affinity-purified. GST, similarly isolated, was used as a negative control in the binding assays. Whole-cell lysates of MET-expressing E. coli BL21 before and after a 3-h incubation at 37°C with shaking and with and without isopropyl β-d-thiogalactoside induction were analyzed and treated as follows. Recombinant proteins or whole-cell lysates were boiled for 5 min in lysis buffer and run on an SDS/15% polyacrylamide gel according to the procedure of Laemmli (44) and blotted onto nitrocellulose membranes, and the membranes were incubated overnight in PBS at 4°C. Membranes were then blocked in 1× PBS/5% milk powder/0.1% Tween 20 for 1 h and overlaid with 10 ml of purified His6-tagged 0.2 μg/ml ZnBD for 2 h at room temperature with shaking, after which diamide (Sigma) was added to a final concentration of 5 mM. The membranes were then incubated for 30 min. The diamide step was performed to cross-link the disulfide bonds that formed. Blots were washed with 1× PBS/0.1% Tween 20 three times for 30 min each and blocked again for 1 h as previously described. Blots were incubated overnight at 4°C with rat polyclonal antiserum (1:4,000 in 1× PBS/5% milk powder/0.1% Tween 20) prepared against recombinant ZnBD of GhCesA1 (39). Blots were again washed three times for 10 min in 1× PBS/0.1% Tween 20 and exposed to goat anti-rat IgG coupled to horseradish peroxidase (Sigma) at a 1:10,000 dilution for 1 h at room temperature. After another three 10-min washes, blots were detected by ECL (GE Healthcare), with SuperSignal West Pico substrate (Pierce), as described by the manufacturer.
Acknowledgments
We thank Thea Wilkins (University of California) for supplying 17-DPA cotton fiber RNA. This work was supported by U.S. Department of Energy Grant DE-FG-03-963ER (to D.P.D.).
Abbreviations
- AD
activating domain
- CesA
cellulose synthase catalytic subunit
- DPA
days postanthesis
- MET
metallothionein
- PCD
programmed cell death
- Pm-SuSy
plasma membrane sucrose synthase
- sLLVY-NH-Mec
succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin
- ZnBD
zinc-binding domain.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY667624).
Extensive screening also revealed two other putative interacting proteins, an α-tubulin and a lipid-transfer protein, but analysis of these interactions is still incomplete.
References
- 1.DeLange E. A. L. In: Cotton Physiology. Mauney J. R., Stewart J. M., editors. Memphis, TN: Cotton Foundation; 1986. pp. 325–349. [Google Scholar]
- 2.Kim H. J., Triplett B. A. Plant Physiol. 2001;127:1361–1366. [PMC free article] [PubMed] [Google Scholar]
- 3.Meinert M., Delmer D. P. Plant Physiol. 1977;59:1088–1097. doi: 10.1104/pp.59.6.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potikha T. S., Collins C. C., Johnson D. I., Delmer D. P., Levin A. Plant Physiol. 1999;119:849–858. doi: 10.1104/pp.119.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaller A. Planta. 2004;220:183–197. doi: 10.1007/s00425-004-1407-2. [DOI] [PubMed] [Google Scholar]
- 6.Solomon M., Belenghi B., Delledonne M., Menachem E., Levine A. Plant Cell Physiol. 1999;11:431–443. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., et al. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 8.Delmer D. P. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 9.Doblin M. S., Kurek I., Jacob-Wilk D., Delmer D. P. Plant Cell Physiol. 2002;43:1407–1420. doi: 10.1093/pcp/pcf164. [DOI] [PubMed] [Google Scholar]
- 10.Pear J., Kawagoe Y., Schreckengost W., Delmer D. P., Stalker D. Proc. Natl. Acad. Sci. USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holland N., Holland D., Helentjaris T., Dhugga K., Xoconostle-Cazares B., Delmer D. P. Plant Physiol. 2000;123:1313–1323. doi: 10.1104/pp.123.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richmond T. A., Somerville C. R. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawagoe Y., Delmer D. P. Genet. Eng. (N.Y.) 1997;19:63–87. doi: 10.1007/978-1-4615-5925-2_4. [DOI] [PubMed] [Google Scholar]
- 14.Kurek I., Kawagoe Y., Jacob-Wilk D., Doblin M., Delmer D. Proc. Natl. Acad. Sci. USA. 2002;99:11109–11114. doi: 10.1073/pnas.162077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arioli T., Peng L., Betzner A. S., Burn J., Wittke W., Herth W., Camilleri C., Hofte H., Plazinski J., Birch R., et al. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 16.Rudolph U., Gross H., Schnepf E. Protoplasma. 1989;148:57–69. [Google Scholar]
- 17.Robinson D. G., Quader H. Eur. J. Cell Biol. 1981;25:278–288. [PubMed] [Google Scholar]
- 18.Paredez A. R., Somerville C. R., Ehrhardt D. W. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 19.Haigler C. H., Ivanova-Datcheva M., Hogan P., Salnikov V. V., Hwang S., Martin K., Delmer D. P. Plant Mol. Biol. 2001;47:29–51. [PubMed] [Google Scholar]
- 20.Baleja J. D., Marmorstein R., Harrison S. C., Wagner G. Nature. 1992;356:450–453. doi: 10.1038/356450a0. [DOI] [PubMed] [Google Scholar]
- 21.Ledger S. E., Gardner R. C. Plant Mol. Biol. 1994;25:877–886. doi: 10.1007/BF00028882. [DOI] [PubMed] [Google Scholar]
- 22.Kagi J. H. R., Kojima Y. Experientia Suppl. 1987;52:25–61. doi: 10.1007/978-3-0348-6784-9_3. [DOI] [PubMed] [Google Scholar]
- 23.Moon J., Parry G., Estelle M. Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwechheimer C., Schwager K. Plant Cell Rep. 2004;23:353–364. doi: 10.1007/s00299-004-0858-z. [DOI] [PubMed] [Google Scholar]
- 25.Ardley H. C., Robinson P. A. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 26.Cuervo A. M., Dice J. F. Front. Biosci. 1998;3:25–43. doi: 10.2741/a264. [DOI] [PubMed] [Google Scholar]
- 27.Becraft P. W., Li K., Dey N., Asuncion-Crabb Y. Development (Cambridge, U.K.) 2002;129:5217–5225. doi: 10.1242/dev.129.22.5217. [DOI] [PubMed] [Google Scholar]
- 28.Wang C., Barry J. K., Min Z., Tordsen G., Rao A. G., Olsen O.-A. J. Biol. Chem. 2003;278:34467–34474. doi: 10.1074/jbc.M300745200. [DOI] [PubMed] [Google Scholar]
- 29.Margis R., Margis-Pinheiro M. Trends Plant Sci. 2003;8:58–62. doi: 10.1016/S1360-1385(02)00011-0. [DOI] [PubMed] [Google Scholar]
- 30.Woltering E. J. Trends Plant Sci. 2004;9:469–472. doi: 10.1016/j.tplants.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Jabs T., Dietrich R. A., Dangl J. L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 32.Vasak M. M., Kägi J. H. R. In: Encyclopedia of Inorganic Chemistry. King R. B., editor. Vol. 4. New York: Wiley; 1994. pp. 2229–2241. [Google Scholar]
- 33.Jiang L., Maret W., Vallee B. L. Proc. Natl. Acad. Sci. USA. 1998;95:3483–3488. doi: 10.1073/pnas.95.7.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maret W. J. Nutr. 2000;130:1455S–1458S. doi: 10.1093/jn/130.5.1455S. [DOI] [PubMed] [Google Scholar]
- 35.Roesijadi G., Bogumil R., Vašák M., Kägi H. R. J. Biol. Chem. 1998;273:17425–17432. doi: 10.1074/jbc.273.28.17425. [DOI] [PubMed] [Google Scholar]
- 36.Jacob C., Maret W., Vallee B. L. Proc. Natl. Acad. Sci. USA. 1998;95:3489–3494. doi: 10.1073/pnas.95.7.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maret W. Biochemistry. 2004;43:3301–3309. doi: 10.1021/bi036340p. [DOI] [PubMed] [Google Scholar]
- 38.Brown R. M., Jr., Willison J. H. M., Richardson C. M. Proc. Natl. Acad. Sci. USA. 1976;73:4565–4569. doi: 10.1073/pnas.73.12.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng L., Xiang F., Roberts E., Kawagoe Y., Greve L. C., Kreuz K., Delmer D. P. Plant Physiol. 2001;126:981–992. doi: 10.1104/pp.126.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradford M. Anal. Biochem. 1976;72:249–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 41.Reinheckel T., Ullrich O., Sitte N., Grune T. Arch. Biochem. Biophys. 2000;377:65–68. doi: 10.1006/abbi.2000.1717. [DOI] [PubMed] [Google Scholar]
- 42.Speranza A., Scoccianti V., Crinelli R., Calzoni G. L., Magnani M. Plant Physiol. 2001;126:1150–1161. doi: 10.1104/pp.126.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan C.-Y., Wilkins T. A. Anal. Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- 44.Laemmli U. K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]