Abstract
Activation of mammalian innate and acquired immune responses must be tightly regulated by elaborate mechanisms to control their onset and termination. MicroRNAs have been implicated as negative regulators controlling diverse biological processes at the level of posttranscriptional repression. Expression profiling of 200 microRNAs in human monocytes revealed that several of them (miR-146a/b, miR-132, and miR-155) are endotoxin-responsive genes. Analysis of miR-146a and miR-146b gene expression unveiled a pattern of induction in response to a variety of microbial components and proinflammatory cytokines. By means of promoter analysis, miR-146a was found to be a NF-κB-dependent gene. Importantly, miR-146a/b were predicted to base-pair with sequences in the 3′ UTRs of the TNF receptor-associated factor 6 and IL-1 receptor-associated kinase 1 genes, and we found that these UTRs inhibit expression of a linked reporter gene. These genes encode two key adapter molecules downstream of Toll-like and cytokine receptors. Thus, we propose a role for miR-146 in control of Toll-like receptor and cytokine signaling through a negative feedback regulation loop involving down-regulation of IL-1 receptor-associated kinase 1 and TNF receptor-associated factor 6 protein levels.
Keywords: immunity, microRNA, LPS, Toll-like receptor
Innate immunity is the first line of host defense, designed to recognize pathogen-associated molecular patterns represented by conserved components of microorganisms unique to the microbial world. The pathogen molecules are detected via a limited number of germ line-encoded receptors expressed in immune cells, such as macrophages and dendritic cells. In the last decade, members of the Toll-like receptor (TLR) family have emerged as the primary evolutionarily conserved sensors of pathogen-associated molecular patterns (1). Binding of the TLRs to their respective ligands initiates a wide spectrum of responses from phagocytosis to production of a variety of cytokines, which in turn shape and enhance the inflammatory and adaptive immune responses.
All TLR receptors trigger signals in a similar fashion because of the presence of Toll and IL-1 receptor (TIR) domains in their cytoplasmic tails. The signaling cascade of TLR4, the founding member of the family and a sensor of LPS, is initiated when adaptor proteins MyD88 and TIR domain-containing adaptor-inducing IFN-β (TRIF) are recruited to the receptor, activating two independent branches of TLR signaling. MyD88 serves as a bridge between TLR4 and IL-1 receptor associated kinase (IRAK1) that then recruits into the complex TNF receptor-associated factor 6 (TRAF6). This chain of events triggers activation of IκB kinase and JNK and, in turn, the downstream NF-κB and AP-1 transcription factors and results in up-regulation of immune-responsive genes (2). The TRIF-dependent branch of signaling leads to the activation of another group of transcription factors, the IRF family, and results in a boost of expression of IFNs and other genes.
TLRs are double-edged swords because abnormal activation of their signaling can be deleterious. Thus, it is not surprising that a distinct group of TLR-activated genes is involved in modulation of TLR signal transduction by interfering with upstream signaling pathways. Several classes of negative regulators of the TLR system have been described in the past: soluble decoy receptors (sTLR2/4), cell-surface transmembrane receptors (ST2, SIGRR, and TRAILR), and numerous intracellular proteins: IRAK-M, suppressor of cytokine signaling 1 (SOCS1), MyD88 short (MyD88s), inhibitor of NF-κB (IκB), and A20 (3).
MicroRNAs (miRNAs) are an evolutionarily conserved class of endogenous ≈22-nt noncoding RNAs involved in posttranscriptional gene repression (4–6). In animals, miRNAs are processed from long primary transcripts [pri-miRNAs (pri-miR)] through an ≈60-bp hairpin precursor step [pre-miRNAs (pre-miR)] into the mature forms by sequential cutting with two RNase III enzymes, Drosha and Dicer (7, 8). Mature miRNAs are then loaded onto the ribonucleoprotein complex dubbed RISC (RNA-induced silencing complex), where they guide the recognition and translational repression or degradation of target mRNAs. pri-miRNAs are transcribed by RNA polymerase II and thus are subject to regulation by the assortment of transcription factors in the cell (9–11). Whereas some miRNAs are widely expressed, others exhibit only limited developmental stage-, tissue-, or cell type-specific patterns (12). In mammals, miRNAs have been associated with diverse biological processes, such as cell differentiation (13–15), cancer (16–18), regulation of insulin secretion (19), and viral infection (20, 21). Studies in plants have shown that miRNAs can be involved in responses to a variety of environmental stresses (22–25). However, understanding of the regulation and function of the bulk of miRNA genes await further investigation.
To examine the potential involvement of miRNAs in regulation of the innate immune response, we analyzed expression of 200 miRNAs after exposure of THP-1 cells to LPS. We show that production of mature forms for several of them (miR-146a/b, miR-132, and miR-155) is induced, and we provide here a detailed profile of expression of one miRNA family from this group (miR-146a/b) in response to various microbial components and proinflammatory mediators. Promoter analysis of the miR-146a gene revealed that NF-κB plays a critical role in induction of its transcription by LPS, TNFα, and IL-1β. In addition, we have determined that TRAF6 and IRAK1 represent potential molecular targets of miR-146 through experiments with 3′ UTR luciferase reporters. These findings suggest that miR-146a/b may function as novel negative regulators that help to fine-tune the immune response.
Results
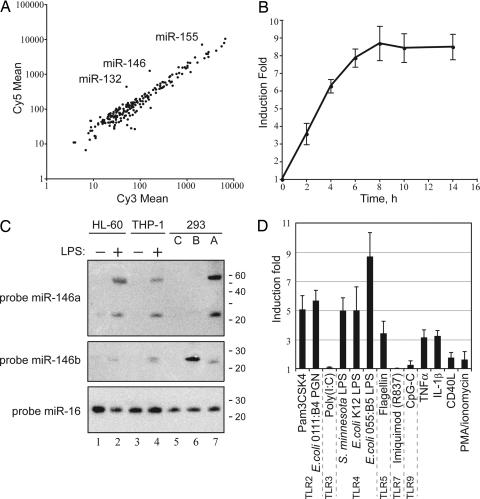
To identify miRNA genes whose expression might be regulated by innate immunity receptors, we analyzed the expression profile of the human acute monocytic leukemia cell line THP-1 using a DNA microarray containing 200 oligonucleotide probes complementary to mature forms of miRNAs of human, mouse, and rat origin. After an LPS challenge, the array revealed up-regulation in the expression of the miR-146, miR-155, and miR-132 genes (Fig. 1A). This result was validated by quantitative RT-PCR (qPCR) analysis of levels of miR-146, miR-155, and miR-132 in LPS-stimulated THP-1 cells using primers that recognize their respective mature forms (data not shown). miR-146 was chosen for detailed analysis in the present study.
Fig. 1.
miRNA miR-146a is an immediate early-response gene induced by various microbial components and proinflammatory mediators. (A) Microarray analysis of miRNA expression in THP-1 cells after stimulation with LPS (E. coli strain 055:B5). The scatter plot shows averaged (n = 4) background-subtracted raw intensities for each probe on both channels for Cy3-labeled control and Cy5-labeled 8-h LPS-treated samples. Each dot represents one miRNA probe. (B) Kinetics of miR-146a up-regulation by LPS. THP-1 cells were stimulated with LPS for the indicated times, and miR-146 expression was analyzed by qPCR and normalized by using 5S RNA levels. (C) Northern blot analysis of miR-146a/b expression. Indicated human cell lines were treated with LPS (E. coli strain 055:B5) for 8 h and probed with DNA oligonucleotides complementary to mature miR-146a (Top) and miR-146b (Middle). To assess the specificity of the probes, we used total RNA from 293 cells transiently transfected with pcDNA3 vector or expression plasmids for either pre-miR-146a or pre-miR-146b (marked as C, A, and B, respectively). (The faint bands in the lanes probed for miR-146b are cross-reactions with mirR-146a bands; the dark band in lane 6 represents miR-146b.) The membrane was reprobed for miR-16 as a loading control (Bottom). Radiolabeled RNA decade marker was used as a molecular weight reference. (D) Analysis of miR-146 expression in response to a panel of innate immunity ligands. THP-1 cells were stimulated with the indicated stimuli for 8 h. miR-146 expression was analyzed by qPCR and normalized by using 5S RNA levels.
The rapid induction of miR-146 in response to LPS, as shown by qPCR, suggested that miR-146 might be an LPS primary-response gene (Fig. 1B). Its expression reached a plateau ≈8 h after LPS challenge.
The human genome contains two miR-146 genes [miR-146a (26) and miR-146b (27)] on chromosomes 5 and 10, respectively, and their mature products differ only by 2 nt in the 3′ region (Fig. 4, which is published as supporting information on the PNAS web site). To study them individually using qPCR proved difficult because primers were unable to distinguish between the mature species (data not shown). Thus, we resorted to Northern blot analysis using DNA oligonucleotide probes complementary to the mature sequences. Hybridization of total RNA from 293/IL1R/MD2/TLR4 cells expressing either pre-miR-146a or pre-miR-146b under the control of the CMV promoter confirmed the ability of the probes to discriminate between the two miR-146 isoforms (Fig. 1C, lanes 5–7). Northern blot analysis of miR-146a expression in two human cell lines of myeloid origin (THP-1 and HL-60) has shown that it is strongly induced after 8 h of LPS treatment (Fig. 1C Top, lanes 1–4). By this method, miR-146b could not be detected before or after induction (Middle). As a control, miR-16 was not affected by LPS (Bottom). Levels of LPS-induced miR-146a comparable to that in THP-1 were observed in several other cell lines of myeloid origin, e.g., human U937, Mono-Mac-6, and mouse WEHI-3 (data not shown). At the same time, human B cell lines Ramos, Bjab, and Namalwa showed no increase in mature miR-146a levels in response to LPS (data not shown).
LPS signals through TLR4 (28), a member of the Toll family of pattern-recognition receptors, which consists of 10 members in humans (29). We investigated whether miR-146a/b expression might be regulated by other TLRs as well as the proinflammatory cytokines IL-1β and TNFα. Ultrapure LPS (from Escherichia coli strain K12 and Salmonella minnesota) consistently produced lower levels of miR-146 up-regulation than did crude preparations of LPS (Fig. 1D), suggesting that other components of the bacterial cell wall in crude preparations of LPS might contribute to miR-146 induction. Indeed, we found that miR-146 expression was significantly induced after exposure of THP-1 cells to peptidoglycan and its synthetic analog, Pam3CSK4, both of which trigger TLR2 signaling (Fig. 1D). Peptidoglycan is characteristic of both Gram-negative and Gram-positive bacteria but is highly enriched in the bacterial cell wall of the latter. Treatment of THP-1 cells with flagellin, a major component of the bacterial flagellar filament and agonist of the TLR5 receptor, also resulted in increased miR-146 expression (Fig. 1D). In contrast, stimulation of TLR3, TLR7, and TLR9 by their respective agonistic ligands did not affect miR-146 expression (Fig. 1D). Proinflammatory cytokines TNFα and IL-1β produced modest increases in miR-146 levels in THP-1 cells, whereas treatment with CD40L or with the phorbol myristyl acetate/ionomycin combination showed no effect.
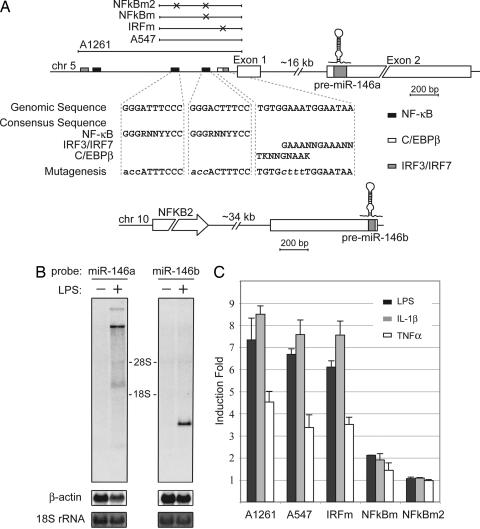
Human miR-146a resides in the LOC285628 gene on human chromosome 5. Analysis of the two ESTs encompassing LOC285628 (GenBank accession nos. BQ430527 and BQ425371) suggests that the gene consists of two exons separated by ≈16 kb of genomic sequence, with the mature miR-146a sequence situated in the second exon (Fig. 2A). Notably, the LOC285628 transcript contains no significant ORF, implying that it probably belongs to a class of noncoding RNAs. Using the 3′- and 5′-RACE technique we confirmed the two-exon structure of the miR-146a primary transcript (pri-miR146a). However, we noticed that its previous characterization was incomplete, and its full length is, in fact, 2,337 bp.
Fig. 2.
miR-146a is an NF-κB-dependent gene. (A) Schematic diagrams of miR-146a (Upper) and miR-146b (Lower) genomic loci on human chromosomes 5 and 10, respectively. Putative binding sites of NF-κB (black), IRF3/7 (gray), and C/EBPβ (white, partially overlaps with IRF3/7) transcriptional factors are shown as boxes. Genomic regions indicated by bars above the miR-146a locus were analyzed in promoter luciferase reporter assay (C). Mutations, disrupting transcription factor binding introduced in the A547 construct, are indicated by X, and the sequences of mutated sites are shown under the locus diagram. (B) Northern blot analysis of pri-miR-146a and pri-miR-146b expression in the THP-1 cell line. Cells were starved for 24 h and then stimulated with LPS for 8 h. Northern blots were hybridized with radiolabeled pre-miR-146a or pre-miR-146b probes, respectively. β-Actin (Northern blot) and 18S rRNA (methylene blue staining) levels are shown as loading controls. (C) miR-146a promoter analysis. Promoter constructs containing genomic fragments of miR-146a locus were transfected into the 293/IL-1R/TLR4/MD2 cell line. Cells were stimulated with TNFα (white bars), IL-1β (gray bars), or LPS (black bars) for 8 h and analyzed by luciferase reporter assay. Promoter regions used in this experiment are depicted schematically in A.
Human miR-146b is located on chromosome 10, yet its sequence is not included in any of the previously cloned ESTs. Our 5′-RACE results indicate that its transcript consists of only one exon with the transcription start site located ≈700 bp upstream of the mature miR-146b sequence (Fig. 2A).
To determine the effect of LPS on pri-miR-146a expression, we analyzed total RNA from LPS-stimulated and control THP-1 cells by Northern blot with a probe corresponding to pre-miR146a. We found that whereas the basal level of pri-miR-146a expression is undetectable in untreated cells, exposure of THP-1 cells to LPS for 8 h resulted in the appearance of four distinct bands. Two of these bands were of high molecular weight (>9 kb), probably corresponding to unspliced miR-146a transcripts, whereas one of the lower bands with the calculated size of 2.3 kb exactly matched the size of pri-miR-146a determined by our RACE experiments (Fig. 2B). Analysis of the same RNAs from THP-1 cells with a pre-miR-146b probe revealed that the primary transcript of this miRNA is also inducible by LPS despite our inability to visualize its mature form.
To understand the pathway by which pri-miR-146a was induced by LPS, we first characterized the miR-146a promoter region using the Genomatix MatInspector software package, focusing on those transcription factors that are common to TLR4, IL-1β, and TNFα signaling. A scan of 1.5 kb of genomic sequence located upstream of the predicted pri-miR-146a start site identified three putative NF-κB (30) and two IRF3/IRF7 consensus binding sites (31) (Fig. 2A). One of the identified IRF3/7 sites overlaps with the site for C/EBPβ (NF-IL6) (32). We examined the role of the predicted transcriptional binding sites in induction of miR-146a expression by generating a series of miR-146a promoter constructs (Fig. 2A) and testing them in a luciferase reporter assay. As shown in Fig. 2C, a miR-146a promoter is found in the upstream 550 bp, and its LPS responsiveness is totally dependent on the NF-κB binding sites. Activation of the miR-146a promoter construct by TNFα and IL-1β was also impaired by mutations in these sites (Fig. 2C). In contrast, we found no role for the IRF3/7 site because mutations of the site had no effect on induction of the luciferase reporter (Fig. 2C).
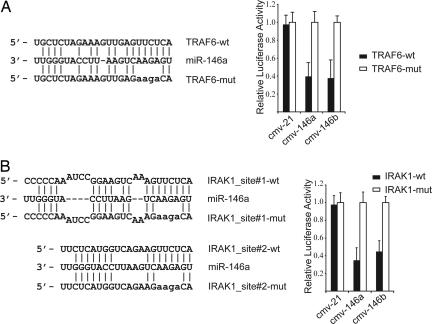
To study the functional consequences of miR-146 induction by LPS we searched for predicted mRNA target sites. 3′ UTRs of mRNAs encoding IRAK1, TRAF6, and COT/Tpl2/MAP3K8 proteins were found to contain miR-146 target sequences by several prediction algorithms (33–36). Whereas COT/Tpl2/MAP3K8 mRNA had only one miR-146 target sequence, TRAF6 and IRAK1 mRNAs contained multiple sites (Fig. 5, which is published as supporting information on the PNAS web site). IRAK1 and TRAF6 are key adapter molecules in TLR and IL-1 receptor signaling cascades, mediating activation of NF-κB and AP-1 pathways. COT/Tpl2 is a serine/threonine protein kinase that is essential for ERK activation downstream of TLRs IL-1 and TNF receptors (37).
To test the possibility that expression of these three miR-146a/b target genes is regulated posttranscriptionally by miR-146, we generated reporter constructs that contain the firefly luciferase gene fused to ≈500 bp of the 3′ UTRs from IRAK1, TRAF6, and COT1 mRNAs containing putative miR-146 target sites (IRAK1-UTR, TRAF6-UTR, and COT1-UTR, respectively) (Fig. 5). These reporter constructs were transiently transfected into 293 cells together with an expression plasmid for either miR-146a or miR-146b under the control of the CMV promoter. We observed a marked reduction in luciferase levels in cells coexpressing miR-146a together with IRAK1-UTR and TRAF6-UTR (Fig. 3A). In contrast, no changes in COT1-UTR reporter plasmid expression were observed in the presence of miR-146a. We also found that miR-146b is capable of repressing the IRAK1-UTR and TRAF6-UTR reporter constructs despite having a 2-nt difference from miR-146a at its 3′ end. Specificity of the miR-146a/b effect on IRAK1-UTR and TRAF6-UTR reporters was suggested by the lack of change of their levels of expression in the presence of an irrelevant miRNA, miR-21. Moreover, mutation of 4 nt in the miR-146a/b target sequences led to a complete abrogation of the negative effect of these miRNAs on expression of IRAK1-UTR and TRAF6-UTR reporter constructs (Fig. 3). These data suggest that the IRAK1 and TRAF6 genes are targets for posttranscriptional repression by miR-146a/b in vivo.
Fig. 3.
IRAK1 and TRAF6 may be molecular targets of miR-146 posttranscriptional repression. Shown is a sequence alignment of miR-146a and its target sites in 3′ UTRs of TRAF6 (A) and IRAK1 (B). Also shown is an analysis of expression of TRAF6-UTR (A) and IRAK1-UTR (B) luciferase reporters in the presence of miR-146a/b or the irrelevant control miR-21. Filled bars correspond to reporter constructs with wild-type miR-146 targeting sites, and open bars correspond to constructs with 4-nt substitutions disrupting base-pairing with the “seed region” of miR-146. In IRAK1-UTR-mut both miR-146-binding sites were mutated as shown.
Discussion
An appropriate innate immune response is required to defend an organism against various pathogens, but if induced either too strongly or for too long the response can be harmful, causing the pathological manifestations of both acute and chronic inflammatory disorders. Thus, cells of the immune system must employ a multilayered control system to keep innate immunity and inflammation in check. Although some of these negative regulation mechanisms have been uncovered in the past (3), we are probably far from unveiling the last of them. Here we report the discovery of a potentially new class of TLR and cytokine receptor signaling regulators: miRNAs.
We have identified by means of expression profiling several miRNA genes, namely miR-146a/b, miR-132, and miR-155, which are induced in the human monocytic cell line THP-1 by LPS, as well as other microbial components and proinflammatory mediators. Through promoter studies we have determined that LPS-mediated elevation of miR-146a expression occurs in an NF-κB-dependent manner. Finally, we have identified IRAK1 and TRAF6 as target genes of miR-146 posttranslational repression, suggesting a novel mechanism of negative feedback regulation of TLR and cytokine receptor signaling.
Induction of miR-146a/b by the TLR system displays a suggestive pattern: TLRs that recognize bacterial constituents and reside on the cell surface (1, 38) (like TLR2, TLR4, and TLR5) trigger miR-146a/b induction; those TLRs that mainly sense viral nucleic acids and localize intracellularly (1, 38) (TLR3, TLR7, and TLR9) have little effect on miR-146a/b expression.
The dual occurrence of miR-146 found in humans is not evolutionarily conserved. Search of the miRNA registry (release 7.1) (39) reveals that the chicken and fish genomes also have two copies of miR-146 but that rodents possess only one (Fig. 4). All miR-146 species that have been cloned differ by only 1 or 2 nt at their 3′ end, a region that is believed to play only a compensatory role in the target recognition mechanism (33). It will be interesting to determine whether these minor variations in miR-146a and miR-146b sequences affect their target specificity and physiological role or whether these two genes have redundant functions and are only differentially regulated at the transcriptional and transcript processing levels.
The results from computational miRNA target prediction algorithms included many more potential targets than we tested, but the TRAF6 and IRAK1 genes yielded particularly high scores. Thus, miR-146, like many mammalian miRNAs, may target a wide spectrum of gene targets, suggesting that it could be involved in regulation of multiple independent physiological processes. It is noteworthy that miR-146 already has been implicated in a number of cellular processes (14, 40, 41), and it will be important to determine whether all of them converge on TRAF6 and IRAK1 and perhaps a few other targets.
It has been reported that miR-146a expression might be involved in cell fate determination in mouse lymphocytes, because its level is substantially increased in T helper 1 cells and decreased in T helper 2 cells relative to its expression in naïve T cells (14). T helper 1 differentiation and secretion of IFNγ by this T cell subset are controlled primarily by the synergistic action of the IL-12 and IL-18 cytokines (42), thus suggesting a possible explanation for the elevated levels of miR-146a in T helper 1 cells. IL-18 receptor triggers NF-κB signaling among other pathways and employs TRAF6 and IRAK1 for its activation (43). On the other hand, miR-146a transcription might also be regulated by the STAT family, which mediates IL-12 effects in T cells (42).
Although the inflammatory response is valuable for dealing with pathogens, it takes a toll on the body and, if unregulated, can lead to serious disease. Thus, understanding the negative regulators of the response is critically important. Most of the studied attenuating mechanisms of inflammation involve negative transcriptional feedback loops, like the proteins IκB and A20 (3). We can now add miRNAs to the list of potential negative regulators of inflammation. This is a feedback system whereby bacterial components induce NF-κB through a MyD88-dependent pathway, resulting in up-regulation of the miR-146 genes, which, upon processing, could down-regulate levels of IRAK1 and TRAF6 proteins, reducing the activity of the pathway. It is noteworthy that in this scenario the TRIF-dependent, antiviral pathway induced by TLR4 and others will remain intact. In agreement with current knowledge on the miRNA mode of inhibition, it seems likely that miR-146 regulatory circuit fine-tunes TLR and cytokine signaling, rather than totally abrogating the signal. Loss of function and overexpression studies in cell culture as well as in animal models will help to identify the exact role for miR-146 in immune and inflammatory responses. Thorough analysis of the regulatory mechanisms maintaining the balance of TLR signaling can identify key pathway molecules and contribute to rational target selection for therapeutic intervention.
Materials and Methods
Reagents.
Peptidoglycan (E. coli strain 0111:B4), CpG oligonucleotide type C, synthetic bacterial lipoprotein Pam3CSK4, ultrapure LPS (S. minnesota and E. coli strain K12), poly(I:C), recombinant flagellin (Salmonella typhimurium), and imiquimod-R837 were purchased from InvivoGen (San Diego, CA). LPS (E. coli 055:B5) and phorbol myristyl acetate were obtained from Sigma (St. Louis, MO), and ionomycin was from Calbiochem (La Jolla, CA). Recombinant human IL-1β was purchased from Cell Sciences (Canton, MA), human CD40L was from R & D Systems (Minneapolis, MN), and human TNFα was from Biosource International (Camarillo, CA).
miRNA Microarray Analysis.
THP-1 cells were stimulated with 1 μg/ml LPS from E. coli strain 055:B5 for 8 h, and total RNA samples were isolated by using the mirVana RNA Isolation kit (Ambion, Austin, TX) according to the manufacturer’s protocol. Forty micrograms of total RNA was enriched for small RNA species, tailed by using the mirVana miRNA Labeling kit (Ambion), and fluorescently labeled by using amine-reactive Cy3 and Cy5 dyes (Amersham Pharmacia, Piscataway, NJ). The fluorescently labeled RNAs from control (Cy3-labeled) and LPS-treated (Cy5-labeled) cells were mixed and hybridized for 14 h with miRNA array slides. Microarrays were prepared by robotic spotting of DNA oligonucleotide probes complementary to 200 miR sequences (mirVana miRNA Probe Set, Ambion) on epoxy-coated slides (Schott Nexterion, Louisville, KY) in quadruplicate. The microarrays were washed as recommended by the manufacturer and scanned by using the GenePix 4200 fluorescent scanner.
Cell Culture, RNA Isolation, and miRNA Quantitative PCR.
THP-1, U937, HL-60, and WEHI-3 were obtained from American Type Culture Collection (Manassas, VA). 293/IL-1R/MD2/TLR4 cells were a kind gift from X. Li (Cleveland Clinic Foundation, Cleveland, OH). BJAB cells were provided by E. Kieff (Harvard University, Boston, MA). Mono-Mac-6 cells were obtained from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). THP-1, U937, HL-60, BJAB, and WEHI-3 cells were grown in RPMI medium 1640 supplemented with 10% FBS, 1× nonessential amino acids, 100 units/ml penicillin, 100 units/ml streptomycin, and 2 mM glutamine in a humidified incubator containing 5% CO2 at 37°C. 293 IL-1R/MD2/TLR4 cells were maintained in DMEM supplemented with 10% FBS, 100 units/ml penicillin, 100 units/ml streptomycin, and 2 mM glutamine. Twenty-four hours before stimulation, 1 × 106 cells were plated in RPMI medium 1640 containing 0.5% FBS. To analyze miRNA expression, cells were treated for 8 h with the following stimuli: 100 ng/ml LPS (E. coli 055:B5), 100 ng/ml Pam3CSK4, 10 μg/ml peptidoglycan, 5 μM CpG oligonucleotide type C, 10 μg/ml ultrapure LPS (from S. minnesota and E. coli strain K12), 25 μg/ml poly(I:C), 100 ng/ml recombinant flagellin (S. typhimurium), 5 μg/ml imiquimod-R837, 10 ng/ml human TNFα, 10 ng/ml human IL-1β, 300 ng/ml human CD-40L, and 50 ng/ml phorbol myristyl acetate in combination with 1 μM ionomycin. Total RNA was isolated by using the mirVana miRNA Isolation kit (Ambion). miRNA expression was measured and quantified by using the mirVana qRT-PCR miRNA Detection Kit (Ambion) or by TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA) according to the manufacturers’ protocol and normalized by 5S rRNA levels.
Northern Blot Analysis and Cloning of miR-146a/b Primary Transcripts.
Cells were starved for 24 h before stimulation in RPMI medium 1640 containing 0.5% FBS and then stimulated with LPS for 8 h. Total RNA was isolated from cells by using the mirVana miRNA Isolation kit (Ambion) or TRI-reagent (Molecular Research Center, Cincinnati, OH). For detection of mature miRNA species, 20 μg of total RNA was loaded on 12% polyacrylamide denaturing gel along with γ-32P-labeled Decade Marker (Ambion) and transferred to GeneScreenPlus membrane (PerkinElmer, Boston, MA) by electrotransfer using semidry Transblot apparatus (Bio-Rad, Hercules, CA). Membranes were hybridized in ULTRAhyb-Oligo solution (Ambion) as recommended by the manufacturer using γ-32P-labeled DNA oligonucleotide probes complementary to mature forms of tested miRNAs. For pri-miR-146a/b detection, 15 μg of total RNA was separated on 1.2% formaldehyde-containing agarose gel and transferred to GeneScreenPlus membrane by capillary transfer. Hybridizations were performed in ULTRAhyb solution (Ambion) with probes corresponding to pre-miR-146a/b. The probes were α-32P-labeled with the HexaLabel DNA Labeling Kit (Fermentas, Hanover, MD). To determine the sequence of pri-miR-146a/b, we carried out 3′- and 5′-RACE using the SMART RACE cDNA amplification kit (CLONTECH, Mountain View, CA) according to the manufacturer’s instructions.
Promoter and 3′ UTR Luciferase Reporter Assays.
To create cmv-146A, cmv-146B, and cmv-21 vectors, genomic fragments (≈300 bp) corresponding to pre-miR-146a/b isoforms and pre-miR-21 were amplified from human genomic DNA and cloned into expression vector pcDNA3 (Invitrogen, Carlsbad, CA). When transfected into 293/IL-1R/TLR4/MD2 cells, these constructs produce mature miRNA, as assessed by Northern blot and qPCR. For promoter luciferase reporter assays, 293/IL-1R/TLR4/MD2 cells were plated at 105 cells per well in 24-well dishes and transfected 24 h later by the calcium phosphate method. Each transfection reaction contained 100 ng of miR-146a promoter fragment in pGL3-basic vector (Promega, Madison, WI) and 200 ng of pcDNA3 as carrier. For normalization of transfection efficiency and extract recovery, all samples included 10 ng of the pCSK-lacZ vector, which constitutively expresses β-galactosidase and is unaffected by NF-κB. Luciferase and β-galactosidase activities were measured as described elsewhere (44). To create 3′ UTR luciferase reporter constructs, fragments (≈500 bp) of 3′ UTRs of IRAK1, TRAF6, and COT1 genes were cloned downstream of CMV-driven firefly luciferase cassette in pMIR-REPORT vector (Ambion). Mutated versions of these constructs, carrying 4-bp substitutions in the miR-146 target sites, were obtained by site-directed mutagenesis. For miRNA target validation studies, 105 293/IL-1R/TLR4/MD2 cells in a 24-well dish were transiently transfected with 10 ng of each firefly luciferase reporter plasmid, 10 ng of pCSK-lacZ vector, and 300 ng of cmv-146A, cmv-146B, or cmv-21.
Supplementary Material
Acknowledgments
We thank Joel Pomerantz for the critical reading of the manuscript and Jose Luis Riechmann, Vijaya Rao, Jaclyn Shingara, and David Brown for help with microarray work. This work was supported by National Institutes of Health Grant GM039458 and by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology. K.-J.C. was supported by Taiwan Merit Scholarship TMS-094-1-A-026.
Glossary
Abbreviations
- miRNA
microRNA
- pre-miR
precursor miRNA
- pri-miR
primary miRNA
- TLR
Toll-like receptor
- TRAF6
TNF receptor-associated factor 6
- IRAK1
IL-1 receptor-associated kinase 1
- qPCR
quantitative RT-PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ658414).
References
- 1.Akira S., Uematsu S., Takeuchi O. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Akira S., Takeda K. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 3.Liew F. Y., Xu D., Brint E. K., O’Neill L. A. Nat. Rev. Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Farh K. K., Grimson A., Jan C., Lewis B. P., Johnston W. K., Lim L. P., Burge C. B., Bartel D. P. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 7.Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 8.Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., Kim V. N. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M. L., Nervi C., Bozzoni I. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 11.O’Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 12.Pasquinelli A. E., Hunter S., Bracht J. Curr. Opin. Genet. Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Chen C. Z., Li L., Lodish H. F., Bartel D. P. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 14.Monticelli S., Ansel K. M., Xiao C., Socci N. D., Krichevsky A. M., Thai T. H., Rajewsky N., Marks D. S., Sander C., Rajewsky K., et al. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esau C., Kang X., Peralta E., Hanson E., Marcusson E. G., Ravichandran L. V., Sun Y., Koo S., Perera R. J., Jain R., et al. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 16.Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 18.He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., Hammond S. M. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 20.Lecellier C. H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., Saib A., Voinnet O. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan C. S., Ganem D. Mol. Cell. 2005;20:3–7. doi: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Jones-Rhoades M. W., Bartel D. P. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 23.Sunkar R., Zhu J. K. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achard P., Herr A., Baulcombe D. C., Harberd N. P. Development (Cambridge, U.K.) 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 25.Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., Voinnet O., Jones J. D. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 26.Cai X., Lu S., Zhang Z., Gonzalez C. M., Damania B., Cullen B. R. Proc. Natl. Acad. Sci. USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 28.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 29.Takeda K., Kaisho T., Akira S. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh S., May M. J., Kopp E. B. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 31.Lin R., Genin P., Mamane Y., Hiscott J. Mol. Cell. Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 34.John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 37.Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J. H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. Cell. 2000;103:1071–1083. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 38.Iwasaki A., Medzhitov R. Nat. Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He H., Jazdzewski K., Li W., Liyanarachchi S., Nagy R., Volinia S., Calin G. A., Liu C. G., Franssila K., Suster S., et al. Proc. Natl. Acad. Sci. USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calin G. A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S. E., Iorio M. V., Visone R., Sever N. I., Fabbri M., et al. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 42.Murphy K. M., Reiner S. L. Nat. Rev. Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 43.Dinarello C. A., Fantuzzi G. J. Infect. Dis. 2003;187(Suppl. 2):S370–S384. doi: 10.1086/374751. [DOI] [PubMed] [Google Scholar]
- 44.Pomerantz J. L., Denny E. M., Baltimore D. EMBO J. 2002;21:5184–5194. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.