Abstract
X-ray crystallographic structures of human DNA polymerase β with nonhydrolyzable analogs containing all atoms in the active site required for catalysis provide a secure starting point for a theoretical analysis (quantum mechanics/molecular mechanics) of the mechanism of chemistry without biasing of modeling assumptions as required in previous studies. These structures provide the basis for a detailed quantum mechanics/molecular mechanics study of the path for the complete transfer of a monophosphate nucleoside donor to the sugar acceptor in the active site. The reaction is largely associative with the main energetic step preceded by proton transfer from the terminal primer deoxyribose O3′ to Asp-256. The key residues that provide electrostatic stabilization of the transition state are identified and compared with those identified by mutational studies.
Keywords: quantum mechanics, molecular mechanics, reaction mechanism
DNA polymerases play a central role in the replication and repair of DNA. High-fidelity proofreading-deficient polymerases typically produce one error per million nucleotides synthesized (1). DNA polymerase β (pol β) is the simplest eukaryotic DNA polymerase and catalyzes the template-directed nucleotidyl transfer reaction during the repair of “simple” base lesions with moderate fidelity, producing approximately one error per 3,000 nt synthesized during base excision DNA repair (2). DNA pol β has been characterized by x-ray crystallography in various liganded states (3–8), and the dynamics were characterized by NMR (9, 10) and time-resolved fluorescence (11). A number of theoretical studies have been carried out with pol β in an effort to understand details of catalysis (12–15). Additionally, computer modeling of conformational changes deduced from x-ray crystallography before and after ligand binding and catalysis have provided insight into substrate discrimination (16–23). These structural and computational studies can be interpreted in the wealth of kinetic and site-directed mutagenesis studies available for pol β (2).
A recent structural study of the precatalytic complex including pol β, DNA template, primer and dNTP analogue, 2′-deoxy-uridine-5′-(α,β)-imido triphosphate, provides a ternary complex structure including a hydroxyl group (O3′) on the primer terminus with two bound Mg ions (24). Both Mg ions are found to be octahedrally coordinated, with each ion bound firmly to a nonbridging oxygen on the dNTP α-phosphate and each with a distinct bound water molecule. The primer O3′ atom, often missing in other x-ray crystal structures and necessary for the nucleophilic attack, is securely bound in the inner coordination sphere of the catalytic Mg ion. As all of the vital components for the reaction are in place in this experimental structure, we have a secure initial reaction system that minimizes the chance of introducing modeling bias at the outset of a theoretical study.
All families of DNA polymerases are believed to catalyze the nucleotidyl transfer reaction through a universal two-metal-ion mechanism (25). Therefore, the detailed reaction pathway we may derive from this study should provide a general picture of nucleotidyl transfer reactions. The nucleotide insertion reaction catalyzed by pol β occurs at a moderate rate (26). Based on transition-state (TS) theory applied to an enzyme-catalyzed reaction (27) and an insertion rate constant of 10 s−1 measured for gapped DNA (28), the apparent free energy of activation is estimated to be 16.1 kcal (1 kcal = 4.18 kJ)/mol at room temperature. Although there is some debate about the extent to which a conformational change or the chemical reaction is the rate-limiting step (29), we may use this value for comparison with our calculated reaction energy barrier.
The development of combined quantum mechanics (QM) and molecular mechanics (MM) methods provides tools for investigating the catalytic process for complex biological systems. The reactive center, where bonds break and form, is treated with an accurate QM approach, whereas the remainder of the enzyme environment is treated with empirical MM (30–32). There have been a number of successful applications of the QM/MM method in the studies of the mechanism of enzyme-catalyzed chemical reactions (refs. 33 and 34 and references therein). In this study, the QM/MM version of the ONIOM method developed by the Morokuma group (35, 36) is used, with the electronic coupling between the QM and MM regions implemented through an electronic embedding approach.
Results and Discussion
We used a constrained dynamics simulation/no-constraint minimization procedure, which resulted in a full-atom model that well represents the initial structure before the reaction. The rmsd of the backbone atoms between the minimized structure and the crystal structure was 0.31 Å. The unconstrained minimized structure with the Amber ff99 force field was further optimized at the ONIOM(B3LYP/3–21G*:Amber) and ONIOM(B3LYP/6–31G*:Amber) levels. A comparison of key active-site distances between optimized structures and crystal structure is presented in Fig. 3, which is published as supporting information on the PNAS web site. Overall, we found that the geometry of the active site optimized by the QM/MM hybrid potential agrees well with the experimental values and significantly better than if optimized with the classical force field. One of the most notable differences is in the coordination between the Mg2+ (binding) and the O1A of the α-phosphate, which changes from a noncoordination distance at 2.79 Å (classical) to a distance at ≈2.1 Å (QM/MM). The positions of some surrounding x-ray crystallographic waters and ions also changed after simulation.
We also investigated the interactions between the critical 3′ hydroxyl group of the primer terminus and the surrounding environment. The optimized structure with the Amber99 force field suggests a H-bonding interaction involving the hydrogen (H3T) of the terminal 3′ hydroxyl group and the O5′ on the incoming nucleotide triphosphate, an interaction also suggested in an earlier study (15). However, such an initial configuration of H3T does not provide a viable pathway for the critical deprotonation of the 3′ hydroxyl group. Exploring the potential energy surface with the QM/MM potential allows us to identify a second energy minimum in which H3T forms a H bond with OD2 of Asp-256. This minimum is ≈4 kcal/mol lower in energy than the energy minimum of the structure with the H3T-O5′(dTTP) H bond. This configuration for H3T suggests a direct proton transfer pathway from the 3′ hydroxyl primer terminus to Asp-256. Proton transfer to an α-phosphate oxygen suggested by a cluster model calculation (12) appears unlikely because such a proton transfer pathway will require large structural rearrangement of the active site, which is hindered by the surrounding residues in the initial structure. A water-mediated proton transfer pathway is also possible if water molecules are able to penetrate and interact with H3T. The proton will subsequently be transferred to the OD1 of Asp-256 (unpublished result) or migrate to the solution (37, 38).
Reaction Mechanism.
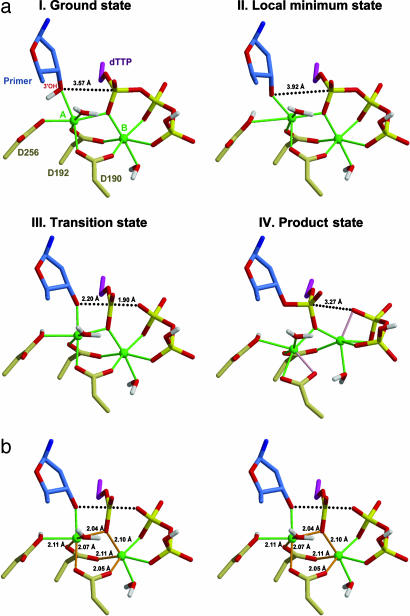
The deprotonation of the terminal hydroxyl group of the growing polynucleotide primer is believed to be an essential initial step for the success of the nucleotidyl transfer reaction. The proton transfer occurred automatically before the formation of O3′-Pα bond when we drove the reaction forward by reducing the O3′-Pα distance. A reverse exploration of the potential energy surface identifies a local minimum of the proton transfer product with an energy 3.5 kcal/mol higher than the reactant state. The existence of the local minimum of the proton transfer product suggests the reaction proceeds by a two-step pathway, in which proton transfer reaction occurs before the nucleotidyl transfer reaction. Because the proton transfer reaction proceeds with a relatively lower energy barrier, the overall activation barrier is determined by the formation of O3′-P1A bond and the breaking of P1A-O3A bond. The rate-determining process in such a two-step reaction will have a lower activation barrier than the single-step process and therefore improves the catalytic proficiency. An illustration of the potential energy profile is presented in Fig. 4, which is published as supporting information on the PNAS web site. The key active-site structures along the reaction path have been obtained in our calculations and are illustrated in Fig. 1. The transfer of the proton to Asp-256 significantly weakens the side-chain coordination with Mg2+(catalytic) and enhances the ligand-metal interaction between O3′ and Mg2+(catalytic). The distance between O3′ and Mg2+(catalytic) decreases from 2.23 to 1.98 Å, whereas the distance between OD2 of Asp-256 and Mg2+(catalytic) increases from 2.07 to 2.93 Å. Concomitantly, the H bond remains between the protonated Asp-256 and deprotonated 3′ hydroxyl group. Although the distance between O3′ and Pα increases slightly from 3.57 to 3.92 Å, the reaction can now proceed smoothly with a better nucleophile in the form of negatively charged O3′. As the O3′-Pα distance decreases along the reaction path, the interaction between Asp-256 and Mg2+(catalytic) also recovers, and thereby contributes to the stabilization of the TS.
Fig. 1.
Critical structures in the QM region determined along the reaction pathway. (a) Structures of ground state, local minimum state, TS, and product state. Blue indicates the sugar of the primer terminus, green indicates Mg ions, and yellow indicates phosphates. A nonmetal coordinated water molecule included in the quantum atoms is deleted for clarity. Note the transfer of the O3′ hydrogen in the ground state to Asp-256 in the local minimum state. Also note the change in the coordination patterns around the Mg ions from the TS state to the product state: newly formed metal interactions in the product state are shown in pink. (b) Stereoview of the TS. The metal interactions likely conserved at TSs for all polymerases are shown in orange. Figure created with Molscript (75).
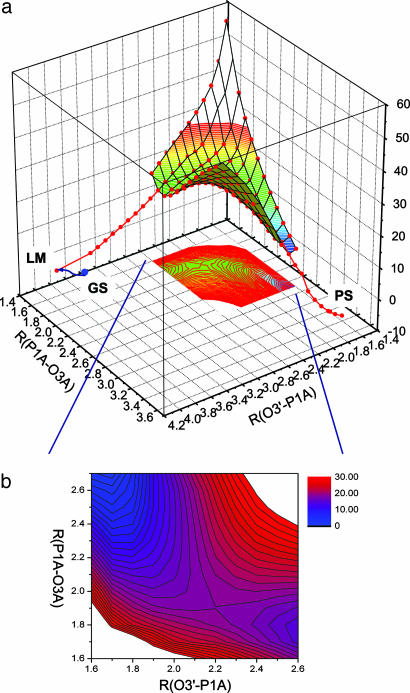
A 2D adiabatic potential energy surface, as a function of the distances associated with the bond-forming and bond-breaking process, was calculated at the ONIOM(B3LYP/6–31G*:Amber) level (Fig. 2). The distance between the primer O3′ and Pα of dTTP (RO3′-P1A) describes the nucleophilic attack process; that between P1Α and α-β bridging oxygen (RP1A-O3A) describes the bond-breaking process.
Fig. 2.
Adiabatic potential energy surface as a function of O3′-P1A and P1A-O3A distances. (a) Three-dimensional plot of the potential energy surface. The reaction path is designated by the red dots; the blue dot is the optimized ground state structure, which is connected to the local minimum state through the proton transfer process by the blue curve. (b) Contour plot of the potential energy surface in the region 1.6 ≤ O3′-P1A ≤ 2.6 Å and 1.6 ≤ P1A-O3A ≤ 2.7 Å. The TS can be identified at RO3′-P1A = 2.2 Å and RP1A-O3A = 1.9 Å. Energies are in kilocalorie per mole.
After the proton transfer reaction, the reaction proceeds via a TS located at RO3′-P1A = 2.2 Å and RP1A-O3A = 1.9 Å with energy E(TS) = 21.5 kcal/mol relative to the reactant state and 18.0 kcal/mol relative to the deprotonated intermediate state. This TS resembles a somewhat associative mechanism of nucleophilic substitution at phosphorus described by Mildvan (39), who suggested a penta-coordinated phosphorane TS. Stable penta-coordinated intermediates have been implied in a number of theoretical studies (37, 38, 40, 41) and in a phosphoryl transfer reaction catalyzed by β-phosphoglucomutase (42–44). However, in our calculations, no stable intermediate state can be identified. The reaction proceeds with the complete formation of O3′-P1A bond, which is followed rapidly by the P1A-O3A bond breaking. The reaction is enabled by a stable cylindrical structure or scaffold comprised of both metals, Asp-190, Asp-192, and the PαO (O1A) (Fig. 1b); this arrangement persists throughout development of the TS (Fig. 1a).
Along the reaction path, as the O3′ draws closer to Pα and Mg2+(catalytic), the H bond between Asp-256 and O3′ weakens and the hydrogen of which rotates to form a new H bond between Asp-256 and Asp-192 as the reaction reaches the TS. Although the H bond between Asp-256 and Asp-192 is distorted, this interaction may provide an important driving force after the TS for the release of Mg2+(catalytic) and pyrophosphate. At the TS, the coordination between O3′ and Mg2+(catalytic) remains strong, as does the O1A and Mg2+(catalytic) interaction; these create significant tension as they join P1A to form a four-member ring. Such nucleophilic substitutions near a tightly bound metal center have also been proposed in the cases of Escherichia coli alkaline phosphatase and P-Ser/P-Thr protein phosphatase (45–49). The tension created through a four-member ring contributes to the transition barrier, but once the reaction passes the TS, the O3′-P1A bond will form quickly, whereas the ligand–metal interaction between O3′ and Mg2+(catalytic) will be weakened significantly. A schematic of interactions of key components around the active site is presented in Fig. 5, which is published as supporting information on the PNAS web site.
Similar TS structures have also been identified at different levels of theory. At the ONIOM(B3LYP/3–21G*:Amber) level, the TS has an energy of 18.2 kcal/mol relative to the reactant state, and at the ONIOM(B3LYP/6–311++G**:Amber)//ONIOM(B3LYP/3–21G*:Amber) level (single point calculations with larger basis set) the TS is only 14.6 kcal/mol above the reactant state. These values are comparable to the apparent experimental free energy of the activation of ≈16 kcal/mol (28).
As the reaction continues to proceed toward the product state, the O3′-Mg2+(catalytic) distance increases to 2.9 Å and the carboxyl group of Asp-190 rotates to restore the coordination needed for Mg2+(catalytic). Interestingly, such an orientation is also found in the crystal structure of a similar complex in which a Na ion takes the place of the catalytic Mg ion, and the O3′ does not coordinate this Na ion (24). When the reaction reaches a stable product state with RO3′-P1A = 1.65 Å, RP1A-O3A = 3.27 Å, and an energy of −5.2 kcal/mol relative to reactant state, Asp-190 no longer strongly interacts with the binding Mg2+. Instead, a new ligand-metal interaction is found between O1A (the original α-β bridging oxygen) and the Mg2+(binding). This geometric change promotes the release of the pyrophosphate and Mg2+(binding) (Fig. 1a).
The “product state” identified here will certainly undergo some further conformational changes, for instance, the relocation of the hydrogen on Asp-256, to prepare for the next catalytic cycle. However, the current calculation captures the most critical steps of the chemistry of the nucleotidyl transfer process.
Energy Decomposition and Residue Contribution.
To establish the roles of different groups in catalysis we applied an energy decomposition analysis that is based on electrostatic interactions. The electrostatic interaction between the atoms directly involved in the reaction and the remaining atoms of the system is a key factor that regulates the reaction rate. That is, the lowering of the activation barrier can be attributed to the stabilization of the TS via electrostatic interactions of the active site with the environment. For the current calculations, the total electrostatic interaction of a given residue with the QM atoms is approximated as
 |
where Qα is the fitted partial charges for QM atoms using the ESP charge-fitting method (50) from the QM calculation, qβ is the partial charge of the MM atom, and Rαβ is the distance between two atoms. Such approximations have been successfully applied in a number of studies (51, 52).
The energy decomposition analysis has been performed here for the calculations at the ONIOM(B3LYP/6–31G*:Amber) level. Because the nucleophilic attack process is the rate-limiting step of the catalytic reaction, our focus is to calculate for each nonquantum unit (usually a residue in the MM region), the difference in electrostatic interaction energy between the proton-transferred intermediate state and TS contributed from each residue. We find that the electrostatic contributions to the TS stabilization sum up to −16.7 kcal/mol: −13.0 kcal/mol can be attributed to the enzyme, 7.0 kcal/mol to the DNA, −7.9 kcal/mol to the surrounding ions, and about −2.8 kcal/mol caused by water molecules, all relative to the proton-transferred intermediate state. The electron density transfer from O3′ to the α-phosphate is rather significant in moving from the proton-transferred state to the TS; therefore, charge distributions of the surrounding environment can have a significant effect on lowering the activation energy barrier. Thus it is reasonable to attempt to link the rate data from mutational changes to the electrostatic stabilization energy of a given residue. However, because mutations can induce structural changes that may affect the activation barrier in addition to any electrostatic energy change, care must be taken in the interpretation of mutation/stabilization energy comparisons. On the other hand, useful correlations in some cases can be found between energy decomposition analysis and site-directed mutagenesis studies. Contributions from selected residues to the stabilization of the TS are listed in Table 1, which is published as supporting information on the PNAS web site. We display the stabilization energies to the TS with reference to both initial state and local minimum state (proton transferred), which allow us to distinguish the contributions to the proton-transfer reaction from the nucleophilic-attack reaction. Also listed are the changes of catalytic activity caused by site-directed mutagenesis studies. Some residues are found to be conserved or partially conserved among X-family DNA polymerases, which are also noted in Table 1. We found that most of the residues with significant contributions were from the charged groups. However, we also notice that the charge pairs, e.g., between DNA phosphate groups and counter ions, can often diminish the net contributions. Based on energy decomposition analysis (kcal/mol), the residues, Arg-149 (−4.6), Arg-183 (−7.1), Arg-254 (3.6), and Asp-276 (2.8), are found to have the most significant contributions because of the orientation of the charged group. A mutation of Arg-183 to alanine will significantly lower the catalytic efficiency (53, 54), which agrees well with the results from energy decomposition analysis. Arg-183, which is found to form a salt bridge with the β-phosphate, lies along the same direction for the phosphoryl transfer with its side-chain guanidium group only 7.4 Å from the α-phosphorous. This residue is conserved in many X-family DNA polymerases (55) because of its role in dNTP binding and maintaining the active site structure and its ability to significantly lower the activation free energy for the catalytic reaction. Arg-149 plays a similar role in that it forms a salt bridge with the γ-phosphate and also stabilizes the charge transfer from α-phosphate to the O3′ of the terminal primer sugar. Its side-chain guanidium group is located 9.3 Å from the α-phosphorous, consequently alanine substitution for Arg-183 has a greater effect on the insertion rate than for Arg-149 (53). Arg-254, however, is crucial for positioning Asp-256 in the active-site structure and also in the conformational changes of the polymerase from open to closed forms. Its presence actually increases the activation energy of reaction, but its role in organizing the active-site structure appears to outweigh the negative impact as the mutation of Arg-254 to alanine or lysine leads to lowering of catalytic activity (56). Asp-276 lies close to the active site; its presence is also found to increase the activation energy of reaction, which is consistent with the finding that mutation D276V increases activity (28). There are two noncharged residues with significant contributions. Phe-272 (1.3) destabilizes the TS through interaction between the backbone carbonyl group and the QM atoms, therefore significant catalytic activity changes upon mutation to leucine are not expected (57). Ser-180 (−1.4) is found to form a H bond with the ligand water of the binding Mg through its side-chain hydroxyl group while its backbone amide also forms a H bond with the β-phosphate of the incoming nucleotide. The S180A mutant is found to exhibit a strong decrease in catalytic activity (53), which may be attributed in part to the importance of the H bond with the water ligand. Ser-180 is also preserved in DNA polymerase λ, which belongs to the X family with pol β (55). Some residues, such as Arg-182 (−1.3), Arg-253 (−1.15), and Arg-333 (−1.3), are found to be conserved in many X-family DNA polymerases (55), and these are often found to play important roles other than stabilizing the TS. Tyr-271 (0.1), although near the active site, does not contribute to the stabilization of the TS, consistent with the mutagenesis studies (58, 59). Several residues with charged side chains (Asp-246, Glu-249, and Arg-258) have a relatively small impact on the stabilization of TS either because they are located away from the active site or are not optimally positioned along the path of the phosphoryl transfer reaction. The corresponding site-directed mutagenesis studies of these residues lead to changes of <10-fold in activity (56, 60, 61). Thr-79 and Met-282 are found to make small contributions to lowering the activation energy, as confirmed by the mutagenesis studies (62, 63). Interestingly, we found several water molecules that contribute significantly to the stabilization or destabilization of TS. One water molecule, which H-bonds with the α-β bridging oxygen, contributes only −1.9 kcal/mol in lowering of the activation barrier; similarly, another nearby water molecule, which interacts with the β-phosphate, provides −1.5 kcal/mol in stabilization energy. Thus, in these calculations, stabilization of the α-β bridging oxygen does not significantly contribute to the overall energy barrier.
To fully predict the mutagenesis effect of any residue would require extensive sampling over configuration space on both the initial state and TS, which is not pursued here. The differences found, however, between the calculated results based on electrostatics and the mutagenesis studies can be used to suggest a more profound role. For instance, the mutation of Gly-274 to proline apparently disrupts dNTP binding through secondary effects. A cis-peptide bond between Gly-274 and Ser-275 would be expected to alter the position of α-helix N upon proline substitution. This helix provides intimate contacts with the nascent base pair (26). Similarly, alanine substitution for Arg-283 results in a dramatic decrease in the rate of correct nucleotide insertion (59), but offers very little TS stabilization on its own (Table 1, −0.89). As with many DNA polymerases, binding of the correct dNTP stabilizes a closed active-site conformation (64). Because alanine substitution for Arg-283 stabilizes the open form (59), residues in the N subdomain are not ideally positioned to stabilize the TS, resulting in a dramatic decrease in reaction rate.
In Table 1, we also list the residue contributions to the stabilization of the TS with the reactant ground state as a reference state. By taking the differences of energy contributions and using different reference states, we found that some residues, such as Arg-40, Lys-280, Lys-27, and Arg-283, also contribute significantly to the initial proton transfer of the H3T from the O3′ atom. Likewise, Lys-234 and Arg-258 promote the proton transfer but not the nucleotidyl transfer process.
The above calculations analyze the interactions between groups of atoms with an optimized wave function for the active site influenced by the surrounding environment. By turning off the electronic embedding in the ONIOM calculation, we can assess the electronic stabilization effect without the enzyme environment, assuming the structure remains unchanged. We performed single point calculations at ONIOM(B3LYP/6–31G*:Amber) using reactant and TS structures without electronic embedding and obtained an energy difference of 49 kcal/mol, which suggests that the two Mg ions and three aspartate groups may not be sufficient to account for all of the catalytic power of the enzyme. Smaller cluster QM calculations may produce a reasonable activation energy barrier, but the catalytic power of the enzyme will not be fully represented in the cluster model. With the complete enzyme-substrate complex, we are able to evaluate the roles of components not available in the cluster calculations, for which the mechanistic chemistry may be substantially different.
Conclusion
In the present study, the reaction mechanism of the nucleotidyl transfer reaction catalyzed by human DNA pol β is investigated by using a combined QM/MM method. We found that the inclusion of the enzyme environment is essential for a more complete understanding of the role of the enzyme in this process, and the use of the hybrid QM/MM potential apparently correctly represents the reactive system. The application of the ONIOM(B3LYP:Amber) method is found to be not only critical for correctly describing the bond breaking and bond-forming process involved in the chemical steps, but also necessary for a more accurate representation of the metal-ligand interactions and H bonds. Using the ONIOM(B3LYP/6–31G*:Amber) hybrid potential, we were able to correctly place the H bond involving the 3′-hydroxyl group of the terminus of primer. The calculations suggest that the Asp-256 plays the role as a general base that is critical for the catalytic activity of the polymerase. The critical role of residue Asp-256 as a general base is also consistent with mutagenesis studies that the D256A mutant has no catalytic activity, whereas the conservative glutamate mutation (D256E) decreases activity 150-fold (56).
Proton transfer from the O3′-hydroxyl group to the neighboring Asp-256 is found to occur before nucleophilic attack. The energy involved for the proton transfer is significantly less than the formation of the O3′-Pα bond. Although the proton transfer is important for advancing the reaction, the major contribution to the energy of activation is determined by the associative process of the 3′-oxygen attacking the α-phosphorus of the incoming dNTP. Hence, bond formation to develop the TS occurs before bond breakage to form the leaving group. It is interesting to compare this result with Mildvan’s method for classification of the mechanism of nucleophilic substitution at phosphorus (39). One can compute that the current nucleotidyl transfer reaction proceeds with ≈50% associative character, because at the TS, RP1A-O3A is 1.9 Å, similar to the distance at the TS that characterizes an SN2 mechanism. Although a stable penta-coordinated phosphorane intermediate cannot be identified, it is possible that it may appear if the coupled motions of the enzyme can provide sufficient stabilization energy. Penta-coordinated phosphorane intermediates are found to be stable in a few model systems (12), but such is not supported by our present calculations. The TS structure observed here is different from those proposed in a cluster model study (12) and an empirical valence bond study on T7 polymerase (37, 38). It is likely that having an initial state provided by more complete x-ray crystal structures accounts for much of the difference in the reaction path detail presented here. Therefore, there is less potential for initial model bias.
The roles of the MM residues in the catalysis were determined by approximating their electrostatic contribution to the TS stabilization. Many of the residues with significant contributions to the lowering of activation energy barrier are a charged group or groups with dipoles interacting closely with the active center. They can be categorized based on their interactions with the reactive center and charge redistributions along the reaction path. The electrostatic stabilization energies (Table 1) provide an estimate of the catalytic response when the residue is mutated. The calculated stabilization energy of each residue is often found to be correlated with the mutagenesis responses although a perfect correlation cannot be expected as some mutations cause profound structural changes that affect the nucleotidyl transfer reaction. Some residues such as Arg-149 and Arg-183 are known to be important in the binding of the incoming nucleotide; they are also critical to the stabilization of the TS in the chemical reaction.
The fact that the calculated activation barrier is in accord with the apparent free energy of activation suggests the proposed kinetic mechanism as the rate-limiting step, which is consistent with several experimental studies (11, 28, 65). However, we must reach this conclusion with several caveats. First, we have calculated the energy surface rather than the free energy surface. Second, as discussed, the forward and reverse processes for the proton-transfer reaction may not be completely converged. Third, a larger basis set would need to be used to draw a secure quantitative conclusion. On the other hand, we are able to establish several key residues/atoms that must be positioned through ligand binding to initiate catalytic Mg binding, thereby supporting an induced fit mechanism for substrate discrimination (i.e., fidelity) (2, 24). Because the fidelity of natural DNA polymerases is primarily dictated by the rate of correct nucleotide insertion (26, 64), our calculations identify and dissect the energetic contribution of key active-site residues role in substrate discrimination. As structures of DNA polymerases exhibiting divergent fidelities are used in similar calculations, strategies to “optimize” catalysis for alternate cellular roles with special requirements may be identified.
Methods
A recent, high-resolution (2.0 Å) x-ray crystal structure of the ternary (gapped DNA-dNTP analog-pol β) complex (24) was used to construct the initial structure for the QM/MM calculation. The refinement of this structure was significantly improved by use of the results of a parallel data collection and refinement (to 1.65 Å) of a substrate complex similar to that of the original 1997 structure (Protein Data Bank ID code 1BPY) (3) (2.2 Å). This structure, while having a Na ion in the catalytic site and missing the O3′ acceptor group, gave an improved rotomer description of the protein for refinement of the structure that contains two Mg ions in the active site and the key O3′ unit securely coordinated to both (24). The dNTP analog, 2′-deoxy-uridine-5′-(α,β)-imido triphosphate, was changed to the canonical Watson-Crick pair, dTTP molecule in the nucleotidyl transfer reaction. Hydrogen atoms were added to the model by using the LEaP program of the Amber 8 package (66, 67). Protonation states on all residues of pol β were based on the estimated values from the PropKa program at pH 7 (http://propka.chem.uiowa.edu). On the basis of the pKa values of triphosphate in solution, which has a pKa value of 6.52 for H(dTTP)3− (68), the total charge of dTTP was set to −3 with one oxygen atom on the γ-phosphate protonated. This assignment prevents excessive negative charge accumulation around the active site and also avoids a highly negatively charged pyrophosphate product. A total of 25 Na ions were added to maintain electrical neutrality of the total system. All of the crystal water molecules were preserved. The system was then solvated in a box of solvent with 21,367 water molecules. The Amber ff99 force field (69) was used. Partial atomic charges for dTTP molecule (Table 2, which is published as supporting information on the PNAS web site) were determined by using the restrained electrostatic potential procedure (70). The TIP3P model was used for the water molecules. Long-range interactions were treated by using the particle mesh Ewald method (71–73).
The dynamic simulations and minimizations were performed by using the SANDER module of the Amber 8 package, as described in Supporting Text, which is published as supporting information on the PNAS web site. The total system in the following QM/MM calculation was defined from the minimized structure (see Fig. 3). The quantum region includes parts of Asp-190, Asp-192, Asp-256, primer terminal nucleotide, dTTP, two Mg ions, and three water molecules for a total of 64 atoms. We refer to the Mg ions as catalytic (coordinates Asp-256) or binding (binds triphosphate). Atoms within 10 Å of the quantum atoms were treated by using a classical MM force field and allowed to move. The remainder of the protein, DNA, and counter ions were fixed. Water molecules within 15 Å of the quantum atoms were also included and fixed during the calculations. A total of 7,378 atoms were included for the QM/MM calculations. The charge for the QM region was −2, and the charge for the MM was +2, therefore the total system remained neutral.
The reaction was investigated with a hybrid QM/MM potential using the ONIOM(MO:MM) method. The quantum region was treated by using density functional theory with the B3LYP exchange-correlation functional and 6–31G* basis set. The remainder of the system was treated by using the Amber ff99 force field. The calculations were performed by using the ONIOM module as implemented in Gaussian 03 (74). Electrostatic interactions between the QM and MM regions were included by using the electronic embedding method, which treats the polarization of the QM region by the MM region with scaled partial atomic charges of MM atoms, and the response of QM region with Merz-Singh-Kollman scheme (50) for charge fitting so as to produce the changing partial charges of the QM atoms.
2D adiabatic potential energy surfaces were calculated as the functions of the O3′-P1A and P1A-O3A distances. These key distances describe the bond-forming and bond-breaking processes. Settings for the potential energy surface scan and convergence criteria are described in Supporting Text.
Supplementary Material
Acknowledgments
We thank the University of North Carolina for access to the necessary computing resources and a reviewer who provided a suggestion for which we are grateful. This work was supported by National Institutes of Health Grant HL-06350 and National Science Foundation Grant ITR/APS:0121361 and in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences in association with National Institutes of Health Grant 1U19CA105010.
Abbreviations
- pol β
polymerase β
- QM
quantum mechanics
- MM
molecular mechanics
- TS
transition state.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Kunkel T. A., Bebenek R. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 2.Beard W. A., Wilson S. H. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 3.Sawaya M. R., Pelletier H., Kumar A., Wilson S. H., Kraut J. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 4.Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Science. 1994;264:1891–903. [PubMed] [Google Scholar]
- 5.Sawaya M. R., Prasad R., Wilson S. H., Kraut J., Pelletier H. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 6.Krahn J. M., Beard W. A., Miller H., Grollman A. P., Wilson S. H. Structure (London) 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 7.Krahn J. M., Beard W. A., Wilson S. H. Structure (London) 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. Structure (London) 2005;13:1225–1233. doi: 10.1016/j.str.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Bose-Basu B., DeRose E. F., Kirby T. W., Mueller G. A., Beard W. A., Wilson S. H., London R. E. Biochemistry. 2004;43:8911–8922. doi: 10.1021/bi049641n. [DOI] [PubMed] [Google Scholar]
- 10.Kirby T. W., DeRose E. F., Beard W. A., Wilson S. H., London R. E. Biochemistry. 2005;44:15230–15237. doi: 10.1021/bi0511742. [DOI] [PubMed] [Google Scholar]
- 11.Kim S. J., Beard W. A., Harvey J., Shock D. D., Knutson J. R., Wilson S. H. J. Biol. Chem. 2003;278:5072–5081. doi: 10.1074/jbc.M208472200. [DOI] [PubMed] [Google Scholar]
- 12.Abashkin Y. G., Erickson J. W., Burt S. K. J. Phys. Chem. B. 2001;105:287–292. [Google Scholar]
- 13.Florian J., Goodman M. F., Warshel A. J. Phys. Chem. B. 2002;106:5739–5753. [Google Scholar]
- 14.Florian J., Goodman M. F., Warshel A. Biopolymers. 2003;68:286–299. doi: 10.1002/bip.10244. [DOI] [PubMed] [Google Scholar]
- 15.Rittenhouse R. C., Apostoluk W. K., Miller J. H., Straatsma T. P. Proteins Struct. Funct. Genet. 2003;53:667–682. doi: 10.1002/prot.10451. [DOI] [PubMed] [Google Scholar]
- 16.Yang G. W., Franklin M., Li J., Lin T. C., Konigsberg W. Biochemistry. 2002;41:2526–2534. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 17.Yang L. J., Arora K., Beard W. A., Wilson S. H., Schlick T. J. Am. Chem. Soc. 2004;126:8441–8453. doi: 10.1021/ja049412o. [DOI] [PubMed] [Google Scholar]
- 18.Yang L. J., Beard W. A., Wilson S. H., Broyde S., Schlick T. Biophys. J. 2004;86:3392–3408. doi: 10.1529/biophysj.103.036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L. J., Beard W. A., Wilson S. H., Broyde S., Schlick T. J. Mol. Biol. 2002;317:651–671. doi: 10.1006/jmbi.2002.5450. [DOI] [PubMed] [Google Scholar]
- 20.Arora K., Schlick T. Biophys. J. 2004;87:3088–3099. doi: 10.1529/biophysj.104.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arora K., Beard W. A., Wilson S. H., Schlick T. Biochemistry. 2005;44:13328–13341. doi: 10.1021/bi0507682. [DOI] [PubMed] [Google Scholar]
- 22.Arora K., Schlick T. J. Phys. Chem. B. 2005;109:5358–5367. doi: 10.1021/jp0446377. [DOI] [PubMed] [Google Scholar]
- 23.Radhakrishnan R., Schlick T. Proc. Natl. Acad. Sci. USA. 2004;101:5970–5975. doi: 10.1073/pnas.0308585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., Wilson S. H. Structure (London) 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brautigam C. A., Steitz T. A. Curr. Opin. Struct. Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 26.Beard W. A., Shock D. D., Vande Berg B. J., Wilson S. H. J. Biol. Chem. 2002;277:47393–47398. doi: 10.1074/jbc.M210036200. [DOI] [PubMed] [Google Scholar]
- 27.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1985. [Google Scholar]
- 28.Vande Berg B. J., Beard W. A., Wilson S. H. J. Biol. Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 29.Joyce C. M., Benkovic S. J. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 30.Warshel A. Annu. Rev. Biophys. Biomol. Struct. 2003;32:425–443. doi: 10.1146/annurev.biophys.32.110601.141807. [DOI] [PubMed] [Google Scholar]
- 31.Gao J. L., Truhlar D. G. Annu. Rev. Phys. Chem. 2002;53:467–505. doi: 10.1146/annurev.physchem.53.091301.150114. [DOI] [PubMed] [Google Scholar]
- 32.Gogonea V., Suarez D., van der Vaart A., Merz K. W. Curr. Opin. Struct. Biol. 2001;11:217–223. doi: 10.1016/s0959-440x(00)00193-7. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Viloca M., Gao J., Karplus M., Truhlar D. G. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 34.Friesner R. A., Guallar V. Annu. Rev. Phys. Chem. 2005;56:389–427. doi: 10.1146/annurev.physchem.55.091602.094410. [DOI] [PubMed] [Google Scholar]
- 35.Vreven T., Morokuma K., Farkas O., Schlegel H. B., Frisch M. J. J. Comput. Chem. 2003;24:760–769. doi: 10.1002/jcc.10156. [DOI] [PubMed] [Google Scholar]
- 36.Dapprich S., Komaromi I., Byun K. S., Morokuma K., Frisch M. J. J. Mol. Struct. 1999;462:1–21. [Google Scholar]
- 37.Florian J., Goodman M. F., Warshel A. J. Am. Chem. Soc. 2003;125:8163–8177. doi: 10.1021/ja028997o. [DOI] [PubMed] [Google Scholar]
- 38.Florian J., Goodman M. F., Warshel A. Proc. Natl. Acad. Sci. USA. 2005;102:6819–6824. doi: 10.1073/pnas.0408173102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mildvan A. S. Proteins Struct. Funct. Genet. 1997;29:401–416. [PubMed] [Google Scholar]
- 40.Dejaegere A., Lim C., Karplus M. J. Am. Chem. Soc. 1991;113:4353–4355. [Google Scholar]
- 41.Uchimaru T., Tanabe K., Nishikawa S., Taira K. J. Am. Chem. Soc. 1991;113:4351–4353. [Google Scholar]
- 42.Lahiri S. D., Zhang G. F., Dunaway-Mariano D., Allen K. N. Science. 2003;299:2067–2071. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- 43.Lahiri S. D., Zhang G. F., Dunaway-Mariano D., Allen K. N. Biochemistry. 2002;41:8351–8359. doi: 10.1021/bi0202373. [DOI] [PubMed] [Google Scholar]
- 44.Tremblay L. W., Zhang G. F., Dai J. Y., Dunaway-Mariano D., Allen K. N. J. Am. Chem. Soc. 2005;127:5298–5299. doi: 10.1021/ja0509073. [DOI] [PubMed] [Google Scholar]
- 45.Egloff M. P., Cohen P. T. W., Reinemer P., Barford D. J. Mol. Biol. 1995;254:942–959. doi: 10.1006/jmbi.1995.0667. [DOI] [PubMed] [Google Scholar]
- 46.Steitz T. A., Steitz J. A. Proc. Natl. Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim E. E., Wyckoff H. W. J. Mol. Biol. 1991;218:449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 48.Chaidaroglow A., Brezinski D. J., Middleton S. A., Kantrowitz E. R. Biochemistry. 1988;27:8338–8343. doi: 10.1021/bi00422a008. [DOI] [PubMed] [Google Scholar]
- 49.Butlerransohoff J. E., Kendall D. A., Kaiser E. T. Proc. Natl. Acad. Sci. USA. 1988;85:4276–4278. doi: 10.1073/pnas.85.12.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Besler B. H., Merz K. M., Kollman P. A. J. Comput. Chem. 1990;11:431–439. [Google Scholar]
- 51.Cisneros G. A., Liu H. Y., Zhang Y. K., Yang W. T. J. Am. Chem. Soc. 2003;125:10384–10393. doi: 10.1021/ja029672a. [DOI] [PubMed] [Google Scholar]
- 52.Liu H. Y., Zhang Y. K., Yang W. T. J. Am. Chem. Soc. 2000;122:6560–6570. [Google Scholar]
- 53.Kraynov V. S., Showalter A. K., Liu J., Zhong X. J., Tsai M. D. Biochemistry. 2000;39:16008–16015. doi: 10.1021/bi0008480. [DOI] [PubMed] [Google Scholar]
- 54.Date T., Yamamoto S., Tanihara K., Nishimoto Y., Liu N., Matsukage A. Biochemistry. 1990;29:5027–5034. doi: 10.1021/bi00473a005. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Diaz M., Bebenek K., Krahn J. M., Blanco L., Kunkel T. A., Pedersen L. C. Mol. Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 56.Menge K. L., Hostomsky Z., Nodes B. R., Hudson G. O., Rahmati S., Moomaw E. W., Almassy R. J., Hostomska Z. Biochemistry. 1995;34:15934–15942. doi: 10.1021/bi00049a008. [DOI] [PubMed] [Google Scholar]
- 57.Li S. X., Vaccaro J. A., Sweasy J. B. Biochemistry. 1999;38:4800–4808. doi: 10.1021/bi9827058. [DOI] [PubMed] [Google Scholar]
- 58.Kraynov V. S., Werneburg B. G., Zhong X. J., Lee H., Ahn J. W., Tsai M. D. Biochem. J. 1997;323:103–111. doi: 10.1042/bj3230103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beard W. A., Osheroff W. P., Prasad R., Sawaya M. R., Jaju M., Wood T. G., Kraut J., Kunkel T. A., Wilson S. H. J. Biol. Chem. 1996;271:12141–12144. doi: 10.1074/jbc.271.21.12141. [DOI] [PubMed] [Google Scholar]
- 60.Dalal S., Kosa J. L., Sweasy J. B. J. Biol. Chem. 2004;279:577–584. doi: 10.1074/jbc.M309607200. [DOI] [PubMed] [Google Scholar]
- 61.Kosa J. L., Sweasy J. B. J. Biol. Chem. 1999;274:35866–35872. doi: 10.1074/jbc.274.50.35866. [DOI] [PubMed] [Google Scholar]
- 62.Maitra M., Gudzelak A., Li S. X., Matsumoto Y., Eckert K. A., Jager J., Sweasy J. B. J. Biol. Chem. 2002;277:35550–35560. doi: 10.1074/jbc.M204953200. [DOI] [PubMed] [Google Scholar]
- 63.Shah A. M., Conn D. A., Li S. X., Capaldi A., Jager J., Sweasy J. B. Biochemistry. 2001;40:11372–11381. doi: 10.1021/bi010755y. [DOI] [PubMed] [Google Scholar]
- 64.Beard W. A., Wilson S. H. Structure (London) 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 65.Arndt J. W., Gong W. M., Zhong X. J., Showalter A. K., Liu J., Dunlap C. A., Lin Z., Paxson C., Tsai M. D., Chan M. K. Biochemistry. 2001;40:5368–5375. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
- 66.Pearlman D. A., Case D. A., Caldwell J. W., Ross W. S., Cheatham T. E., Debolt S., Ferguson D., Seibel G., Kollman P. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 67.Case D. A., Darden T. A., Cheatham I. T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Wang B., Pearlman D. A., et al. AMBER8. San Francisco: Univ. of California; 2004. [Google Scholar]
- 68.Sigel H., Griesser R. Chem. Soc. Rev. 2005;34:875–900. doi: 10.1039/b505986k. [DOI] [PubMed] [Google Scholar]
- 69.Wang J. M., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 70.Cornell W. D., Cieplak P., Bayly C. I., Kollman P. A. J. Am. Chem. Soc. 1993;115:9620–9631. [Google Scholar]
- 71.Sagui C., Darden T. A. Annu. Rev. Biophys. Biomol. Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- 72.Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 73.Darden T., York D., Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 74.Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Montgomery J. A., Jr., Vreven T., Kudin K. N., Burant J. C., et al. Gaussian 03. Wallingford, CT: Gaussian; 2004. Revision C.02. [Google Scholar]
- 75.Kraulis P. J. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.