Abstract
Orthologous gene structures in eight vertebrate species were compared on a genomic scale to detect the birth and maturation of new internal exons during the course of evolution. We found that 40% of new human exons are alternatively spliced, and most of these are cassette exons (exons that are either included or skipped in their entirety) with low inclusion rates. This proportion decreases steadily as older and older exons are examined, even as splicing efficiency increases. Remarkably, the great majority of new cassette exons are composed of highly repeated sequences, especially Alu. Many new cassette exons are 5′ untranslated exons; the proportion that code for protein increases steadily with age. New protein-coding exons evolve at a high rate, as evidenced by the initially high substitution rates (Ks and Ka), as well as the SNP density compared with older exons. This dynamic picture suggests that de novo recruitment rather than shuffling is the major route by which exons are added to genes, and that species-specific repeats could play a significant role in recent evolution.
Keywords: repeats, splicing
In the course of evolution, new genes can be created by duplication or rearrangement of existing genes and in higher organisms by the shuffling of exons from one gene to another (1). The addition of an exon to an existing gene could also take place by the recruitment of an intronic fragment [an exaptation (2)], constituting the de novo formation of a novel exon. This idea has recently gained support from pair-wise comparisons of alternative splicing in humans vs. rodents. Modrek and Lee (3) showed that cassette exons (exons that are either included or skipped in their entirety) that are included at a low frequency in one species are usually not present in the other species, suggesting that alternative splicing is associated with either exon creation or exon loss. Approaching the matter from the other direction, Wang et al. (4) compared human and mouse genomes and defined 2,695 “rodent-specific” exons that are missing in the human genome. They found that most of these exons are spliced at a low frequency, and that they originate from unique intronic sequences.
Notwithstanding these findings, some central questions remain: Are the nonconserved minor spliced exons simply the result of temporary splicing mistakes without evolutionary relevance (i.e., garbage), or are they substrates for selection, destined to become functional in the future? Does the presence of an exon in one species and its absence in the other represent exon creation or exon loss (5)?
To address these questions, we determined the evolutionary course of the birth and maturation of exons by using multiple genome comparisons. We showed here that new exons are born by recruitment of highly repeated sequences as cassettes with low inclusion rates, after which they gradually gain protein-coding capacity and functionality. This trend was observed in both the human and mouse genomes. Our data provide robust and large-scale evidence for the de novo recruitment of intronic fragments as a major mode of exon creation in recent evolution.
Results
Division of Exons According to Their Evolutionary Ages.
We compiled a database composed of >100,000 human exons and classified them according to whether their orthologs could be found in the chimpanzee, dog, mouse, rat, chicken, zebrafish, and fugu genomes. We reasoned that a human exon whose ortholog is present in a given organism must have been “born” before the divergence between humans and that organism. On the other hand, if the ortholog is absent, either it was born after the divergence between humans and that organism, or it was lost in that particular organism. However, if it is also absent in all other more divergent organisms tested, then the latter is unlikely. Based on this rationale, all human exons were divided into five groups according to their divergence from other vertebrates, to classify them according to their birth. About 70,000 exons are common to humans, fugu, and zebrafish and so represent the most ancient group. Over 10,000 exons were shared by human and chicken but were absent in fish. These exons were presumably created after the human–fish but before the human–chicken split. We did not require these human–chicken orthologs to exist in intermediate genomes such as mouse, rat, and dog; in this way, we included ancient exons that could have been lost in some intermediate lineages. Similarly, 20,345, 2,341, and 2,179 exons were assigned to groups that were created before the human–rodent, human–dog, and human–chimpanzee splits, respectively. It should be noted that these groups are mutually exclusive and comprise the entire exon dataset.
A similar number of mouse exons and their orthologs in multiple vertebrate genomes were also extracted, yielding 1,249 exons that are present exclusively in the mouse and rat genomes. We then ascertained the splicing events associated with each human or mouse exon by examining the corresponding EST and mRNA sequences.
The Proportion of Exons That Are Alternatively Spliced Is Higher in Newer Exons.
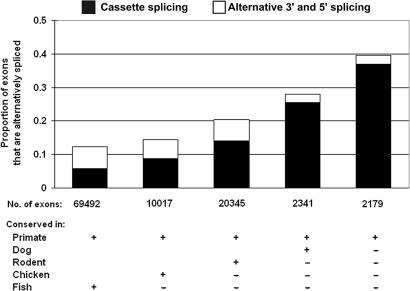
Overall, we found that 15% of primate exons (present in both human and chimpanzee) are alternatively spliced, with 9% being cassette exons. Intriguingly, we found a strong inverse correlation between the evolutionary age and the proportion of cassette exons (Fig. 1, filled areas in each column). Over 35% of the most recently evolved human exons are cassette exons, in contrast to 5% of the most ancient exons. Wang et al. (4) reached a similar conclusion for rodent exons based on a mouse–human comparison. Here, by examining eight vertebrate species, we found that exons with intermediate ages have intermediate proportions of cassette exons, establishing a clear trend for the younger exons to be cassette exons (P < 0.002 for the null hypothesis that the five groups assort randomly with the proportion of cassette exons). Interestingly, the proportion of exons that use alternative 5′ or 3′ splice sites does not show the same trend (Fig. 1, open areas in the columns), because it is very infrequent among the most recent exons (2–3% in exons born after the human–dog and –rodent splits), and it remains relatively constant in older exons (5–6% in exons born before the human–dog and –rodent splits). This different evolutionary course may reflect the fact that exons using alternative 5′ or 3′ splice sites are built from existing exons, whereas cassette exons represent the birth of novel exons. We have focused here on the latter and, to simplify the analysis, we have considered only internal exons, disregarding terminal exons.
Fig. 1.
Recent exons are more likely to be alternatively spliced cassette exons. Human exons were sorted according to whether their orthologs can be found in various vertebrate genomes from chimpanzee to zebrafish. The existence of an orthologous exon in another genome indicates the exon appeared before the divergence of that genome and the human genome. Following this rationale, we divided all human exons into five evolutionary groups. We then examined the frequency and extent of alternative splicing in each human exon group. The columns show the proportion of human exons in each group that are alternatively spliced. The filled area in each column represents cassette exons, and the open area represents exons that use a single alternative splice site. The total number of exons in each group is shown beneath each column.
Turning to the mouse, we observed the same inverse correlation between exon age and alternative splicing (Fig. 6, which is published as supporting information on the PNAS web site). However, here the proportions of alternatively spliced exons are generally lower (by ≈30%) in all evolutionary groups. Because the volume of the mouse EST database is only about two-thirds that of human, we considered the possibility that a detection bias could be the cause of this discrepancy. However, randomly purging a third of the ESTs in the human database to make it the same size as the mouse database did not remove this disparity; the proportion of human cassette exons overall was ≈1.6 times that of mouse.
Inclusion Rates of Cassette Exons Are Lower in Newer Exons.
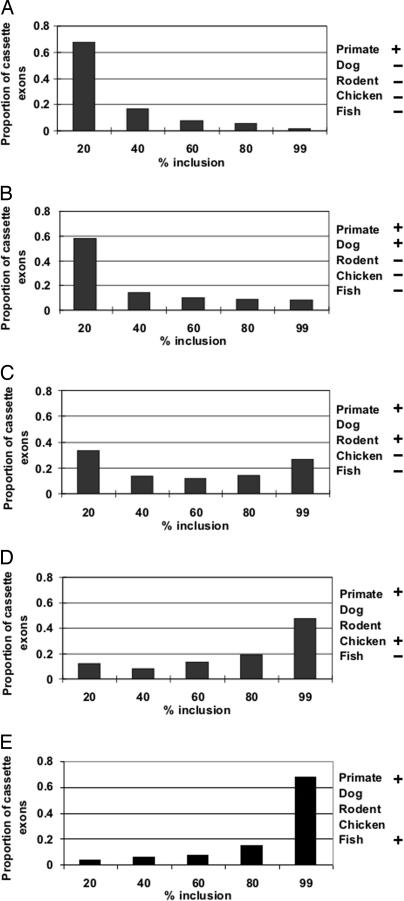
We next asked whether the inclusion rate of human cassette exons increases with evolutionary age, reasoning that the longer an exon has been preserved, the more likely it is to be functional and therefore used. That this is indeed the case can be seen in Fig. 2, which shows the distribution of inclusion rates for the five evolutionary groups. Most of the younger human cassette exons are spliced inefficiently (1–20% of the time; Fig. 2A). This situation gradually changes as the exons get older (Fig. 2 A–E), such that in the most ancient group, the inverse distribution was found; now the majority of cassette exons are included 80–99% of the time (Fig. 2E). The same phenomenon was observed for mouse exons, suggesting this progression may be general for all mammals (Fig. 7, which is published as supporting information on the PNAS web site). These results, together with those of Fig. 1, confirm the findings derived from human–mouse comparisons that nonconserved exons are mostly minor exons (3, 4). But beyond that, our data suggest an evolutionary course for cassette exons. When exons first appear, they tend to be cassette exons and are only occasionally included in the final transcripts. Such minor young exons provide substrates for further evolution and are subject to selection (4, 6–9). Presumably, some of these exons would confer an adaptive advantage to the organism (e.g., a new useful protein domain) and would be preserved by selection. The inclusion rates of these exons would subsequently increase and possibly reach constitutive status given enough time.
Fig. 2.
The recent human exons are mostly spliced at low inclusion rates, and the splicing efficiency increases with time. Human cassette exons were divided into five groups, as described in Methods. For each cassette exon in the dataset, we counted the number of ESTs in which the exon is included (Ni) or excluded (Ne). Percent inclusion is defined as Ni/(Ni + Ne) × 100%. The figure shows the histograms of percent inclusion for the five exon groups; the different groups are composed of exons of increasing conservation from top to bottom, the top representing the most recently evolved exons. The upper bounds of the five bins used are shown beneath each column.
The Majority of Recently Born Exons Originated from Highly Repeated Sequences.
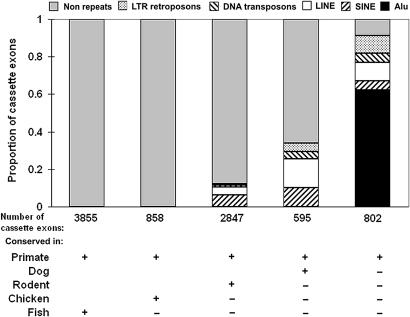
We noticed that many of the most recent human exons resemble genomic interspersed repeat sequences. To systematically examine whether highly repeated sequences contribute to the birth of exons, we used RepeatMasker to classify the exons and their flanking sequences. Strikingly, >90% of the most recent (primate-specific) cassette exons overlap with repeats (Fig. 3). In particular, 67% overlap with short interspersed nuclear elements (SINEs), and 62% overlap with Alu elements, a primate-specific class of SINE repeats. Remarkably, a substantial number of recent constitutive exons also overlap with Alus (372, 28%) or other classes of repeats (386, 29%). Among all recent exons, those that overlap with an Alu have a lower rate of inclusion on average (52%) than those that do not (82%; Fig. 8, which is published as supporting information on the PNAS web site). It has previously been estimated that 5% of all cassette exons overlap with Alus (10), and that Alu sequences can be exonized through a small number of mutations (11, 12). Consistent with the idea of exonization (13), we found the degree of sequence overlap with recent cassette exons is extensive; among all of the recent cassette exons that overlap with Alus, 90% fall entirely within those Alus (Fig. 9, which is published as supporting information on the PNAS web site). Overall, 40% of all recent (alternative and constitutive) exons are completely overlapped by Alus. This proportion far exceeds the expected probability (9%) that a random region of 120 nt (the average size of an internal exon) would fall within an Alu (P < 10−10).
Fig. 3.
Recent cassette exons consist primarily of “exonized” interspersed repeat elements. RepeatMasker was used to find highly repeated sequences in and around the cassette exons in our dataset. Cassette exons were divided into five groups, as described in the text and the legend to Fig. 1. The patterned areas show the proportions of cassette exons that have significant similarity to different types of repeats. The total number of cassette exons in each group is shown beneath each column.
Although our data point to Alus as playing an important role in creating new exons, other repeats such as long interspersed elements, DNA transposons, and LTR transposons (but not simple repeats) have been exonized as well (Fig. 3). Because exons in more divergent evolutionary groups are examined, the proportion of cassette exons that overlap with repeats (especially SINEs) diminishes rapidly. For example, for exons common to dog as well as primates, only 34% and 10% overlap with all repeats and SINEs, respectively, compared with 90% and 67% for the most recent exons (Fig. 3). As expected, the SINEs exonized in this group of exons are not the primate-specific Alus but are mammalian-wide interspersed repeats (data not shown). In sharp contrast to the mammalian-specific exons, the two most ancient groups of exons (before the human–fish and human–chicken splits) do not overlap with any repeats (Fig. 3).
We examined rodent-specific exons in a similar manner. Here 65% of the most recent rodent cassette exons originate from repeats and 28% from lineage-specific SINEs (Fig. 10, which is published as supporting information on the PNAS web site), compared with 90% for primate repeats and 62% for Alus (Fig. 3). The difference (P < 10−10) can be explained by two factors: (i) Alus comprise a higher fraction of the human genome (10.7%) than mouse-specific repeats of the mouse genome (7.6%; ref. 14). (ii) Alu sequences are rich in 3′ splice site-like sequences (Fig. 11, which is published as supporting information on the PNAS web site) and so are poised for exonization; rodent-specific repeats have a lower density of such sites (Fig. 11). LTRs present the inverse situation; their contribution to recent cassette exons in the mouse is twice that of primates, in keeping with the 2-fold greater abundance of LTRs in the mouse compared with primates.
The Proportion of Exons Located in Untranslated mRNA Regions Is Higher in Newer Exons.
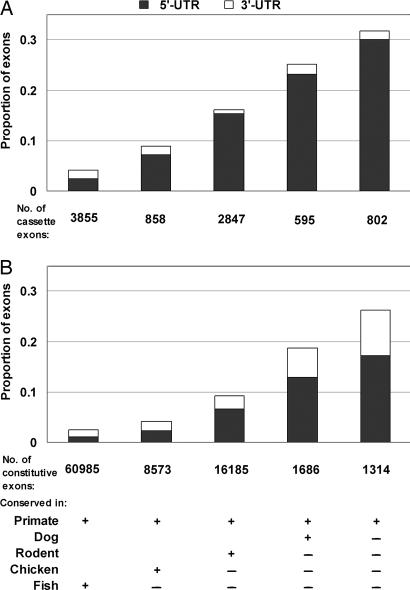
There is a strong propensity for the recent primate-specific exons to lie within UTRs. More than 30% of recent cassette exons occur as 5′-UTRs; this proportion decreases steadily with age and is only 1–2% in ancient exons (Fig. 4A). There are many fewer recent cassette exons in the 3′-UTRs compared with 5′-UTRs; the former may be selected against, because they could trigger nonsense-mediated RNA decay (making the normal stop codons in the preceding exons appear premature), or because they could provide microRNA targets. Constitutive exons also exhibit these trends (Fig. 4B). Notwithstanding the abundance in UTRs, a substantial proportion of recent exons comprise protein-coding regions, indicating that such new exons are effecting protein changes. Among the recent constitutive exons, about half of Alu-overlapping exons lie within UTRs, whereas exons that do not overlap with any repeat lie mostly in coding regions (Fig. 12, which is published as supporting information on the PNAS web site). These data suggest that many recent exons first arise in noncoding regions where their presence is better tolerated and from where they may later evolve into protein-coding exons. This scenario provides a mechanism by which simpler proteins get more complex by adding domains to their termini, a pattern of domain accretion that has often been observed (ref. 15; Fig. 2).
Fig. 4.
Many recent internal exons lie in the UTRs of genes. The protein-coding properties of cassette (A) or constitutive exons (B) from the different evolutionary groups were determined by using well annotated mRNA sequences. These exons were divided into three classes: (i) pure 5′-UTRs; (ii) coding exons, partial 5′-UTRs (containing translation start sites) plus pure coding exons plus partial 3′-UTRs containing stop codons; and (iii) pure 3′-UTRs. The column height shows the total proportion of noncoding exons, with the filled area indicating the exons in the 5′UTR and the open areas the 3′ UTR.
Newer Exons Are Evolving at a Higher Rate.
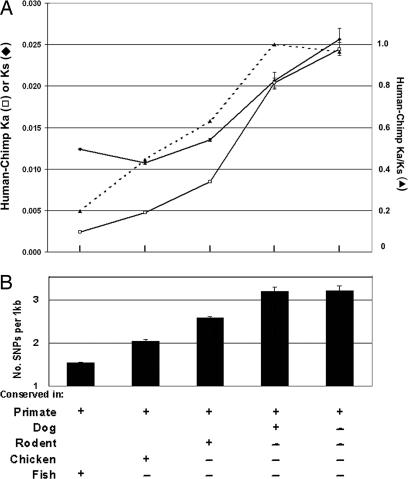
If the newly born exons lack any function, they may exhibit an unconstrained and therefore rapid rate of evolution compared with more mature exons. To measure these evolutionary rates, we calculated the nonsynonymous and synonymous amino acid substitution rates (Ka and Ks, respectively) between orthologous human and chimpanzee coding exons for the five different evolutionary groups. A Ka/Ks ratio of <1 is generally considered evidence for purifying selection, i.e., pressure against mutational change of a functional sequence; whereas a ratio near 1 indicates little or no such pressure, i.e., a lack of function. The Ka/Ks ratio (the average Ka divided by the average Ks for each group) was close to 1 for the most recent exons and steadily decreased to 0.2 in the most ancient exons (Fig. 5A). An examination of Ka and Ks individually is also revealing. Ka increases by an order of magnitude in the most recent exons compared with the most ancient, signifying that the recent exons are undergoing a much more rapid evolution of protein sequences (Fig. 5A). These results are consistent with the finding of Xing and Lee (8) that minor alternative exons are evolving faster. Interestingly, Ks values were also higher for these recent exons (2-fold), in keeping with the idea that the recent exons are rapidly evolving at the nucleotide level as well (i.e., independent of protein-coding capacity). These nucleotide changes are presumably creating exonic splicing enhancers that increase exon inclusion (16–18). The change in Ks with time supports the growing realization that synonymous codon changes are not always neutral, and so the Ka/Ks ratio is an imperfect gauge of protein selection (9, 18, 19).
Fig. 5.
Recent human exons are evolving much faster than ancient exons. (A) The synonymous and nonsynonymous substitution rates, Ks (open squares) and Ka (filled diamonds), respectively, between human and chimpanzee exon pairs and the Ka/Ks ratio (dotted line) are shown for each evolutionary group. The number of coding exons used to calculate the Ka and Ks were 67,476, 8,801, 14,080, 730, and 509 for the five evolutionary groups, respectively. (B) Recent human exons have a higher SNP density compared with ancient exons. SNPs from human dbSNP database were mapped to human exons in different evolutionary groups, and the density of SNPs was measured as described in Methods.
We next asked whether the rapid evolution of a new exon was also characteristic of its host gene taken as a whole; for instance, if the new exon disrupted the former gene function, an entirely new function might be created. We found the reverse to be true; Ka and Ks values for the other exons in the genes harboring the exon in question show no such difference in rate, regardless of the age of the classifying exon (Fig. 13, which is published as supporting information on the PNAS web site). This result is consistent with the notion that new exons initially correspond to functionless inclusions in minor splicing isoforms and gradually gain function with time, and that this function is most often related to the function of the host gene.
Evolutionary rates can also be reflected by SNP density, which reflects the intraspecies polymorphism level. We therefore examined the distribution of SNPs among the human exons of the different evolutionary groups. A decreasing trend with evolutionary age was observed; SNP density is 2-fold higher in the most recent compared with the most ancient exons (Fig. 5B), similar to the difference in Ks above. This result provides further evidence that newly born exons are less functional and are evolving rapidly.
Recruitment of Exons from Intronic Sequences vs. Exon Shuffling.
Our method for classifying exons according to their time of appearance relies on the absence or presence of an exon in an aligned region between two species. An ancient exon that had been shuffled into an intron in one species but not the other would be an old exon in a new context but would appear as a new exon. However, most new exons overlap with repeats, making it unlikely that they represent ancient shuffled exons and leading to the conclusion that most new exons do not arise by exon shuffling. To test the possibility that new human exons of unique sequence represent shuffled exons, we compared 100 such human exons against the mouse EST database. None exhibited significant similarity (E values <10−3). In sharp contrast, among 100 randomly selected ancient exons, 83 yielded hits with E values <10−5 (Fig. 14, which is published as supporting information on the PNAS web site). These results suggest that exon shuffling is at most a minor contributor to the current population of new exons.
Expression Profiles of the Genes That Contain the Recently Born Exons.
Finally, an EST expression profile of the 1,790 genes that contain the 2,179 recent exons showed approximately equal distribution among 30 tissues, with the exception of testis, which exhibited an EST abundance twice the average of all other tissues (Fig. 15, which is published as supporting information on the PNAS web site). These data eliminated the concern that these exons appear only in highly specialized genes or tissues and so are not representative of the whole genome.
Discussion
One plausible explanation for the fact that exons not conserved between any two species tend to be cassette-spliced is that these exons are simply the results of “temporary” splicing mistakes and will eventually be lost in evolution. Our data strongly argue against this possibility by showing a continuous and presumably ongoing course of exon evolution in terms of improved splicing, reduced repeat content, and increased protein fixation. Taken together, our results support the following scenario. Many exons are created from introns, first in the form of cassette exons; such exons may be selected against to the extent that they disrupt the normal function of the resident genes. Two mechanisms mitigate the selection pressure against these exons. First, the new exons are spliced inefficiently and therefore appear in only a minority of transcripts; second, these exons are preferentially located in UTRs, leaving the proteins intact. These exons will later become incorporated into protein-coding regions. Highly repeated sequences are a rich source of such new exons. The new exons evolve rapidly at both the protein level and the RNA level; some of these exons eventually become advantageous to the organism through the evolution of a new protein function (20). The splicing efficiency of such advantageous exons then increases through positive selection pressure, eventually reaching constitutivity in some cases.
It would be interesting to document the mutational events that lead to the creation of an exon from an intronic sequence (11). Comparative genomics might be used for this purpose, using a species that is the right distance from human to be informative. The chimpanzee is probably too close, and the mouse is probably too far in this regard. The macaque could be the best choice, but at present, there is not enough splicing information (mRNA or EST data) to determine whether a candidate sequence is used as an exon.
Our finding that a majority of the most recent exons originate from highly repeated sequences contrasts with that of Wang et al. (4), who concluded that recent rodent exons, defined as missing in humans and pigs, are derived mostly from unique intronic sequences. Two possible biases in their study may explain the disparity. First, we estimate these authors used ≈700,000 pig ESTs (for ≈10,000 exons in our reconstruction of the procedure) for the ancestral reference to differentiate gain or loss of exons in the mouse genome compared with human. This number may be insufficient to capture the majority of alternatively spliced cassette exons; by comparison, the human dbEST comprises ≈7,500,000 ESTs. To test this idea, we randomly reduced the number of human ESTs to 750,000 and found only 15% of the alternatively spliced exons detected by our original database (data not shown). Thus, a substantial proportion of the exons collected by Wang et al. (4) could be ancient exons that have been lost in the human genome rather than new exons gained in the mouse (which are actually present but were undetected in the pig genome). Second, to parse the intron–exon structures, Wang et al. (4) aligned full length mouse cDNAs from the RefSeq database to the mouse genomic sequence. The RefSeq cDNA database imparts a strong curation bias toward abundant and repeat-free transcripts (21). Exons spliced at low inclusion rates and repeat-overlapping exons are therefore underrepresented in this database and were precluded (4). These types of exons represent major classes found in our analysis.
Others have previously noted a connection between human Alu sequences and alternative splicing. Sorek et al. (11) found 26 exons derived from Alu sequences and showed that just a few mutations sufficed for exonization. Krull et al. (13) documented the time course during primate evolution of four examples of Alu exonization. Zheng et al. (22) and Sorek et al. (10) found numerous examples of alternatively spliced exons that overlapped with repeat sequences of diverse types; almost no constitutively spliced exons did so. Repeat elements have also been shown to evolve into transcriptional regulatory elements (23) and to specify protein domains (24). In one case, the DNA-binding domain contributed by a recently exonized transposon repeat was shown to be retained and functional in the chimeric protein product (25). The results reported here show that highly repeated sequences are the most important source of new exons in both humans and the mouse. At least two reasons help explain this predominance: (i) Alu repeats contain motifs similar to the splice consensus sequences, so few changes are required to effect exonization (Fig. 11 and ref. 11); and (ii) the ability of these sequences to transpose allows them to move to more hospitable environments for splicing, e.g., away from intronic silencing elements (26, 27). Such “preexon shuffling” could represent a rich source of variation that complements mutation, as has been argued for experimental DNA shuffling (28). The shuffling of preexisting exons, on the other hand, was found here not to be a major contributor to the appearance of new exons in genes.
It is possible that the exaptation of intron sequences as new exons is a major route to new genes, along with gene duplication. However, because we would not have detected new genes that arose by gene duplication, we cannot compare the relative contribution of these two processes. Moreover, it has not yet been directly shown to what extent newly exapted repeated sequences in fact go on to contribute functional phenotypes or even the proportion that become fixed. To get a rough preliminary estimate of the latter, we plotted the cumulative number of new exons as a function of divergence time (29). All of the exon data fell close to a straight line (60 new exons per million years, R2 = 0.97; Fig. 16, which is published as supporting information on the PNAS web site), including the point for the most recent exons. If the great majority of the most recent exons were not to become fixed, we would have expected that point to be high relative to older exons. Although this result is consistent with the idea that most exons do eventually become fixed, it is far from persuasive, because divergence times may be inexact, and there are few data points provided by this work.
This work suggests that highly repeated sequences, rather than being parasitic invaders and junk, play an important evolutionary role in the evolution of new genes. The documentation of a number of Alu exonization events led Sorek et al. (11) to propose that exaptation of Alus may have “promoted speciation of the human lineage.” Our data support this idea and extend it to additional classes of repeats and to other mammals.
Methods
Compilation of Human and Mouse Exon–Intron Structures.
Human and mouse exon–intron structures were determined by aligning mRNA/EST sequences from the UniGene database (ftp://ftp.ncbi.nih.gov/repository/UniGene/Homo_sapiens and download Hs.seq.all.gz) to assembled genomic sequences (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens) using sim4. Only ESTs that span at least two exon–exon joints were considered. A perl script was written to parse exon/intron borders from the alignment.
Orthologous Exons in Other Genomes.
We downloaded a human-referenced eight-genome alignment from the University of California, Santa Cruz Genome Bioinformatics Site (http://hgdownload.cse.ucsc.edu/goldenPath/hg17/multiz8way).
These genomes are aligned by using multiz (30) and blastz (31). We then mapped each exon to the human genome by using megablast (32). The corresponding segment of a second species was then extracted according to the coordinates of the human exon if it was present in the University of California, Santa Cruz (UCSC) alignment, and if the AG and GT dinucleotides bordering the exon in the second species were conserved (a negligible number of orthologous exons were rejected by this second criterion). This approach maximized the possibility of finding orthologous exons and not simply homologous sequences, because the exon alignments are within the syntenic context of the UCSC whole-genome alignment. For rodent-specific exons, we downloaded another multiple genome-alignment from UCSC (http://hgdownload.cse.ucsc.edu/goldenPath/mm7/multiz17way) that uses the mouse genome as the reference genome.
Division of Exons According to Their Evolutionary Ages.
The evolutionary group to which a human exon is assigned depends on the most divergent genome where its ortholog exists. The order (from the most to the least divergent from human) of the eight genomes was set to be fugu/zebrafish, chicken, mouse/rat, dog, and chimpanzee/human, where “/” means equivalence. Classes were formed by pairing human exons with those of another single species or single group. Only human exons also represented in the chimpanzee were used. A fish or rodent exon was considered a member of that class if it was present in either of two equivalent species. Classifications using the mouse genome as a reference were carried out similarly. Annotated listings of all exons in each grouping are at http://cubweb.biology.columbia.edu/lac2/exonsbyage.
Repeats.
We used RepeatMasker (www.repeatmasker.org) to mask the interspersed repeat sequences, always using the most sensitive setting (“-s” parameter in the command line). The type and location of repeats were determined by examining the “*.out” output of RepeatMasker. An exon was considered to overlap with a repeat if it shared at least one nucleotide with any type.
Noncoding Exons.
To determine their protein-coding properties, we first mapped exons to well annotated RefSeq mRNA sequences. For those exons that were not present in RefSeq, we mapped the two flanking exons from the EST sequences to RefSeq and then inferred the protein-coding property of the exon in question from the protein-coding properties of the flanking exons.
Ka and Ks Calculation.
We calculated Ka, Ks, and their variances between human and chimpanzee following the approach developed by Li (33). The phases of exons were determined by aligning exons to the RefSeq database (ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot) and parsed by examining the annotation of RefSeqs. Exons that failed to be aligned, were in noncoding regions, or had ambiguous phases were discarded.
SNPs.
Chromosomal coordinates of all SNPs in the dbSNP build 124 database were obtained from the University of California, Santa Cruz genome bioinformatics site. We then mapped SNPs to exons according to their coordinates. SNPs derived from transcribed sequences (ESTs or cDNA sequences) or without exact coordinates were ignored. The SNP density of each group of exons was defined as the total number of SNPs falling into these exons divided by the total number of nucleotides comprising these exons.
Comparing Human Exons Against the Mouse Transcriptome.
We randomly selected 100 new exons in the most recent evolutionary group that do not overlap with repeats as well as 100 such exons in the most ancient group. Blastn (using default parameters) was used to compare these exons with mouse ESTs downloaded from the National Center for Biotechnology Information dbEST database. For each exon, the E value of the best hit was recorded.
Expression Profile.
We parsed expression profile information of genes that contain the recent exons by examining the “Hs.profiles” file in the UniGene database. Expression levels were gauged by the normalized number of ESTs in each pool.
Supplementary Material
Acknowledgments
We thank M. Arias, D. Kelley, and M. Tobias for useful discussions and J. Thornton for a careful and critical reading of the manuscript. This work was funded by a grant from the National Institutes of Health (to L.A.C.).
Abbreviation
- SINE
short interspersed nuclear element.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Long M., Deutsch M., Wang W., Betran E., Brunet F. G., Zhang J. Genetica. 2003;118:171–182. [PubMed] [Google Scholar]
- 2.Gould S., Vrba E. Paleobiology. 1982;8:4–15. [Google Scholar]
- 3.Modrek B., Lee C. J. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 4.Wang W., Zheng H., Yang S., Yu H., Li J., Jiang H., Su J., Yang L., Zhang J., McDermott J., et al. Genome Res. 2005;15:1258–1264. doi: 10.1101/gr.3929705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ast G. Nat. Rev. Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 6.Resch A., Xing Y., Alekseyenko A., Modrek B., Lee C. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing Y., Lee C. J. Trends Genet. 2004;20:472–475. doi: 10.1016/j.tig.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Xing Y., Lee C. Proc. Natl. Acad. Sci. USA. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xing Y., Lee C. Bioinformatics. 2005;21:3701–373. doi: 10.1093/bioinformatics/bti613. [DOI] [PubMed] [Google Scholar]
- 10.Sorek R., Ast G., Graur D. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 12.Lev-Maor G., Sorek R., Shomron N., Ast G. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 13.Krull M., Brosius J., Schmitz J. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 14.Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 15.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Hurst L. D., Pal C. Trends Genet. 2001;17:62–65. doi: 10.1016/s0168-9525(00)02173-9. [DOI] [PubMed] [Google Scholar]
- 17.Pagani F., Raponi M., Baralle F. E. Proc. Natl. Acad. Sci. USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parmley J. L., Chamary J. V., Hurst L. D. Mol. Biol. Evol. 2006;23:301–309. doi: 10.1093/molbev/msj035. [DOI] [PubMed] [Google Scholar]
- 19.Xing Y., Lee C. Gene. 2006;370:1–5. doi: 10.1016/j.gene.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi Y., Sakata H., Makino Y., Urabe I., Yomo T. J. Mol. Evol. 2003;56:162–168. doi: 10.1007/s00239-002-2389-y. [DOI] [PubMed] [Google Scholar]
- 21.Pruitt K. D., Tatusova T., Maglott D. R. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng C. L., Fu X. D., Gribskov M. RNA. 2005;11:1777–1787. doi: 10.1261/rna.2660805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bejerano G., Lowe C. B., Ahituv N., King B., Siepel A., Salama S. R., Rubin E. M., Kent W. J., Haussler D. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 24.Gotea V., Makalowski W. Trends Genet. 2006;22:260–267. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Cordaux R., Udit S., Batzer M. A., Feschotte C. Proc. Natl. Acad. Sci. USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H., Chasin L. A. Mol. Cell Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairbrother W. G., Chasin L. A. Mol. Cell Biol. 2000;20:6816–6825. doi: 10.1128/mcb.20.18.6816-6825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtzman A. L., Govindarajan S., Vahle K., Jones J. T., Heinrichs V., Patten P. A. Curr. Opin. Biotechnol. 2001;12:361–370. doi: 10.1016/s0958-1669(00)00228-7. [DOI] [PubMed] [Google Scholar]
- 29.Hedges S. B. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 30.Blanchette M., Kent W. J., Riemer C., Elnitski L., Smit A. F., Roskin K. M., Baertsch R., Rosenbloom K., Clawson H., Green E. D., et al. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz S., Kent W. J., Smit A., Zhang Z., Baertsch R., Hardison R. C., Haussler D., Miller W. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., Schwartz S., Wagner L., Miller W. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 33.Li W. H. J. Mol. Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.