Abstract
Congenital hypertrophy/hyperplasia of the retinal pigmented epithelium is an ocular lesion found in patients harboring mutations in the adenomatous polyposis coli (APC) tumor suppressor gene. We report that Apc-deficient zebrafish display developmental abnormalities of both the lens and retina. Injection of dominant-negative Lef reduced Wnt signaling in the lens but did not rescue retinal differentiation defects. In contrast, treatment of apc mutants with all-trans retinoic acid rescued retinal differentiation defects but had no apparent effect on the lens. We identified Rdh5 as a retina-specific retinol dehydrogenase controlled by APC. Morpholino knockdown of Rdh5 phenocopied the apc mutant retinal differentiation defects and was rescued by treatment with exogenous all-trans retinoic acid. Microarray analyses of apc mutants and Rdh5 morphants revealed a profound overlap in the transcriptional profile of these embryos. These findings support a model wherein Apc serves a dual role in regulating Wnt and retinoic acid signaling within the eye and suggest retinoic acid deficiency as an explanation for APC mutation-associated retinal defects such as congenital hypertrophy/hyperplasia of the retinal pigmented epithelium.
Keywords: colon cancer, retina, APC
Heterozygous mutations in the tumor suppressor adenomatous polyposis coli (APC) result in familial adenomatous polyposis, a syndrome characterized by the presence of hundreds of adenomatous colonic polyps that can ultimately progress to frank adenocarcinoma (1). Familial adenomatous polyposis patients also exhibit extracolonic manifestations of APC mutation, including congenital hypertrophy/hyperplasia of the retinal pigmented epithelium and in some cases a failure of ventral retinal development known as retinal coloboma (2). Similarly, mice with targeted Apc gene disruption develop congenital hypertrophy/hyperplasia of the retinal pigmented epithelium lesions and retinal coloboma (2, 3). The mechanisms underlying APC mutation-induced ocular defects in humans and mice remain undefined.
Extensive research regarding APC function suggests that aberrant WNT/β-catenin signaling accounts for the development of colon carcinomas in individuals harboring APC mutations (4, 5). Specifically, truncating mutations in APC are thought to alleviate blockade of β-catenin/LEF transcriptional activity, thus promoting cell proliferation and inhibiting cellular differentiation (5). Consistent with a potential role for APC in abnormal eye development, the WNT/β-catenin signaling cascade is critical for specification of lens tissue and regulation of lens growth and differentiation (6, 7). WNT signaling also plays a role in various aspects of retinogenesis (8–10). For example, in zebrafish Wnt11 and Fz5 coordinate to allow retinogenesis to proceed by suppressing signals that inhibit eye field specification (11).
An additional APC function includes its recently described role in regulating the production of retinoic acid (RA) in the intestine (12, 13). It is well established that RA is required during eye development (14). Specifically, development and differentiation of the photoreceptor cell layer and the retinal pigmented epithelium depend on retinoid signaling (15, 16). The ventral retina, in particular, is highly dependent on RA. For example, Hyatt et al. (17) demonstrated that zebrafish treated with exogenous RA displayed an enhancement of ventral retina characteristics, whereas knockdown of an enzyme involved in RA biosynthesis (bcox) hindered the establishment of the ventral retina (18). In addition, recent studies show that targeted deletion of the retinoid receptor RXRα within the mouse retinal pigmented epithelium (RPE) caused RPE abnormalities, including the appearance of congenital hypertrophy/hyperplasia of the retinal pigmented epithelium-like lesions similar to those present in mice bearing mutated APC (19, 20).
Although the above studies establish a role for WNT and RA in directing ocular development, they do not delineate whether perturbations in WNT or RA signaling underlie ocular abnormalities seen after APC mutation. We report here that Apc mutant zebrafish harbor ocular abnormalities similar to those present in mice and humans with APC mutation. Furthermore, we describe a dual role for Apc in ocular morphogenesis. Apc regulation of canonical WNT/β-catenin signaling appears active in the developing lens. In contrast, Apc control of RA production via retinol dehydrogenase 5 appears confined to the retina and is required for retinal differentiation.
Results and Discussion
Consistent with reports in humans and mice, we found that homozygous apc mutant zebrafish harbor retinal coloboma (Fig. 1A). This defect was present in 100% (n = 75) of homozygous apc mutant embryos but was absent in apc heterozygous siblings (n = 50) (Fig. 1A). We also noted the presence of a protruding lens phenotype (Fig. 1A). Histologic analysis of apc mutant eyes exposed a lack of retinal organization, particularly in the ventral retina, in comparison to wild-type retinas. Specifically, the RPE was underdeveloped in the ventral retina, as was the ventral portion of the photoreceptor cell layer (Fig. 6, which is published as supporting information on the PNAS web site). In addition to ventral retina defects, the remaining RPE and photoreceptor cell populations appeared undifferentiated (Fig. 6). Confirming this histological analysis, whole-mount in situ hybridization (Fig. 1A) for the interphotoreceptor retinoid binding protein (irbp), a marker of photoreceptor and retinal pigmented epithelial cells, revealed failed retinal differentiation of these cell layers (21). Wild-type apc larvae expressed irbp (Fig. 1A; 100% positive; n = 70) whereas apc mutant retinas lacked irbp expression (0% positive; n = 39). In addition, other photoreceptor cell markers, such as zpr1, were lost in apc mutants (Fig. 7, which is published as supporting information on the PNAS web site). Loss of irbp and zpr1 expression was in contrast to early markers of retinal progenitors such as crx, otx2, neurod, pax6.2, and atoh7, all which were present in apc mutant retinas (Fig. 8, which is published as supporting information on the PNAS web site). Furthermore, apc mutant eyes showed no increase in apoptotic cells as assessed by acridine orange staining (data not shown). These findings indicate that, although apc mutant zebrafish specify retinal progenitor cells, terminal differentiation of the retina is not achieved.
Fig. 1.
Coloboma and retinal differentiation are not rescued by DN-LEF. (A) Light microscopic (LM) analysis of wild-type or apc mutant larvae at 72 hpf revealed the presence of retinal coloboma (arrow) in apc mutants. Whole-mount in situ hybridization indicates that IRBP expression is present in wild-type but not in apc mutant larvae at 72 hpf. At 72 hpf, wild-type TOPdGFP larvae have little GFP expression in the eye, whereas apc mutant larvae display robust GFP expression in the lens (arrowhead). (B) Injection of DN-LEF reduces expression of TOPdGFP in the apc mutant lens in comparison to control-injected mutants. (C) Quantitative RT-PCR for GFP (white bars) or IRBP (black bars) using total RNA from 72-hpf wild-type (wt), apc mutant (mt), apc mutants injected with 8 pg of DN-LEF (bars marked with “8”), or APC mutants injected with 40 pg of DN-LEF (bars marked with “40”). Error bars represent standard deviation.
To determine whether the lack of retinal differentiation, as well as other ocular defects in apc mutant zebrafish, result from aberrant WNT/β-catenin activation, we produced an apc mutant TOPdGFP transgenic zebrafish line (22, 23) that permits interrogation of Apc function while simultaneously analyzing WNT/β-catenin signaling. Quantitative RT-PCR revealed that apc mutant TOPdGFP embryos [72 h postfertilization (hpf)] express 3-fold more GFP than wild-type apc TOPdGFP siblings (Fig. 1C), thereby confirming that WNT/β-catenin signaling is hyperactive within whole embryos after apc mutation. However, whole-mount in situ hybridization examining GFP expression within the eyes of homozygous apc mutant TOPdGFP larvae, in comparison to wild-type siblings, at 72 hpf showed increased GFP levels primarily within the mutant lens (Fig. 1 A and B). Increased GFP was also noted within the developing hindbrain (Fig. 1A). Consistent with previous reports, GFP levels within the RPE were detectable; however, these levels did not appear to change significantly (Fig. 1A) in the apc mutants (22).
The above observations indicate WNT/β-catenin signaling in lens, but not necessarily retina, of the APC mutant fish. Furthermore, it is possible that the TOPdGFP reporter constructs does not reflect the activity of all TCF/LEF family members. To examine the role of WNT signaling in the apc mutant retinas more closely, we sought to rescue irbp expression by injecting dominant-negative (DN) LEF, a known inhibitor of WNT/β-catenin signaling (24). Although injection of DN-LEF into apc mutant TOPdGFP embryos resulted in a dose-dependent decrease in GFP expression both globally and within the lens (Fig. 1 B and C), it failed to restore irbp expression (Fig. 1C). These data indicated that disregulation of WNT signaling may not underlie retinal differentiation defects resulting from mutation in apc.
Given these findings, we considered the possibility of an additional function for apc in the zebrafish retina. The presence of severe ventral retinal defects and the absence of cellular differentiation in apc mutant embryos were highly reminiscent of retinoid deficiency (14, 15, 17, 18, 25). This finding, coupled with our previous findings that apc regulates RA production in the intestine, led us to hypothesize that retinoid deficiency underlies the defects present in apc mutant retinas. To test this hypothesis, we attempted to restore differentiation of the RPE and photoreceptor cell layers in apc mutants by treatment with vehicle (DMSO) or all-transretinoic acid (ATRA). Histologic analysis showed that ATRA treatment rescued photoreceptor and RPE morphology but did not appear to rescue lens protrusion in the apc mutants (Figs. 2A and 6). This same treatment restored the expression of the photoreceptor cell differentiation marker irbp in 45% of mutant embryos (n = 42) (Fig. 2B). In contrast, only 2% (n = 45) of mutant embryos treated with vehicle expressed irbp. Similar rescue was seen with zpr1 (Fig. 7). In addition to irbp, RT-PCR also showed that the RPE-specific marker trp2 was reduced in APC mutants and restored by RA treatment (Fig. 2C).
Fig. 2.
RA rescues APC mutants. (A) Toluidine blue staining of wild-type or APC mutant eyes treated with vehicle (V) or ATRA (RA) at 72 hpf. Arrows indicate the dorsal retina; double arrows indicate ventral retina. (B) The percent of apc mutants treated with vehicle (white bar) or ATRA (black bar) that stained positively for IRBP by whole-mount in situ hybridization. (C) Quantitative RT-PCR for IRBP (white bars) and TRP2 (black bars) using total RNA from 72-hpf wild type (WT), APC mutant (MT), and APC mutants treated with ATRA.
Because Apc appears to control RA biosynthesis in the intestine through transcriptional induction of intestinal specific retinol dehydrogenases (rdhs) (12, 26), we performed RT-PCR using a panel of zebrafish adult tissue RNAs with primers specific for several candidate, eye-specific rdhs, including rdh5 (GenBank accession no. DQ000308), prrdh1 (GenBank accession no. AY306006), prrdh2 (GenBank accession no. AY306007), rdh10 (GenBank accession no. NM_201331), and rdh12 (GenBank accession no. BC076473) (Fig. 9, which is published as supporting information on the PNAS web site). According to this analysis, all of the rdhs we surveyed demonstrated some expression in the adult eye, with rdh5 being the most highly expressed in the eye. Whole-mount in situ hybridization for rdh5, prrdh1, prrdh2, rdh10, and rdh12 in wild-type and apc mutant larvae at 72 hpf revealed expression of each of these rdhs in various regions of the eye in wild-type larvae (Fig. 3A). In addition, the expression of several rdhs was decreased in apc mutants (Fig. 3A). Most notably, rdh5 was robustly expressed in the RPE of wild-type larvae yet undetectable in apc mutants (Fig. 3A). The RPE-specific expression of rdh5 was confirmed by histologic sectioning after whole-mount in situ hybridization (Fig. 3B).
Fig. 3.
Rdh5 expression is decreased in apc mutants. (A) Whole-mount in situ hybridization on wild-type or apc mutant larvae at 72 hpf for rdh5, prrdh1, prrdh2, rdh10, or rdh12. rdh5 and rdh12 expression are absent in apc mutants. (B) After whole-mount in situ hybridization on 96-hpf larvae with an antisense probe for rdh5 we performed histologic sectioning, which revealed the RPE-specific expression of rdh5. (C) HCT116 human colon cancer cells were transfected with empty vector or rdh5 and treated with retinol, and then RA was extracted and quantified by comparison with the internal extraction standard, TTNPB. Error bars represent standard deviation.
The best-described role for rdh5 in mammals is the conversion of 11-cis-retinol to 11-cis-retinal during regeneration of visual chromophores (27). However, rdh5 is also capable of producing retinoids suitable for RA signaling (27). Consistent with this latter activity, we found that HCT116 cells transfected with rdh5 produced ≈3-fold more RA after retinol addition than vector-only control cells (Fig. 3C). Given these results, we focused on rdh5 as a candidate rdh downstream of apc in the RPE.
We sought to knock down rdh5 function in vivo by injection of antisense morpholino oligonucleotides. RT-PCR with primers specific for rdh5 confirmed that splicing of the rdh5 transcript was blocked by morpholino injection (Fig. 10, which is published as supporting information on the PNAS web site). Consistent with rdh5 as an in vivo RDH, analysis of rdh5 morphant embryos revealed the presence of retinal coloboma (Fig. 4A) and ventral retinal abnormalities, including the abnormal development of RPE and photoreceptor cells (Fig. 4A and Fig. 11, which is published as supporting information on the PNAS web site). A translation-blocking morpholino against rdh5 gave a similar phenotype (data not shown). Similar to that seen in apc mutants, whole-mount in situ hybridization demonstrated an absence of irbp in 92% of rdh5 morphants (n = 61) (Fig. 4B). irbp expression was restored in 35% of rdh5 morphants (n = 40) after treatment with ATRA (Fig. 4B). In contrast, only 6% (n = 49) of rdh5 morphant embryos treated with vehicle expressed irbp (Fig. 4B). Furthermore, histologic analysis of the RPE and photoreceptor layer suggested that these cell layers were partially rescued in response to ATRA treatment (Figs. 4A and 11). Rdh5 knockdown, therefore, recapitulated the differentiation defects present in apc mutants. However, the morphological defects, such as disruption of laminar patterning that occurred in the apc mutant, were more severe than those present in rdh5 morphants. This difference fits with the specific expression domain of rdh5, which appears restricted to the RPE. Defects within the apc mutant retinas in other specific retinal cell layers may be due to apc control of additional rdhs such as rdh12, which appeared limited to the retinal ganglion cell layer.
Fig. 4.
Rdh5 controls RA production in the RPE and rescues apc mutants. (A) Light microscopic analysis of rdh5 morphants reveals the presence of retinal coloboma (arrow). Toluidine blue staining of vehicle-treated or ATRA-treated rdh5 morphant eyes reveals rescue of ventral retinal morphology by ATRA treatment (arrows indicate the ventral retina). (B) Irbp (arrowheads) expression is present at 72 hpf in wild-type larvae, absent in rdh5 morphants, and partially rescued after ATRA treatment, as determined by whole-mount in situ hybridization. (C) Irbp expression (arrowheads) is present at 72 hpf in wild-type larvae, absent in APC mutants, and partially restored in apc mutants injected with rdh5.
The absence of rdh5 expression from apc mutant retinas combined with the similarities between rdh5 morphant and apc mutant retinas supports a model wherein apc controls retinal differentiation via regulation of rdh5 expression. Consistent with this idea, we found that injection of a plasmid encoding rdh5 into apc mutants restored irbp expression in 27% of embryos (n = 44), whereas 0% of control-injected apc mutant embryos expressed this marker (n = 32) (Fig. 4C). We also noted two domains of irbp expression in the ventronasal region of the retina and area thought to harbor early, differentiating photoreceptor cells (Fig. 4C).
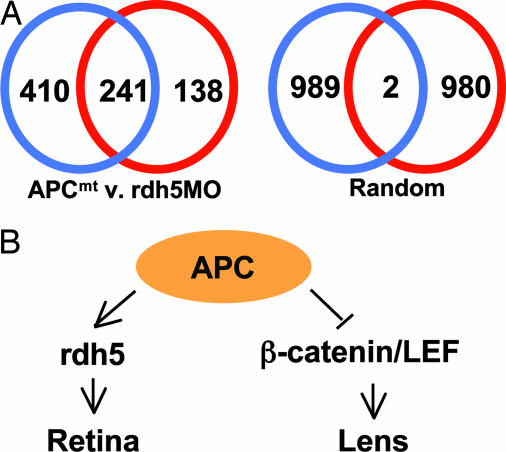
In an effort to understand the overall contribution of apc and rdh5 to eye development, we next determined the transcriptional profiles of the apc mutant and the rdh5 morphant by performing oligo-based microarray analysis using total RNA harvested from whole embryos at 72 hpf compared with wild type at 72 hpf. Our analysis initially surveyed 17,433 independent zebrafish genes. The resulting data confirmed the presence of numerous statistically significant gene expression changes of 2-fold or greater in apc mutants (651 genes total) and rdh5 morphants (379 genes total). More importantly, a comparison of the two gene sets revealed a profound overlap in the expression patterns of apc mutants and rdh5 morphants. Remarkably, 64% of the genes showing expression changes in rdh5 morphants were also differentially expressed in apc mutants (Fig. 5A). A similar control analysis comparing 1,000 genes that were randomly selected for each group showed no significant overlap (Fig. 5A). Consistent with data presented above, irbp expression was lost in both apc mutant and rdh5 morphant embryos (Table 1, which is published as supporting information on the PNAS web site). In addition, a number of opsins, which mark photoreceptor cell differentiation, were absent in both apc mutants and rdh5 morphants. This overlap indicates that the apc/rdh5 pathway represents a prominent effector pathway controlled by apc in the developing embryo. However, 63% of the expression changes in apc mutants were not accounted for by knockdown of rdh5. The apc mutant-specific changes fit with the additional functions of apc in regulating signal pathways such as WNT within the eye. However, because these analyses were performed by using whole embryos, some of the apc mutant-specific changes likely reflect the roles of apc in non-eye tissues. For example, we reported previously intestinal differentiation defects in apc morphant and mutant zebrafish (16, 28). Extraocular gene expression changes within the apc mutant would not be expected in rdh5 morphants in that rdh5 expression is specific to the retina. Consistent with these findings, intestinal rdhs rdh1 and rdh1l were absent in apc mutant embryos but not rdh5 morphant embryos (data not shown).
Fig. 5.
Microarray analysis delineates the apc–RA pathway. (A) The extent of overlap of gene sets showing at least 2-fold, statistically significant (P < 0.05) expression changes in apc mutants (blue circle) and rdh5 morphants (red circle) was determined by using the microarray analysis software GeneSpring. The “APCmt v. rdh5MO” diagram represents the number of statistically significantly disregulated genes in each mutant or morphant. A control analysis conducted by using 1,000 randomly selected genes for each condition showed little overlap between the two groups. (B) The schematic diagram shows the proposed genetic relationship between apc and rdh5 in the retina.
A number of studies suggest diverse roles for APC in regulating cell physiology. For example, early studies of APC function identified APC as binding to microtubule tips, where it is reported to promote and stabilize microtubule polymerization both in vitro and in vivo (28–30). In this manner APC is proposed to contribute to microtubule attachment to chromosomes, thereby participating in chromosomal segregation during mitosis (31, 32). These findings suggest that mutations in APC may contribute to the chromosomal instability frequently observed in colorectal tumorigenesis (33, 34).
Our findings support a model wherein Apc executes dual roles in regulating developmental signaling pathways in the eye. Apc control of RA production is critical for retinal development and differentiation, whereas Apc regulation of the WNT/β-catenin pathway appears active in the lens (Fig. 5B). These findings fit well with the established roles for RA in the development of the retina (14, 16, 25) and in RPE abnormalities found in Apc and Rxrα knockout mice (19, 20). Loss of RA production in the developing retina offers an explanation for the presence of retinal defects, such as congenital hypertrophy/hyperplasia of the retinal pigmented epithelium, commonly present in familial adenomatous polyposis patients. In addition, our data indicate that Apc regulation of RPE differentiation occurs independent of Apc control of WNT/β-catenin signaling. In this respect, other studies support β-catenin-independent functions of APC in cellular differentiation. For example, recent studies using mice show that loss of Apc impairs T cell differentiation during thymus development. These defects were not fully recapitulated by introduction of stabilized β-catenin (35). In addition, Dang et al. (36) demonstrated that APC and cdx2 regulate Kruppel-like factor 4, which suppresses proliferation in a β-catenin-independent manner. Interestingly, Samowitz et al. (37) reported that, in contrast to APC mutations, β-catenin mutations appear in smaller adenomas and are rarely seen in invasive carcinomas. Taken together the current data suggest that activation of WNT/β-catenin signaling alone does not fully recapitulate the consequences of APC mutation.
Materials and Methods
Zebrafish Stocks and Embryo Culture.
Wild-type and apc mutant Danio rerio (zebrafish) were maintained on a 14-h:10-h light:dark cycle. Fertilized embryos were collected after natural spawnings and allowed to develop at 28.5°C. Control and experimental embryos were raised in 0.003% phenylthiourea to inhibit pigment formation (38). Homozygous apc mutants were identified by PCR using genomic DNA isolated from clipped tails (23). PCR primers used were as follows: forward, 5′-atccactaataatgttgcagctgat-3′; reverse, 5′-ctgatgaaaactccaccgttttatg-3′.
Whole-Mount in Situ Hybridizations and Immunofluorescence.
Zebrafish embryos were fixed in sucrose-buffered 4% paraformaldehyde, rinsed in PBS, dehydrated in methanol, and stored at −20°C. Digoxigenin-labeled riboprobes for irbp, rdh5, prrdh1, prrdh2, rdh10, and rdh12 were generated by linearization of pCRII (Invitrogen, Carlsbad, CA) containing the corresponding cDNA followed by in vitro transcription with T7 or Sp6 RNA polymerase (Roche, Indianapolis, IN). The riboprobe for GFP was a generous gift from R. Dorsky (University of Utah). Whole-mount in situ hybridizations were carried out as described (26). Embryos were cleared in 70% glycerol in PBS and photographed by using an Olympus DP12 digital camera. Whole-mount immunofluorescence was carried out on 72-hpf zebrafish embryos. Embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C and then rinsed briefly and washed for 1 h in distilled water to permeabilize. Embryos were dehydrated to 100% methanol and stored at −20°C. After rehydration to 1× PBS, embryos were permeabilized with 0.1% collagenase (Sigma, St. Louis, MO) in PBS for 30 min at room temperature. The embryos were washed three times for 15 min in PBST (PBS with 0.1% Tween 20) followed by fixation in 4% paraformaldehyde in PBS for 10 min. After washing three times for 15 min in PBST, blocking was carried out by incubating the embryos in FBS-PBST (10% heat-inactivated FBS/0.1% Tween 20/1% DMSO in PBS) for 2 h at room temperature. Primary antibody (zpr1, 1:50; Zebrafish International Resource Center, University of Oregon, Eugene, OR) diluted in FBS-PBST was applied overnight at 4°C and then rinsed extensively in PBST. Embryos were incubated in secondary antibody (goat anti-mouse Alexa Fluor 488, 1:200; Molecular Probes, Eugene, OR) and TO-PRO-3 nuclear stain (1:500; Invitrogen) for 4 h at room temperature followed by extensive rinses in PBST. After dehydration to 100% ethanol, embryos were infiltrated and embedded by using an Immuno-Bed kit (Polysciences, Warrington, PA). Sectioning was carried out by using a 2055 microtome (Leica), and confocal images were acquired by using an IX81 FV300 Confocal Microscope (Olympus).
RT-PCR.
Single-stranded cDNA was synthesized from 1 μg of total RNA using SuperScript III (Invitrogen). PCR primers used were as follows: rdh5 forward, 5′-GACCGGTTGTGACTCTGGTT-3′; rdh5 reverse, 5′-TGCTTTCGAAGTCCTCGATT-3′; prrdh1 forward, 5′-CAGCGTCAAAGACCGACATA-3′; prrdh1 reverse, 5′-ATCGACATGGTGACGTTGAA-3′; prrdh2 forward, 5′-ACTCTGCTGCCTCTGGACAT-3′; prrdh2 reverse, 5′-GACGCTGCTCATGACGATAA-3′; rdh10 forward, 5′-GAATATCGCCACCGAGTTGT-3′; rdh10 reverse, 5′-GGGCTTCAATTGTCGGTAGA-3′; rdh12 forward, 5′-GCTGGTGTCATGATGTGTCC-3′; rdh12 reverse, 5′-CTCTGGCAGTAGGCAAGTCC-3′. β-Actin primers were as described previously (26).
Quantitative RT-PCR.
Single-stranded cDNA was synthesized from 1 μg of total RNA using SuperScript III (Invitrogen). PCR was performed by using a LightCycler instrument and software, version 3.5 (Roche Diagnostics). Primers were as follows: irbp forward, 5′- ACATGTTTGGGGACTTCGAG-3′; irbp reverse, 5′-ATCTCCTGCCTGTGAGCTGT-3′, GFP forward, 5′-CCAGATCCGCCACAACATCG-3′; GFP reverse, 5′-GTCCATGCCGAGAGTGATCC-3′. PCRs were performed in duplicate by using the LightCycler FastStart DNA Master SYBR Green I kit (Roche). PCR conditions were as follows: 35 cycles of amplification with 10 s of denaturation at 95°C, 5 s of annealing at 58°C, and 10 s of extension at 72°C. A template-free negative control was included in each experiment.
Morpholino and Microinjection Experiments.
Antisense morpholino oligos were obtained from Gene Tools (Philomath, OR). The rdh5 MO splicing-blocking morpholino (5′-ACTTAAGCTCACCTTTATCTCCAAC-3′), rdh5 MO2 translation-blocking morpholino (5′-ACTCATACATCGCTTCTACCTCCTG-3′), and control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′) were solubilized to 1 mM in 1× Danieau buffer. For microinjections, 0.5 nl of 0.5–1.0 mM morpholino was injected into zebrafish embryos at the one- to two-cell stages.
For microinjection experiments, 8–50 pg of plasmid DNA containing DN-LEF (a kind gift from D. Ayer, University of Utah) or full-length rdh5 were injected at the one-cell stage.
RA Rescue Experiments.
To rescue rdh5 morphants and APC mutants by application of RA, embryos were incubated in 500 nM ATRA in DMSO at 30 hpf and 50 hpf for 1 h. Embryos were then washed in embryo water. Control embryos were treated over these periods with an equal volume of DMSO.
ATRA Extraction and HPLC Analysis.
HCT116 cells were transfected with a vector (pDEST40) containing rdh5 cDNA. Cells were then treated with retinol, and ATRA production was quantified as previously described (26).
Histological Analyses.
Embryos were fixed in 10% neutral buffered formalin, rinsed in PBS, and embedded in glycol methacrylate (Polysciences). Five-micrometer sections were cut by using a Leica microtome and stained with toluidine blue. Sections were analyzed by using an Axiovert100 microscope (Zeiss), and pictures were taken by using a Magnafire color camera (Olympus).
Microarray Analysis.
Microarray analyses were performed by using total RNA harvested by TRIzol reagent (Invitrogen) from apc mutants or rdh5 morphants and compared with total RNA from wild-type embryos at 72 hpf. Zebrafish microarrays were obtained from Agilent, and data analysis was performed by using GeneSpring software. The generation of cRNA probes and hybridization to slides were performed according to the manufacturer’s protocol (Agilent, Palo Alto, CA) and as described previously (12).
Supplementary Material
Acknowledgments
We thank Drs. Anna-Pavlina Haramis and Hans Clevers (Netherlands Institute for Developmental Biology, Hubrecht Laboratory and Centre for Biomedical Genetics, Utrecht, The Netherlands) for providing the apc mutant zebrafish used in this study; Dr. Richard Dorsky for providing the TOPdGFP zebrafish; the DNA peptide resource, the DNA sequencing resource, the microarray resource, and the centralized zebrafish animal resource at the University of Utah; and Drs. Ed Levine, David Virshup, Stephen Prescott, and Frank Fitzpatrick for helpful discussions. This work was supported by grants from the American Cancer Society, the National Cancer Institute, and the Huntsman Cancer Foundation (to D.A.J.).
Abbreviations
- RA
retinoic acid
- rdh
retinol dehydrogenase
- ATRA
all-transretinoic acid
- APC
adenomatous polyposis coli
- irbp
interphotoreceptor retinoid binding protein
- RPE
retinal pigmented epithelium
- hpf
hours postfertilization
- DN
dominant-negative.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ000308).
References
- 1.Fodde R., Smits R., Clevers H. Nat. Rev. Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 2.Kermane A., Tachfouti S., El Moussaif H., Mohcine Z. Bull. Soc. Belge Ophtalmol. 2004:59–64. [PubMed] [Google Scholar]
- 3.Marcus D. M., Rustgi A. K., Defoe D., Brooks S. E., McCormick R. S., Thompson T. P., Edelmann W., Kucherlapati R., Smith S. Arch. Ophthalmol. 1997;115:645–650. doi: 10.1001/archopht.1997.01100150647013. [DOI] [PubMed] [Google Scholar]
- 4.Polakis P. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 5.Radtke F., Clevers H. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 6.Stump R. J., Ang S., Chen Y., von Bahr T., Lovicu F. J., Pinson K., de Iongh R. U., Yamaguchi T. P., Sassoon D. A., McAvoy J. W. Dev. Biol. 2003;259:48–61. doi: 10.1016/s0012-1606(03)00179-9. [DOI] [PubMed] [Google Scholar]
- 7.Smith A. N., Miller L. A., Song N., Taketo M. M., Lang R. A. Dev. Biol. 2005;285:477–489. doi: 10.1016/j.ydbio.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Jin E. J., Burrus L. W., Erickson C. A. Mech. Dev. 2002;116:173–176. doi: 10.1016/s0925-4773(02)00128-4. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrmann S., Stark M. R., Heller S. Gene Expression Patterns. 2003;3:659–662. doi: 10.1016/s1567-133x(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 10.Liu H., Mohamed O., Dufort D., Wallace V. A. Dev. Dyn. 2003;227:323–334. doi: 10.1002/dvdy.10315. [DOI] [PubMed] [Google Scholar]
- 11.Cavodeassi F., Carreira-Barbosa F., Young R. M., Concha M. L., Allende M. L., Houart C., Tada M., Wilson S. W. Neuron. 2005;47:43–56. doi: 10.1016/j.neuron.2005.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jette C., Peterson P. W., Sandoval I. T., Manos E. J., Hadley E., Ireland C. M., Jones D. A. J. Biol. Chem. 2004;279:34397–34405. doi: 10.1074/jbc.M314021200. [DOI] [PubMed] [Google Scholar]
- 13.Nadauld L. D., Shelton D. N., Chidester S., Yost H. J., Jones D. A. J. Biol. Chem. 2005;258:30490–30495. doi: 10.1074/jbc.M504973200. [DOI] [PubMed] [Google Scholar]
- 14.Hyatt G. A., Dowling J. E. Invest. Ophthalmol. Vis. Sci. 1997;38:1471–1475. [PubMed] [Google Scholar]
- 15.Marsh-Armstrong N., McCaffery P., Gilbert W., Dowling J. E., Drager U. C. Proc. Natl. Acad. Sci. USA. 1994;91:7286–7290. doi: 10.1073/pnas.91.15.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyatt G. A., Schmitt E. A., Fadool J. M., Dowling J. E. Proc. Natl. Acad. Sci. USA. 1996;93:13298–13303. doi: 10.1073/pnas.93.23.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyatt G. A., Schmitt E. A., Marsh-Armstrong N., McCaffery P., Drager U. C., Dowling J. E. Development (Cambridge, U.K.) 1996;122:195–204. doi: 10.1242/dev.122.1.195. [DOI] [PubMed] [Google Scholar]
- 18.Lampert J. M., Holzschuh J., Hessel S., Driever W., Vogt K., von Lintig J. Development (Cambridge, U.K.) 2003;130:2173–2186. doi: 10.1242/dev.00437. [DOI] [PubMed] [Google Scholar]
- 19.Perrault I., Hanein S., Gerber S., Barbet F., Ducroq D., Dollfus H., Hamel C., Dufier J. L., Munnich A., Kaplan J., Rozet J. M. Am. J. Hum. Genet. 2004;75:639–646. doi: 10.1086/424889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori M., Metzger D., Picaud S., Hindelang C., Simonutti M., Sahel J., Chambon P., Mark M. Am. J. Pathol. 2004;164:701–710. doi: 10.1016/s0002-9440(10)63157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stenkamp D. L., Cunningham L. L., Raymond P. A., Gonzalez-Fernandez F. Mol. Vis. 1998;4:26. [PubMed] [Google Scholar]
- 22.Dorsky R. I., Sheldahl L. C., Moon R. T. Dev. Biol. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- 23.Hurlstone A. F., Haramis A. P., Wienholds E., Begthel H., Korving J., Van Eeden F., Cuppen E., Zivkovic D., Plasterk R. H., Clevers H. Nature. 2003;425:633–637. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 24.Hovanes K., Li T. W., Munguia J. E., Truong T., Milovanovic T., Lawrence Marsh J., Holcombe R. F., Waterman M. L. Nat. Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 25.Mey J., McCaffery P., Drager U. C. J. Neurosci. 1997;17:7441–7449. doi: 10.1523/JNEUROSCI.17-19-07441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadauld L. D., Sandoval I. T., Chidester S., Yost H. J., Jones D. A. J. Biol. Chem. 2004;279:51581–51589. doi: 10.1074/jbc.M408830200. [DOI] [PubMed] [Google Scholar]
- 27.Duester G. Eur. J. Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- 28.Munemitsu S., Souza B., Muller O., Albert I., Rubinfeld B., Polakis P. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- 29.Nathke I. S., Adams C. L., Polakis P., Sellin J. H., Nelson W. J. J. Cell Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zumbrunn J., Kinoshita K., Hyman A. A., Nathke I. S. Curr. Biol. 2001;11:44–49. doi: 10.1016/s0960-9822(01)00002-1. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan K. B., Burds A. A., Swedlow J. R., Bekir S. S., Sorger P. K., Nathke I. S. Nat. Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 32.Bienz M. Nat. Cell Biol. 2001;3:E67–E68. doi: 10.1038/35060140. [DOI] [PubMed] [Google Scholar]
- 33.Lengauer C., Kinzler K. W., Vogelstein B. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 34.Fodde R., Kuipers J., Rosenberg C., Smits R., Kielman M., Gaspar C., van Es J. H., Breukel C., Wiegant J., Giles R. H., Clevers H. Nat. Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 35.Gounari F., Chang R., Cowan J., Guo Z., Dose M., Gounaris E., Khazaie K. Nat. Immunol. 2005;6:800–809. doi: 10.1038/ni1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang D. T., Mahatan C. S., Dang L. H., Agboola I. A., Yang V. W. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samowitz W. S., Powers M. D., Spirio L. N., Nollet F., van Roy F., Slattery M. L. Cancer Res. 1999;59:1442–1444. [PubMed] [Google Scholar]
- 38.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish. Eugene: Univ. of Oregon Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.