Abstract
The structure of a Bacillus subtilis YphC/GDP complex shows that it contains two GTPase domains that pack against a central domain whose fold resembles that of an RNA binding KH-domain. Comparisons of this structure to that of a homologue in Thermotoga maritima reveals a dramatic rearrangement in the position of the N-terminal GTPase domain with a shift of up to 60 Å and the formation of a totally different interface to the central domain. This rearrangement appears to be triggered by conformational changes of the switch II region in this domain in response to nucleotide binding. Modeling studies suggest that this motion represents transitions between the “on” and “off” states of the GTPase, the effect of which is to alternately expose and bury a positively charged face of the central domain that we suggest is involved in RNA recognition as part of the possible role of this enzyme in ribosome binding.
Keywords: EngA, RNA interaction, conformational change
Members of the GTPase superfamily are critical components of many signaling pathways where the conformational changes associated with GTP binding and hydrolysis play important roles in processes ranging from cell division, cell cycling, signal transduction, mRNA translation, and hormone signaling (1). The structure of several small GTPases has shown that they share a common fold in which a central β-sheet is flanked by α-helices with strong sequence identity between the different GTPase subfamilies being located in five motifs (G1–G5), which are involved in nucleotide and metal binding, with very little sequence identity elsewhere (2). Members of this enzyme family cycle between an “on” state after the binding of GTP and an inactive “off” state after hydrolysis of the nucleotide to GDP, with the subsequent release of GDP returning the enzyme to an empty, inactive state (3). The ability of these GTPases to carry out their function while in the “on” state is suggested to depend on conformational changes between the structures of the different nucleotide complexes that are generated after rearrangements in two distinct regions termed the switch I and switch II regions, which include motifs G2 and G3, respectively (1, 4).
Genomic studies have shown that bacteria possess 11 universally conserved GTPases, many of which have been proposed to act through interactions with the ribosome (5). For example, the GTPase Era has been implicated in ribosome maturation through binding to the 16S ribosomal RNA of the small 30S ribosomal subunit via a C-terminal KH-domain, which contains a consensus VIGXXGXXI motif previously implicated in RNA recognition (6–8).
Among these universally conserved bacterial GTPases, EngA is unique as it contains two GTPase domains joined by a variable length acidic linker, the only such protein described to date (5, 9). Genome database analysis has shown that the EngA family is restricted to bacteria and a number of important parasites such as Plasmodium and Eimeria, but absent in man, yeast and fungi. The structure of a putative EngA homologue of Thermotoga maritima (TmDer) has revealed a domain architecture in which a C-terminal domain adopts a fold reminiscent of an RNA binding KH-domain, which is flanked by both GTPase domains (10).
In Bacillus subtilis, an EngA homologue is encoded by the gene YphC and knockout studies have suggested that like TmDer this enzyme is essential for bacterial survival with YphC mutants containing YphC under the control of a LacI-repressible and IPTG inducible promoter displaying an increase in cell length, nucleoid condensation and abnormally curved cell shape (9, 11). Furthermore, studies on Escherichia coli RrmJ, a heat shock controlled rRNA methyltransferase that modifies the 23S rRNA in intact 50S ribosomal subunits has shown that null mutants of this gene display severe growth defects, which are restored by the overexpression of EngA (12). Together, these data suggest that EngA forms interactions with the ribosome to act as a cellular messenger.
In this paper, we report the crystal structure of a B. subtilis YphC/GDP complex to a 2.5-Å resolution. The structure reveals that changes within the switch II region associated with GTP binding and subsequent hydrolysis result in dramatic movements of the first GTPase domain, exposing and covering the positively charged β-sheet face of the KH-like domain, suggesting that the differences in conformation reflect the RNA binding “on” and “off” states of the enzyme, respectively.
Results
Overall Fold of YphC.
The structure of YphC from B. subtilis has been solved to 2.5Å resolution revealing a three domain architecture consisting of two GTP binding domains which are both bound to a GDP cofactor, packing on either side of a central C-terminal domain. (Fig. 1A and B). The overall structure is composed of 401 residues forming 16 β-strands (82 residues) and 13 α-helices (143 residues) with overall dimensions of 70 × 55 × 40 Å. The N-terminal GTP binding domain (D1, residues 3–166) is composed of six β-strands (β1–β6) and five α-helices (α1-α5) (Fig. 2), with the secondary structural elements forming a folding pattern that closely resembles the TRAFAC class of GTPases (2). The two GTPase domains are joined by a linker region between α5 from D1 and β7 of D2 (166–175), which is rich in acidic residues and variable in length among different members of the EngA family (Figs. 1A and 2). The second GTP binding domain (D2, residues 175–352) is composed of seven β-strands (β7–β13) and six α-helices (α6–α11) (Figs. 1A and 2), with a similar structural arrangement as D1. The switch I region and several residues of the switch II region are disordered within D1 (residues 29–41 and 57–67), but whereas the switch I region is disordered within D2 (204–214), the switch II region is ordered. The C-terminal central domain is composed of a three-stranded β-sheet (β14, β15, and β16) flanked on only one side by two α-helices (α12 and α13).
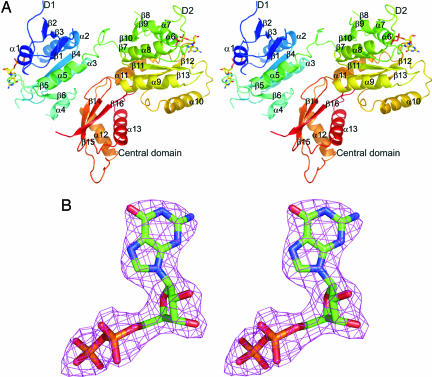
Fig. 1.
The crystal structure of the YphC/GDP complex and representative electron density. (A) A stereo diagram showing the structure of the YphC/GDP complex with each element of secondary structure labeled and the D1, D2 and central domains identified. The bound GDP molecules are shown in stick format. (B) A stereo diagram showing the final 2Fo − Fc map, contoured at 1 σ, surrounding the GDP cofactor in the second GTPase domain of YphC.
Fig. 2.
A sequence alignment of YphC homologues in B. subtilis (B. sub), Staphylococcus aureus (S. aur), Listeria innocua (L. inn), Streptococcus pneumoniae (S. pne), E. coli (E. col), Salmonella typhimurium (S. typ), Vibrio cholerae (V. cho), Pseudomonas aeruginosa (P. aer), Chlamydophila caviae (C. cav), Campylobacter jejuni (C. jej), Helicobacter pylori (H. pyl), and Thermotoga maritima (T. mar). The elements of secondary structure and the sequence numbering for B. subtilis YphC are shown above the alignment with cylinders representing α-helices and arrows representing β-strands. The fully conserved residues are shown by a black box with reverse type. The position of the five motifs (G1–G5), which can be identified in enzymes belonging to the GTPase family, are highlighted by gray boxes underneath the aligned sequences. The switch I and II regions within each GTPase domain are underlined and enclosed by a box.
Comparison of the Two GTP Binding Domains.
Domains D1 and D2 share ≈25% sequence identity with an overall rms deviation of 1.7 Å over 101 C-α atoms. There is clear sequence similarity between the switch I regions of these two domains, both of which are disordered in the structure. In contrast, their switch II regions are strikingly different, with D2 containing an additional three residues and being significantly more basic including a DTAGxR(R/K)GK motif a characteristic common to the EngA family (Fig. 2). Examination of the structure shows that Lys-236 from the switch II region forms interactions with the terminal phosphate of GDP.
Analysis of the electrostatic surface potential of D1 calculated by the program GRASP (13) reveals that it has an overall acidic charge. In particular, a region that is rich in negatively charged residues forms the interface between D1 (Asp-132, Glu-140, Asp-154, Asp-157, and Glu-161) and a complementary positively charged surface on the central domain (Arg-353, Lys-391, Arg-433, Arg-435, and Lys-436), with a number of these residues being conserved in character across the EngA family. Analysis of the electrostatic surface on D2 reveals a large patch of negative charge formed by residues from β10, α10, and its C-terminal loop and a large area of positive charge formed by residues from one face of α8 and α9 (Lys-245, Arg-250, Lys-253, Arg-257, Lys-276, and Arg-277) and residues from the switch II region (Arg-234, Lys-235, Lys-236, and Lys-238).
Structural Comparisons Between TmDer and YphC.
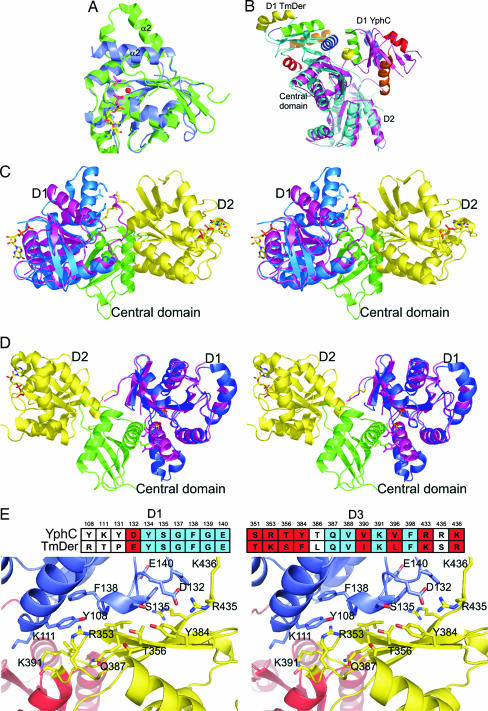
YphC and TmDer share ≈35% sequence identity (Fig. 2) and the fold of all three domains in the two structures is broadly similar. More widely within the EngA family the only significant insertions lie at the linker region between the D1 and D2 domains and at the C terminus. In the structure of TmDer, although not added in the crystallization mixture, it was realized that GDP was bound to D2, which has been suggested to arise as a result of the high affinity of this domain for the nucleotide (10). Refinement of the latter structure also identified the presence of two phosphate molecules bound to D1 whose position lies close to the predicted β and γ phosphates of GTP on the basis of structure comparison to other GTPases, leading to the suggestion that this domain might represent a conformation similar to that of a GTP bound “on state.” Because GDP is bound to both GTPase domains within YphC, we presume that this represents the “off” state of the enzyme. A structural superposition of either D1 or D2 in YphC with D2 in TmDer reveals that they all share a similar conformation, reflecting their common GDP bound state. However, superposition of any of these domains onto domain D1 in TmDer reveals that, although their core folds are similar, there are differences involving shifts of residues on α2, which forms part of the switch II region (Fig. 3A). Given the difference in the nature of the ligands bound to these two domains (GDP in D1 and D2 of YphC and D2 of TmDer and two phosphate molecules in D1 of TmDer), this finding suggests that this conformational change is a response to the binding of GDP versus a GTP mimic. More globally, the most dramatic difference between the structures of YphC and TmDer is that, although the positions of the equivalent D2 and central domains share the same relative orientation, domain D1 is observed to be dramatically repositioned with a shift of ≈60 Å and a rotation such that the opposite face of this domain is presented to the central domain (Fig. 3B and Movie 1, which is published as supporting information on the PNAS web site).
Fig. 3.
A comparison of the structure of YphC and TmDer to illustrate the differences in domain orientation. (A) The superposition of the D1 domains from YphC (blue) and TmDer (green) showing the difference in position of the switch II region which includes α2 in the GDP bound (YphC) and phosphate bound (TmDer) structures. The position of the GDP molecules in YphC (multicolored) and the two phosphate ions (purple) and the putative cation binding site (red) are shown. (B) The superposition of TmDer (blue) and YphC (magenta) showing the close structural similarity of both the D2 and the central domains. In contrast D1, adopts two different orientations with respect to the central and D2 domains in YphC and TmDer, the strands are blue in TmDer and magenta in YphC with the helices in both being colored red, blue, green, yellow and orange for helices 1, 2, 3, 4, and 5, respectively. (C) Superposition of YphC (purple) and the D1 domain from TmDer (blue) based on the overlap of their D1 domains alone, showing the steric clash between the switch II region of the TmDer on state D1 and the linker between D1 and D2 in YphC. (D) Superposition of TmDer and the D1 domain from YphC (magenta) based on the overlap of their D1 (blue) domains alone, showing the steric clash between the switch II region of the YphC off state D1 and the central domain. (E) A stereo representation of the structure of B. subtilis YphC around the interface between D1 and the central domain with residues in D1, D2, and the central domain colored blue, red, and yellow, respectively. Drawn in atom format are the 24 residues whose surface is predicted to change significantly on the transition between the on and off states. The sequence alignment between TmDer and B. subtilis YphC of the 24 residues that form the interface between D1 and the central domain in the GDP bound off conformation is displayed above the stereo interface diagram with those residues that show sequence identity and similarity colored red and blue, respectively.
Destabilising the D1/Linker Interface Stimulating Off to On Transitions.
The off state seen in the YphC/GDP complex is stabilized by the overall negative charge on D1, a conserved feature of the EngA family, packing against the conserved positively charged face of the central domain. This interface involves α3, α4, α5, and β4 of D1-forming packing interactions with β14, β15, and β16 of the central domain. However, superposition of domain D1 from TmDer (on state) onto D1 in the structure of full-length YphC suggests that there would be a steric clash between the switch II region of this domain and the acidic linker between D1 and D2 in YphC, as a consequence of the conformational differences between their respective D1 domains. Thus, the binding of GTP to D1 in YphC would lead to a shift in its switch II region, which could act as a trigger for the destabilization of this interface promoting the large-scale domain rearrangement (Fig. 3C and Movie 1).
Destabilizing the D1/Central Domain Interface Stimulating On to Off Transitions.
The on state domain arrangement seen in TmDer involves the switch II region in D1 packing against α12 of the central domain. The superposition of D1 from YphC (off state) onto D1 in the structure of full-length TmDer suggests that there would be a steric clash between this switch II region and α12 in the central domain in TmDer. Thus, the hydrolysis of GTP to GDP in D1 of TmDer would lead to a shift in its switch II region, promoting the destabilization of the D1/central domain interface, stimulating the on to off transition (Fig. 3D and Movie 1).
Conservation of the Interface Between the D1 and Central Domains.
To provide further evidence to support the suggestion that the difference in conformation between YphC and TmDer is biologically relevant we have analyzed the pattern of sequence conservation for those residues which are responsible for forming the interface between D1 and the central domain of the putative GDP off state seen in YphC. The overall level of sequence identity or similarity (R/K, T/S, D/E, V/I/L, and Y/F) amongst residues that are solvent exposed is 24% and 34%, respectively, if residues involved in nucleotide recognition are excluded. In contrast, of the 24 residues that are predicted to change their surface area significantly in the “on” to “off” transition as a result of being buried at the interface between the D1 and central domains, 10 are identical (42%) between the B. subtilis YphC and T. maritima TmDer, and 19 (79%) of the 24 are either identical or similar (R/K, T/S, D/E, V/I/L, and Y/F; Fig. 3E). Furthermore, analysis of the surface of D1 in TmDer reveals that, with the exception of the nucleotide-binding region, this is the only area where significant sequence conservation is displayed. The much greater level of sequence conservation in this region strongly supports the view that the proposed conformational transition seen here occurs in the members of the EngA GTPase family.
Discussion
The structure determination of the B. subtilis YphC/GDP complex and its comparison to its homologue in T. maritima, TmDer, has revealed a dramatic rearrangement in their domain orientations. Analysis of the overall electrostatic surface potential for YphC in the off state reveals a lack of any large continuous positively charged patch that might be implicated in forming interactions with the negatively charged RNA of the ribosome. In contrast, analysis of either TmDer or YphC modeled into a TmDer on conformation reveals that the movement of D1 in YphC exposes a positively charged surface which extends over the central domain and the D2 GTPase domain (Fig. 4A). Analysis of the central domain of YphC shows it adopts a αββαβ topology in which a β-sheet is flanked on one side by two α helices. A structural homology search using the programs DALI (14) and MSDfold (15) reveals that the fold of this domain is reminiscent of a KH-domain (16) (Fig. 4B), members of which include the ribosomal S3 domain and the C-terminal domain of the Era GTPase, which has been implicated in forming interactions with RNA (7, 17). In addition, the structures of other RNA-binding proteins that adopt a KH-like fold, but lack a classical KH motif, have shown that interactions also occur via the positively charged face of the β-sheet seen for example in the U1A splicesome, the heterogeneous nuclear ribonucleoprotein and the C-terminal domain of the S3 ribosome subunit (18–20) (Fig. 4B). Interestingly sequence analysis of several EngA homologues reveals the conservation of the positively charged character of the β-sheet which is exposed as part of the domain rearrangement. Modeling studies superimposing the central domain of YphC onto the β-sheet of the C-terminal domain from the Thermus thermophilus ribosomal S3 subunit reveal that the ribosomal RNA would be positioned across the contiguous positively charged surface over the central domain and the D2 GTPase domain, tempting speculation that this is the site of RNA recognition when YphC is in its GTP bound “on” state (Fig. 4C).
Fig. 4.
Analysis of the electrostatic surface of YphC in the off and on states and its possible role in RNA binding. (A) The GDP bound off state and GTP bound on state (modeled on the basis of the TmDer structure) of YphC as both a cartoon representation with D1, D2, and the central domain colored magenta, yellow and green, respectively, and with its corresponding electrostatic surface. (B) The central domain of YphC and the N- and C-terminal domains of the Thermus thermophilus S3 ribosomal subunit shown as both a cartoon representation and their corresponding electrostatic surfaces. In the case of the S3 C-terminal domain, the position of the nearby RNA is also shown. (C) A diagram showing the electrostatic surface of YphC modeled in an on conformation to show the possible location of an RNA-binding site. The RNA has been positioned based on the structural similarity of the central domain of YphC to the C-terminal domain of the Thermus thermophilus ribosomal S3 subunit.
Structural comparisons have shown that this difference is most likely triggered by local conformational changes in D1 resulting from differences in interactions in the various nucleotide complexes that form part of the cycling between the “on” and “off” states of this GTPase. Furthermore, given the suggested role for the EngA family to which YphC and TmDer belong in ribosome binding (12), the observation that the domain rearrangement covers and uncovers a putative RNA binding surface has prompted us to speculate that this conformational change is a critical component of the function of this family of enzymes. Currently, the role of each of the two GTPase domains in YphC is not fully understood. However, the observation that mutagenesis of residues in D1 abolishes GTPase activity, whereas the equivalent changes in D2 are without apparent effect, suggests that the former represents the primary site of GTP hydrolysis (10). Whether domain D2 is involved in GTP binding and what role this domain plays in modulating enzyme activity are yet to be determined.
Experimental Procedures
A MAD data set to 2.5 Å was collected on frozen crystals of selenomethionine (SeMet) incorporated B. subtilis YphC at the Daresbury synchrotron radiation source as reported (21). Initial phases for the structure of the YphC/GDP complex crystal were obtained by using the program SOLVE (22) with 10 of the 12 expected selenium atoms being identified after refinement of the heavy atom sites. The program RESOLVE (23) was then used to carry out density modification with the resulting phase set having a figure of merit of 0.71. The electron density map from RESOLVE was of a sufficient quality to allow for 401 of the expected 436 residues to be built through iterative cycles of model building and refinement in REFMAC5 (24) with four loops being disordered in the structure. Continuous density for the GDP cofactor could be identified in each of the two GTPase domains, allowing them to be unambiguously placed and refined. The final model has Rfactor and Rfree values of 0.22 and 0.27, respectively. Analysis using the program PROCHECK (25) showed that no non-glycine residues were in the generously allowed or disallowed regions. Refinement statistics are given in Table 1. Figs. 1, 3, and 4 were produced by using the graphics program PyMol (www.pymol.org), with the electrostatic surfaces being calculated in GRASP (13). Fig. 2 was produced by using the program Alscript (26).
Table 1.
Refinement statistics for the YphC/GDP complex
| Resolution range, Å | 20.0–2.5 |
| No. of reflections | 15,393 |
| Rfactor/Rfree* | 22.0/27.4 |
| No. of nonhydrogen atoms | |
| Protein | 3,198 |
| Average temperature factor, Å2 | |
| Protein | 43 |
| Cofactors | 41 |
| Zn ion rms, | 49 |
| rms deviations from ideal values | |
| Bond lengths, Å | 0.01 |
| Bond angles, ° | 1.3 |
| Ramachandran plot (non-Gly, non-Pro residues), %† | |
| Residues in most favored regions | 90.6 |
| Residues in additionally allowed regions | 9.4 |
*R factor = Σhkl (|Fobs| − |Fcalc|)/Σhkl|Fobs|. Rfree was calculated on 5% of the data omitted randomly.
†The Ramachandran plot was calculated by using the program PROCHECK (24).
Supplementary Material
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council.
Glossary
Abbreviation:
- TmDer
T. maritima EngA.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic structure coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2HJG).
References
- 1.Bourne H. R., Sanders D. A., McCormick F. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 2.Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 3.Vetter I. R., Wittinghofer A. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 4.Knudsen C., Wieden H. J., Rodnina M. V. J. Biol. Chem. 2001;276:22183–22190. doi: 10.1074/jbc.M102186200. [DOI] [PubMed] [Google Scholar]
- 5.Caldon C. E., Yoong P., March P. E. Mol. Microbiol. 2001;41:289–297. doi: 10.1046/j.1365-2958.2001.02536.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen X., Court D. L., Ji X. Proc. Natl. Acad. Sci. USA. 1999;96:8396–8401. doi: 10.1073/pnas.96.15.8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma M. R., Barat C., Wilson D. N., Booth T. M., Kawazoe M., Hori-Takemoto C., Shirouzu M., Yokoyama S., Fucini P., Argrawal R. K. Mol. Cell. 2005;18:319–329. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Hang J. Q., Meier T. I., Zhao G. Eur. J. Biochem. 2001;268:5570–5577. doi: 10.1046/j.1432-1033.2001.02493.x. [DOI] [PubMed] [Google Scholar]
- 9.Hwang J., Inouye M. J. Biol. Chem. 2001;276:31415–31421. doi: 10.1074/jbc.M104455200. [DOI] [PubMed] [Google Scholar]
- 10.Robinson V. L., Hwang J., Fox E., Inouye M., Stock A. M. Structure (Cambridge, U.K.) 2002;10:1649–1658. doi: 10.1016/s0969-2126(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 11.Morimoto T., Loh P. C., Hirai T., Asai K., Kobayashi K., Moriya S., Ogasawara N. Microbiology. 2002;148:3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- 12.Tan J., Jakob U., Bardwell J. C. J. Bacteriol. 2002;184:2692–2698. doi: 10.1128/JB.184.10.2692-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholls A., Sharp K. A., Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 14.Holm L., Sander C. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 15.Krissinel E., Henrick K. Acta Crystallogr. D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 16.Burd C. G., Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 17.Grishin N. V. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovine L., Oubridge C., Avis J. M., Nagai K. Structure (London) 1996;4:621–631. doi: 10.1016/s0969-2126(96)00066-4. [DOI] [PubMed] [Google Scholar]
- 19.Katahira M., Miyanoiri Y., Enokizono Y., Matsuda G., Nagata T., Ishikawa F., Uesugi S. J. Mol. Biol. 2001;311:973–988. doi: 10.1006/jmbi.2001.4862. [DOI] [PubMed] [Google Scholar]
- 20.Wimberly B. T., Brodersen D. E., Clemons W. M., Jr., Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 21.Xu L., Muench S. P., Roujeinikova A., Sedelnikova S. E., Rice D. W. Acta Crystallogr. F. 2006;62:435–437. doi: 10.1107/S1744309106011456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger T. C. J. Synchrotron Rad. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G., Vagin A., Dodson E. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski R. A., McArthur M. W., Moss D. S., Thornton J. M. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 26.Barton G. J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.