Abstract
Chromatin at centromeres is distinct from the chromatin in which the remainder of the genome is assembled. Two features consistently distinguish centromeres: the presence of the histone H3 variant CENP-A and, in most organisms, the presence of heterochromatin. In fission yeast, domains of silent ‘heterochromatin’ flank the CENP-A chromatin domain that forms a platform upon which the kinetochore is assembled. Thus, fission yeast centromeres resemble their metazoan counterparts where the kinetochore is embedded in centromeric heterochromatin. The centromeric outer repeat chromatin is underacetylated on histones H3 and H4, and methylated on lysine 9 of histone H3, which provides a binding site for the chromodomain protein Swi6 (orthologue of Heterochromatin Protein 1, HP1). The remarkable demonstration that the assembly of repressive heterochromatin is dependent on the RNA interference machinery provokes many questions about the mechanisms of this process that may be tractable in fission yeast. Heterochromatin ensures that a high density of cohesin is recruited to centromeric regions, but it could have additional roles in centromere architecture and the prevention of merotely, and it might also act as a trigger for kinetochore assembly. In addition, we discuss an epigenetic model for ensuring that CENP-A is targeted and replenished at the kinetochore domain.
Keywords: centromere, RNA interference, heterochromatin, CENP-A, kinetochore
1. Centromeric chromatin
In many organisms, centromeric chromatin is cytologically distinct from the remainder of the chromosomes. In metaphase spreads of colchicine-treated human chromosomes, the centromere is seen as the primary constriction on chromosomes, and the chromosomes remain cohesed at this position. During interphase, centromeres are seen as DAPI-bright regions in the nucleus, indicating the presence of a constitutively condensed chromatin, known as heterochromatin (Bernard & Allshire 2002). Heterochromatin has phenotypic consequences for genes placed in its vicinity. Placement of genes near to centromeric heterochromatin as a result of chromosomal rearrangement or insertion results in their silencing. This phenomenon—position effect variegation—was first observed in Drosophila, and is seen in organisms from yeast to humans (Reuter & Spierer 1992; Dillon & Festenstein 2002; Richards & Elgin 2002; Schotta et al. 2003).
Centromeric DNA is not conserved in sequence between organisms, yet similar features appear—reams of repetitive elements, such as alpha satellite in humans, minor satellite in mouse, AATAT and TTCTC satellites in Drosophila, interspersed with complex A+T rich repeats and mobile elements (Cleveland et al. 2003; Sun et al. 2003). Consequently, the sequence requirements for functions remain ill-defined.
The other defining trait of centromere regions, in addition to heterochromatin, is the presence of a Histone H3 variant, CENP-A (also known as cid, Cenp-a, Cnp1). It is localized to the inner kinetochore plate, and is underlain by the centromeric heterochromatin. (Warburton et al.. 1997; Blower et al. 2002; Cleveland et al. 2003). Even at the simple point centromeres of budding yeast that require only 125 bp DNA for full segregation function, there is a unique form of chromatin (Cottarel et al. 1989; Cleveland et al. 2003) A CENP-A/Cse4-containing nucleosome is associated with the kinetochore. It is flanked by phased (histone H3-containing) nucleosomes, and the DNA has differential nuclease sensitivity. Thus, there is no chromatin with features of silent chromatin at budding yeast centromeres, and the Sir3/Sir4-based silent chromatin at the telomeres and mating type locus (Perrod & Gasser 2003) differs from silent chromatin or heterochromatin in other eukaryotes, which involves chromodomain proteins such as HP1 and Swi6.
2. Fission yeast centromeres
Fission yeast centromeres span 35–110 kb and are composed of a central core region of non-repetitive DNA (cnt), flanked by inverted repeat regions—the innermost repeats (imr) and the outer repeats (otr), as shown in figure 1 (Takahashi et al. 1992; Steiner et al. 1993; Wood et al. 2002). The DNA is encompassed in two distinct chromatin domains: the central core region, consisting of the cnt and imr sequences, upon which the kinetochore is assembled and the heterochromatic outer repeat domains (silent chromatin). Thus the fission yeast centromeres resemble those of vertebrates, with the kinetochore embedded in a sea of silent chromatin or heterochromatin. The two domains are cytologically distinct (Kniola et al. 2001). Similarity has been noted between fission yeast outer repeats and transposable elements (Halverson et al. 1997). In other species, mobile elements are also seen in centromeric regions, raising the possibility that remnants of transposable elements might now contribute to centromere structure and function. Marker genes placed within the fission yeast centromere are transcriptionally silenced (Allshire et al. 1994, 1995). The degree and properties of the silencing vary with position in the centromere; strong silencing is observed at the outer repeats, while that in the central core is less robust.
Figure 1.
Schematic of centromere 1. Centromere 1 spans approximately 35 kb and consists of a central core (cnt) of non-repetitive sequence flanked by innermost repeats (imr) and outer repeats (otr: made up of dg and dh elements, black and white arrows), which together form an almost perfect inverted repeat around the central core. Insertion of marker genes anywhere in the centromere results in their transcriptional silencing. The quality of silencing varies, depending on the insertion site: strong repression occurs at the outer repeat regions, while silencing in the central core is less robust. The centromere is divided into two domains: the central core domain (cnt and imr) and the outer repeat domain. Different classes of mutants affect silencing in each domain, and each is associated with a distinct set of proteins. Short vertical lines represent tRNA genes (Kuhn et al. 1991; Takahashi et al. 1991) which, intriguingly, occur at the transition between the domains. The central core region has a unique chromatin structure as indicated by the smear pattern which results upon partial digestion with micrococcal nuclease. CENP-Acnp1 replaces histone H3 in the central region, and upon this chromatin platform the kinetochore is assembled. Proteins such as Mis6, Mis12, Mal2 and Sim4 are specifically associated with the central core region. Mutants in these genes disrupt the unusual chromatin structure, and (where tested) cause alleviation of central core silencing. The outer repeats are packaged in nucleosomes, which are underacetylated on the N-terminal tails, owing to the action of the histone deacetylases Clr3, Clr6 and Sir2. This allows di-methylation of lysine 9 of histone H3 by the histone methyltransferase Clr4, providing a binding site for the chromodomain proteins Swi6 (HP1) and Chp1. This Swi6-containing heterochromatin is responsible for the recruitment of a high density of cohesin to the outer repeat region, which is important for proper biorientation of centromeres at mitosis, and may have other roles (see figure 5). The assembly of heterochromatin is dependent on the RNAi machinery and siRNAs derived from centromeric transcripts. This involves the RNAse III-like endonuclease Dicer, the RITS complex (Chp1, Tas3, Ago1 and siRNAs), the RNA dependent-RNA polymerase Rdp1, and the UVDDB protein Rik1. For clarity, not all components are shown in the diagram. See the text for further details and, for a more comprehensive review of the genes and proteins involved in the assembly of the two domains, see Pidoux & Allshire (2004) and Ekwall (2004).
3. Central core chromatin and the kinetochore
The central core DNA is AT rich; cnt1 and cnt3 share a 3.3 kb element that is 99% identical and cnt2 has an element with 48% identity over 1.5 kb (Wood et al. 2002). The left and right imr inverted repeats are unique to each centromere. The central core region is packaged in a unique chromatin structure (Polizzi & Clarke 1991; Takahashi et al. 1992). While bulk chromatin exhibits a canonical ladder pattern upon limited digestion with micrococcal nuclease, hybridization with a central core probe gives a smear pattern. This unique pattern is not attributable to DNA sequence since it does not form when the DNA is introduced into budding yeast or if it is placed on a chromosome arm (Marschall & Clarke 1995). It forms only when the central core is in a functional context: only on centromere plasmids with segregation function is the unique chromatin seen over the central core, and it is lost in mutants that affect central core silencing and function. The other major difference between central core chromatin and the rest of the genome is the presence of the histone H3 variant CENP-Acnp1 which replaces (totally or partially) histone H3 (Takahashi et al. 2000; Mellone & Allshire, unpublished). Presumably, this CENP-Acnp1 is assembled into specialized nucleosomes, and their structure or organization cause the atypical nuclease sensitivity. Upon this specialized central core chromatin, the kinetochore is assembled.
Transcriptional silencing at the central core reflects the assembly of a fully functional kinetochore complex rather than canonical heterochromatin. The silencing is weaker than in the outer repeat regions, and in order to perform an alleviation of silencing screen, it was necessary to use a promoter-crippled arg3+ gene inserted into the central core of cen1 (Pidoux et al. 2003). Sim (silencing in the middle of the centromere) mutants identified in the screen include CENP-Acnp1 itself and the kinetochore protein Sim4, which is probably the homologue of CENP-H. Sim4 and other kinetochore proteins, such as Mis6, Mis12, Mal2, and Ams2 identified in various screens, are specifically associated with the central core region by chromatin immunoprecipitation (Saitoh et al. 1997; Goshima et al. 1999; Jin et al. 2002; Chen et al. 2003). In all these mutants, the specialized central core chromatin is disrupted and replaced by a ladder pattern, and in many CENP-Acnp1 association is reduced.
An intriguing and vital question in understanding centromere function is how CENP-Acnp1 nucleosomes are incorporated at the central core and only at the central core. This is unlikely to be directed by sequence alone. mis6 mutants fail to incorporate newly synthesized GFP-Cnp1, prompting the proposal that it is a loading factor for CENP-Acnp1 (Takahashi et al. 2000). However, its mammalian and budding yeast homologues, CENP-I and Ctf3, respectively, do not appear to share this property (Measday et al. 2002; Nishihashi et al. 2002). As noted above, several other central core mutants also affect the levels of CENP-Acnp1 at the central core (Pidoux et al. 2003; Chen et al. 2003). While some of these genes may turn out to encode CENP-A specific chromatin assembly factors, it is unlikely to be the whole story, and does not address the issue of incorporation site. One idea is that it is the presence of a functional kinetochore that directs where incorporation occurs (Mellone & Allshire 2003; figure 2). How might this be achieved? The role of the centromere is to segregate chromosomes through interaction with spindle microtubules (MTs). Tension is exerted across properly bi-oriented sister centromeres. In this scenario, tension would induce a ‘mark’, which would be read as a signal to incorporate CENP-A. CENP-A loading could be contemporaneous with the tension at metaphase, or, more likely, the mark would be in the form of a modification or conformational change that would be read in the next cell cycle, probably in S phase, when the signal would prompt the incorporation of CENP-A nucleosomes with the travelling replication fork. Alternatively, chromatin remodelling machinery could strip out H3 nucleosomes and replace them with CENP-A nucleosomes at marked sites post-replication. Such a model is attractive as it serves to link the two major roles of the centromere—chromosome segregation and the maintenance of the site of kinetochore formation cell cycle after cell cycle and generation after generation—in an epigenetic mechanism that depends on kinetochore integrity rather than on the DNA sequence.
Figure 2.

Targeting and incorporation of CENP-A at functional centromeres. (a) At functional centromeres, CENP-A nucleosomes (white circles) form a platform for kinetochore assembly (grey lozenges) which allows proper interaction with spindle microtubules at mitosis. Tension is generated across correctly bi-oriented centromeres at metaphase. We propose that this tension induces an epigenetic ‘mark’ (*) to direct the incorporation of new CENP-A at the active centromere. CENP-A could conceivably be incorporated in a direct response to tension at metaphase, or, more probably, the mark would be read in the next cell cycle, at S phase. (b) Random partitioning would mean that some marked CENP-A nucleosomes (or other kinetochore component) remained on each chromatid as the replication fork passed. These would signal the incorporation of fresh CENP-A nucleosomes in the gaps, rather than H3-containing nucleosomes. Alternatively, H3-nucleosomes would be incorporated at replication, and the mark read post-replication to signal exchange of H3-nucleosomes for CENP-A nucleosomes by specialized chromatin remodelling activities. (c) At defective centromeres, the assembly of imperfect kinetochores (white lozenges) would cause reduced/absent MT interaction and tension at mitosis. The centromeres would fail to receive the epigenetic mark (and segregation would be impaired). (d) At unmarked centromeres and at euchromatin (e), H3-containing nucleosomes would be incorporated at S phase by default. Subsequent failure to properly biorient would lead to further CENP-A reduction and eventual loss of centromere function. This epigenetic mechanism would ensure that CENP-A is replenished at active centromeres but prevent ectopic incorporation at euchromatic sites. It guarantees long-term propagation of centromeres but also permits plasticity.
4. Centromeric heterochromatin
Histones H3 and H4 in the nucleosomes of outer repeat chromatin are underacetylated on lysines in their N-terminal tails, compared with euchromatin (Ekwall et al. 1997; Mellone et al. 2003). This correlates with transcriptional silencing: a ura4+ gene placed within the outer repeats is strongly silenced, with very few colonies forming on media lacking uracil and many forming on media containing the counter-selective drug FOA (Allshire et al. 1995). Hypoacetylation is important for centromere function, as brief treatment with the histone deacetylase inhibitor TSA causes a heritable increase in acetylation of centromeric chromatin, expression of marker genes and a concomitant defect in chromosome segregation (Ekwall et al. 1997). The hypoacetylated state of the outer repeat chromatin is owing to the activities of three histone deacetylases: Clr3, Clr6 and Sir2 (Nakayama et al. 2001; Bjerling et al. 2002; Shankaranarayana et al. 2003). Clr3 acts to deacetylate lysine 14 of histone H3, and Sir2 deacetylates lysine 9 of histone H3, while Clr6 has a broader specificity, deacetylating several of the lysines in the histone H3 and H4 N termini.
In addition to their hypoacetylation, outer repeat nucleosomes differ from their euchromatic counterparts in their methylation status. Lysine 9 of histone H3 is methylated in heterochromatic regions—the centromeric outer repeats and the silent mating type locus. Expressed regions lack lysine 9 methylation but are methylated on lysine 4 of histone H3 (Hall et al. 2002). The histone methyltransferase Clr4 is responsible for the methylation of histone H3 K9 (Rea 2000). This modification creates a binding site for the chromodomain protein Swi6 (Ekwall et al. 1995; Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001). Both these proteins have orthologues in Drosophila that were identified in screens for suppression of position effect variegation. Su(var)3-9 is a histone H3 K9 methyltransferase and Su(var)2-5 encodes heterochromatin protein 1 (HP1), the counterpart of Swi6 (Reuter & Spierer 1992; Dillon & Festenstein 2002; Richards & Elgin 2002; Schotta et al. 2003). The Suvar3-9 and HP1 proteins interact and are located at the heterochromatic pericentromeric regions in Drosophila and mammals (James et al. 1989; Wreggett et al. 1994; Aagaard et al. 1999; Minc et al. 1999). Likewise, in fission yeast, the absence of either Swi6 or Clr4 causes alleviation of outer repeat (and mating type) silencing, and Swi6 is specifically located at these regions (Allshire et al. 1995; Ekwall et al. 1995; Partridge et al. 2000; Nakayama et al. 2000; Noma et al. 2001). These mutants have increased chromosome loss, lagging chromosomes on late anaphase spindles and hypersensitivity to MT-destabilizing drugs. Assays that detect alleviation of silencing have been used in genetic screens to pinpoint other factors required for the formation of heterochromatin (Ekwall et al. 1996, 1999; Volpe et al. 2002, 2003); many of these are involved in the RNA interference pathways (see below). Another chromodomain protein, Chp1, is required for methylation of histone H3 lysine 9 and binds dimethylated H3 peptide in vitro (Partridge et al. 2000, 2002). It is found in the RITS complex along with Tas3, Ago1 and siRNAs (Verdel et al. 2004). Rik1 is a UVDDB-like protein which is required for lysine 9 methylation (Neuwald & Poleksic 2000, Nakayama et al. 2001). The centromere suppressor of position effect mutants, csp7 to 13, lose methylation of lysine 9 and fail to recruit Swi6 and Rad21-cohesin; csp9 is an allele of ago1 (Ekwall et al. 1999; Volpe et al. 2002, 2003).
The outer repeat dg elements are 97% identical between centromeres (Wood et al. 2002). Such conservation implies a requirement for specific sequences. In an ectopic silencing assay, a 1.6 kb region of the outer repeat region was inserted at a euchromatic site (ade6 locus) next to a ura4+ gene. The ura4+ gene became transcriptionally silenced and gained heterochromatic marks and features: hypoacetylation, methylation of histone H3 lysine 9 and the recruitment of Swi6 and Rad21-cohesin (Partridge et al. 2002). The region could be further narrowed down to 1 kb, suggesting that a short specific sequence is able to act as a nucleation site for heterochromatin assembly. Similar observations have been made for a short sequence from the mating type locus (Ayoub et al. 2000).
5. Role of RNAi in the formation of heterochromatin
The observations that marker genes inserted into centromeric heterochromatin in fission yeast were transcriptionally silenced, and no steady-state transcripts were detected coming from the outer repeat regions, suggested that the centromeres were transcriptionally inert (Fishel et al. 1988). Ironically, it transpires that the formation of silent heterochromatin is actually dependent on transcripts from the outer repeat regions and the RNA interference machinery (Reinhart & Bartel 2002; Volpe et al. 2002; Grewal & Moazed 2003; Schramke & Allshire 2004; Ekwall 2004). Mutants in components of the RNAi machinery, Argonaute (Ago1), Dicer (Dcr1) and RNA-dependent RNA polymerase (Rdp1), alleviate silencing of marker genes in the centromeric outer repeats and have lagging chromosomes. In these mutants, but not in the wild type, transcripts derived from the top and the bottom strands are detectable. The transcripts are partially complementary and could potentially form dsRNA that would be a substrate for the RNase III-related endonuclease Dicer. Nuclear run-on assays indicate that the transcripts are actually produced in wild-type cells but are normally not detectable, presumably because of processing (Volpe et al. 2002). Interestingly, mouse centromeric satellite repeats are also transcribed (Lehnertz et al. 2003).
Observations in several organisms suggest that the activities of the RNAi machinery can have three outcomes (figure 3; Grewal & Moazed 2003; Schramke & Allshire 2004; Ekwall 2004): the degradation of messenger RNA homologous to siRNAs (post-transcriptional gene silencing, (PTGS), involving the RISC complex that includes Argonaute); chromatin-based silencing (either DNA methylation in plants, methylation of histone H3 lysine 9 and recruitment of chromodomain proteins such as Swi6 in fission yeast, or, both in human cells; Kawasaki & Taira 2004); and translational inhibition. Some proteins appear to have roles in only one of these processes, while others have roles in both mRNA degradation and the chromatin-dependent silencing pathway. The chromodomain protein Swi6 operates only in the chromatin-based pathway, RNAi components such as Ago1, Dcr1 and Rdp1 affect both pathways (e.g. Ago1 is thought to be a component of both RISC and RITS; Volpe et al. 2002; Schramke & Allshire 2003; Verdel et al., 2004). The phenotypes of a clr4Δ mutant unexpectedly suggest that it may have roles in both pathways.
Figure 3.
Silencing by RNA interference. Observations in several organisms suggest that the RNA interference machinery causes silencing by three different routes. This is illustrated for silencing by a plasmid-expressed hairpin RNA homologous to a 280 bp region of the ura4 gene in fission yeast. Box: dsRNA is cleaved by Dicer to produce siRNAs. Middle: silencing by mRNA degradation or post-transcriptional gene silencing (PTGS). siRNAs are incorporated into the RNA-induced silencing complex (RISC, which includes Argonaute—Ago1 in fission yeast), which causes degradation of mRNA homologous to the siRNAs. Left: chromatin-based silencing. In plants this involves DNA methylation to silence genes. In fission yeast, siRNAs incorporated into the RITS complex (Ago1, Chp1 and Tas3) lead to methylation of lysine 9 of histone H3, binding of Swi6 and recruitment of cohesin at homologous regions. The assembly of heterochromatin requires the histone deacetylases Clr3, Clr6 and Sir2, and the histone methyltransferase Clr4, Rik1 and Rdp1. In wild-type cells, spreading of heterochromatin occurs beyond the region of homology. In swi6Δ cells, lysine 9 methylation occurs only at the region of homology, indicating that Swi6 is required for spreading. Right: siRNA-dependent translational inhibition contributes to silencing in some organisms, but it is not known whether this occurs in fission yeast.
How is the transition made from acetylated euchromatin to the heterochromatic state? Two models can be envisaged for how siRNAs elicit the formation of heterochromatin. In one, homologous DNA is the target for siRNAs (Jones et al. 1999; Mette et al. 2000). The evidence for this DNA–RNA view comes from experiments in plants in which dsRNAs homologous to a promoter region lead to chromatin-based silencing. More recently, it has been shown that transfection of human cells with several siRNAs results in chromatin-based silencing of a target gene (Kawasaki & Taira 2004). Since promoters themselves are not expected to be transcribed, it is suggested that the siRNAs may be targeting DNA. The second model—which we favour—invokes RNA–RNA pairing between the siRNAs and homologous nascent transcripts (figure 4). Experiments in fission yeast (Schramke & Allshire unpublished) indicate that transcription is required to induce chromatin-based silencing. Whether the requirement is for the nascent transcript or the act of transcription is not yet known. In this scenario, nascent transcripts would attract homologous siRNAs assembled in the RITS complex (Chp1, Ago1, Tas3), along with histone modifying enzymes—the histone deacetylase activities and the histone methyltransferase Clr4. This would be analogous to the situation with other chromatin modifying activities that are known to track along with elongating RNA polymerase II, for example, the SET1 and SET2 histone H3 lysine 4 and lysine 36 methyltransferases (Hampsey & Reinberg 2003). Thus, heterochromatin would spread along with RNA polymerase as in travels down the chromatin fibre.
Figure 4.
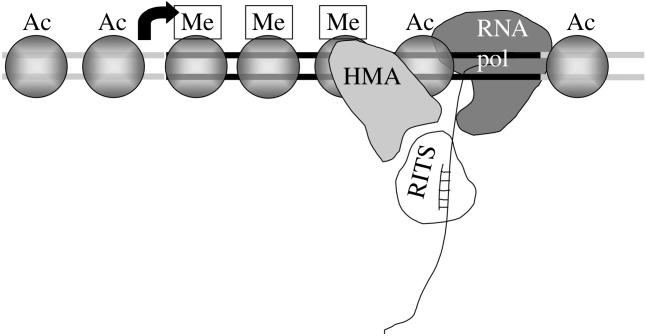
Chromatin-based silencing could involve siRNAs targeting nascent RNA transcripts. In this model, siRNAs in the RITS complex would home in on their target by hybridization with the nascent transcript produced by RNA polymerase II. Histone modifying activities (HMA), including histone deacetylases (Clr3, Clr6, Sir2) and histone methyltransferase (Clr4), would piggyback on the RITS complex. As the polymerase traverses the region, deaceylation and methylation of nucleosomes would occur, leading to the assembly of repressive heterochromatin. Ac is acetylation; Me is methylation. In an alternative model (not shown), targeting involves the hybridization of siRNAs to DNA.
If the repetitive elements found at centromeres in fission yeast, mammals and Drosophila are indeed the remnants of retrotransposons, it might be expected that retrotransposons and remnant solo LTRs that lie outside the centromere would also be coated with repressive heterochromatin. In fission yeast, TF1 and TF2 LTRs do indeed bear the hallmarks of heterochromatin, lysine 9 methylation and, Swi6 binding. When ago1, dcr1 or rdp1 are deleted, transcripts from both top and bottom strands of the LTRs are detected. These patches of heterochromatin have an effect on nearby genes, since mutation of RNAi components cause their derepression, as does the deletion of individual LTRs (Schramke & Allshire 2003).
Thus the RNAi machinery can act to bring about the formation of silent chromatin outside the context of centromeric DNA. Does sequence matter, or does any double-stranded RNA have the potential to induce heterochromatin? Expression from a plasmid of a hairpin RNA to a portion of the ura4 gene induced silencing and heterochromatin formation on the ura4+ gene in the genome but failed to produce these effects on a genomic copy of ura4 that lacked this portion of the gene (Schramke & Allshire 2003). This suggests that lysine 9 methylation, Swi6 recruitment, and thus silent chromatin, can potentially be formed on any piece of DNA simply by production of the appropriate dsRNA. The ability to produce ‘synthetic’ heterochromatin will be valuable in the dissection of heterochromatin assembly and centromere function.
In wild-type cells expressing the ura4 hairpin, sequences upstream of the homologous region also become modified with lysine 9 methylation. This spreading phenomenon fails to occur in cells lacking Swi6, suggesting that Swi6 itself is required for the propagation of silent chromatin marks (Schramke & Allshire 2003), consistent with other observations (Hall et al. 2002; Shankaranarayana et al. 2003). In endogenous centromeres, heterochromatin is formed not only at regions homologous to the centromeric transcripts but throughout the outer repeat region, suggesting that here too, spreading occurs. A model can be envisaged in which the Clr3 and Clr6 histone deacetylases are recruited by the RITS complex/siRNAs to a homologous nascent transcript held by elongating RNA polymerase II, and deacetylate nucleosomes on the adjacent chromatin. This would enable Clr4 to act to methylate histone H3 lysine 9, providing a binding site for Swi6. From this nucleation site, the Sir2 NAD+-dependent HDAC and Clr4 HMTase would sequentially modify the adjacent chromatin, allowing Swi6 recruitment and spreading of silent chromatin (Ekwall 2004). The ability of Swi6 to dimerize might also contribute to heterochromatin propagation (Cowieson et al. 2000). In addition, the fission yeast CENP-B homologues, which display similarity to transposases, play a role in nucleation of heterochromatin in fission yeast (Nakagawa et al. 2002), while CENP-B itself binds α-satellite DNA at centromeres (Cleveland et al. 2003).
How do the observations of ectopic silencing, which imply sequence specificity, square with the ability to induce synthetic heterochromatin with a non-centromeric hairpin? Perhaps it is the potential to produce dsRNA that is important, rather than the actual sequence itself. Investigation of the details of transcript requirements will be informative.
6. Epigenetic features of centromeres
There is strong evidence to indicate that centromeres, including those of fission yeast, are epigenetically regulated (Karpen & Allshire 1997; Sullivan et al. 2001; Amor & Choo 2002). Variegation of marker gene silencing occurs: the ade6 gene inserted at the central core produces red (repressed state), white (expressed state), sectored and pink colonies on indicator plates (Allshire et al. 1994). Thus, distinct metastable states can be adopted by normal centromeres and propagated in genetically identical cells. Transient treatment of cells with the histone deacetylase inhibitor TSA causes a heritable increase in acetylation of outer repeat histones, loss of marker gene silencing and defective centromere function (Ekwall et al. 1997). These defective acetylated lineages are able to regain the functional state at a low frequency.
Extensive series of experiments using fragments of centromeric DNA have been performed to define sequence requirements for centromere function (Niwa et al. 1986, 1989; Hahnenberger et al. 1989, 1991; Matsumoto et al. 1990). However, unlike in budding yeast where the 125 bp of DNA confers mitotic and meiotic segregation function, in fission yeast it has been impossible to define a short stretch of DNA that embodies the centromere. The overall conclusion of these studies is that an entire central core and an extensive region of an outer repeat (dg) are the minimal requirements for segregation function. Addition of inverted repeat elements is beneficial in some cases. However, it appears that context is important, and there is evidence for a modular nature of centromeres as parts of a central core can substitute for others. Introduction into cells of certain minimal constructs produces two classes of transformant—those with ‘stable’ plasmids and those with ‘unstable’ plasmids (Steiner & Clarke 1994). The differences were not owing to mutations, and the passage of each type of plasmid through bacteria and reintroduction into fission yeast again produced both the stable and the unstable class. This suggests that the plasmids have to achieve, through a stochastic process, the correct chromatin conformation to create a functional centromere. Once attained, this specific chromatin environment would then promote the recruitment of CENP-Acnp1. A 2.1 kb ‘centromere enhancer’ element from the outer repeats (essentially dg) was defined as being particularly important for centromere function (Baum et al. 1994). However, further experiments indicated that this element was actually dispensable and could be substituted with another outer repeat element. In these experiments, if a ‘non-functional’ minimal centromere plasmid was propagated for many generations with selection, in a low proportion of cases, functional centromere plasmids were created when the selection was removed (Ngan & Clarke 1997). This suggests that if given enough time, the minimal constructs are sometimes able to adopt a functional centromere architecture; this presumably involves the recruitment of proteins and the assembly of specialized centromeric chromatin. Note that, in all cases, outer repeat and central domain sequences were required to form a functional centromere, indicating that the transcribed outer repeats provide some role in kinetochore assembly over the central domain. We imagine that this would include both heterochromatin, with its hypoacetylated lysine 9 methylated histones, and a region of CENP-Acnp1 chromatin with the characteristic morphology.
7. What are the roles of heterochromatin in centromere function?
Both central core and outer repeat regions are required for de novo formation of a functional centromere when naked DNA is transformed into fission yeast. On the other hand, genes encoding central core proteins such as mis6, mis12, mal2, cnp1 and sim4 are essential, yet virtually all outer repeat proteins are non-essential (for instance swi6, clr4, chp1, rik1, tas3, ago1, dcr1 and rdp1 RNAi components). What, then, is the function of the twin outer repeat domains?
One known function of the outer repeats is in cohesion. swi6 mutants prematurely lose cohesion at the centromere, but show no defect in cohesion along the chromosome arms. Consistent with this, chromatin immunoprecipitation indicates that in a swi6 mutant, Rad21-Cohesin is lost specifically from the heterochromatic outer repeats (and other heterochromatin sites) but other sites are unaffected (Bernard et al. 2001; Nonaka et al. 2002). swi6 and rad21 mutants are synthetically lethal. In a rad21 mutant at the permissive temperature, there is enough residual cohesin function to ensure segregation function, though there is a high incidence of lagging chromosomes. In the absence of Swi6, centromeric cohesion is lost, but arm cohesion is adequate to ensure a sufficient level of chromosome bi-orientation. In the absence of both Swi6 and Rad21, the lack of centromeric cohesion and the reduced arm cohesion are so debilitating that the accurate chromosome slips below a level that is compatible with viability. Swi6 has been shown to interact physically with another subunit of cohesin, Psc3 (Nonaka et al. 2002). Thus, Swi6-containing heterochromatin is responsible for recruiting a high density of cohesin to the centromeric outer repeats. Interestingly, this link seems to be conserved since vertebrate cells lacking dicer display disrupted heterochromatin and defective centromeric cohesion (Fukagawa et al. 2004).
Fission yeast centromere 1 is almost perfectly symmetrical (Wood et al. 2002): the imr sequences are perfect inverted repeats (98% identical), as are the outer repeats; nucleotide changes on one side are often mirrored on the other. The imr sequences of centromeres 2 and 3 are also perfect inverted repeats, though there are different numbers of otr repeats on the left and right sides of the two centromeres. The symmetry has prompted speculation that the two sides of the centromere interact in some way to form a higher order structure such as a loop (Fishel et al. 1988; Takahashi et al. 1992; Clarke et al. 1993). Might heterochromatin and the cohesin it recruits have a role in the architecture of such a loop? Possibly it has a role in holding together not only sister chromatids, but in intramolecular synapsis of the left and right sides of the centromere. What would the purpose of such a loop be? Perhaps this architecture would ensure that the central core is presented optimally for kinetochore assembly (figure 5). Mutants in RNAi, heterochromatin and cohesin components have a high incidence of lagging chromosomes (chromatids) on late anaphase spindles (Ekwall et al. 1995, 1996; Hall et al. 2003; Volpe et al. 2003). In larger eukaryotes, lagging chromosomes have been shown to be merotelically oriented—this is when at single kinetochore is simultaneously attached to MTs emanating from both spindle poles (reviewed in Pidoux & Allshire 2003). Kinetochore stretching occurs, and there is failure or delay in segregating to the poles. Merotely is a major contributor to aneuploidy in mammalian tissue culture cells (Cimini et al. 2001). Fission yeast kinetochores are each associated with two to four MTs (Ding et al. 1993) and the behaviour of lagging chromosomes observed in living cells is consistent with merotelic attachment (Pidoux et al. 2000). The hypothetical loop structure may be important for the imposition of rigidity on the kinetochore, ensuring that the multiple MT binding sites on each kinetochore are clamped together so that they all face the same direction and thus promote monopolar (amphitelic) attachment. Another protein which may influence centromere orientation is Pcs1 (Rabitsch et al. 2003). Its homologue in budding yeast, Csm1, is part of the monopolin complex required for mono-orientation of sister kinetochores in meiosis I, and is thought to act in clamping the MT-attachment sites together. Pcs1 does not seem to be required in fission yeast meiosis I, but its absence in mitosis causes lagging chromosomes, suggesting that it could play a role in clamping adjacent MT attachment sites together on a kinetochore.
Figure 5.

Speculative models for the role of outer repeat heterochromatin in centromere specification and architecture. (a) The de novo assembly of kinetochores might be facilitated by heterochromatic ‘signposts’ which instruct the cell to incorporate CENP-A. (b) The Swi6-containing heterochromatin of the outer repeats is known to recruit a high density of cohesin for sister-chromatid cohesion. It might also have a role in intramolecular synapsis of the two sides of a single centromere to form a hypothetical loop structure. This specialised architecture might be required to present the central core in a favourable configuration for kinetochore assembly, and in ensuring a rigid structure in which multiple MT-binding sites are clamped together so they are all oriented towards the same spindle pole, thus reduce the likelihood of merotelic attachment. The diagram represents a 3-dimensional structure. The ends of each sister chromatid are indicated (* and *, § and §). Chromosome arms are shown in light grey; outer repeats, dark grey; central core, black; kinetochore, white boxes; MTs, thin black lines. Intermolecular heterochromatin/cohesin between sister chromatids is indicated by the horizontal stippled lines. Intramolecular heterochromatin/cohesin, holding the two sides of each centromere together is indicated by vertical stippled lines.
As we have seen, cells with compromized outer repeat heterochromatin have chromosome segregation defects but they are viable. Yet when plasmids are transformed into cells, both central core and outer repeat elements are essential for segregation function. Perhaps the outer repeats are required initially in the establishment of a functional centromere to tell the cell where to deposit CENP-A for the assembly of the kinetochore (figure 5). Then the essential function of the outer repeats may be over, and its roles may be limited to those important but not absolutely essential roles described above—cohesion (arm cohesion suffices) and prevention of merotely (lagging chromosomes occur, but accurate segregation is at a level compatible with viability). Obviously, in non-experimental situations, the cell is not going to encounter naked DNA upon which to assemble a kinetochore. Perhaps the outer repeats function in a kind of ‘reset’ mechanism that can boost centromere architecture if the structure becomes weakened, or there is reduced CENP-A for some reason.
These ideas are testable now that the ability to make ‘synthetic’ heterochromatin exists (Schramke & Allshire 2003). Can a centromere form when a central core is placed between two blocks of synthetic heterochromatin, for instance? Once a centromere is assembled, can outer repeat sequences be removed, or the trigger for synthetic heterochromatin switched off? The pioneering experiments of Mitsuhiro Yanagida and Louise Clarke should be revisited in the light of our knowledge about protein domains of the centromere, the nature and genesis of heterochromatin and the functional significance of CENP-A, and the specialized central core chromatin.
The kinetochore is formed over the central core chromatin, and this is where MTs would be expected to contact the centromere; indeed the microtubule-associated protein (MAP) Dis1 has been localized to this region by ChIP. However, another MAP, Alp14, is located at the imr and otr regions. While both proteins associate with the centromere in a mitosis dependent fashion, only Alp14's association is fully dependent on MTs (Garcia et al. 2001; Nakaseko et al. 2001). Perhaps these differences are a reflection of multiple types of MT interaction with the kinetochore.
In Drosophila, CENP-Acid and histone H3 are interspersed on experimentally produced extended chromatin fibres, yet in fixed cells they appear in cytologically distinct domains as juxtaposed cylinders or layers (Blower et al. 2002). The difference between the two observations is explicable by a model in which the centromeric chromatin loops or coils so that all the CENP-A domains are in register, forming a surface for kinetochore assembly and MT interaction. If the fission yeast centromere does indeed form a loop structure as we propose, it might represent a single kintochore unit, interacting with two to four mts, whereas higher eukaryotic centromeres, which are known to be modular structures (Zinkowski et al. 1991; Blower et al. 2002), would consist of re-iterated units.
8. Conclusions and speculation
In fission yeast, there are two distinct protein domains: the heterochromatin and kinetochore domains. Proteins that associate with each domain are required for the full segregation function of the centromere, and both domains are required for de novo centromere formation. The kinetochore is assembled over the central core CENP-Acnp1-containing chromatin. Putative CENP-Acnp1 specific chromatin assembly factors could be aided by an epigenetic mechanism reliant on segregation competence, which would operate to replenish CENP-Acnp1 in the kinetochore domain. The assembly of Swi6-containing heterochromatin over the outer repeat regions are dependent on the RNAi machinery. This centromeric heterochromatin is responsible for recruiting a high density of cohesin, and could have supplementary related roles in the formation of a higher order loop structure and in the prevention of merotely. In addition, we believe that the heterochromatin domain may act as a trigger in kinetochore formation in fission yeast. Could such a model apply to higher eukaryotes? At mammalian centromeres, CENP-A and HP1 are in cytologically distinct locations—the inner kinetochore plate and the underlying heterochromatin, respectively (Cleveland et al. 2003). The formation of neocentromeres on rearranged human chromosomes is an intriguing phenomenon (Warburton 2004); neocentromeres form at places not normally associated with centromere activity and in at least one case have been shown by sequencing to be devoid of alpha satellite DNA. Faced with the need to make a centromere, a region with the potential to recruit CENP-A and nearby to one with the potential to form heterochromatin would be favoured. It is not implausible that these could be places where there is AT-rich DNA and non-coding antiparallel transcripts.
Acknowledgments
We thank members of the Allshire lab for discussions. The research carried out in the Allshire lab is funded by the Wellcome Trust. R.C.A. is a Wellcome Trust Principal Research Fellow.
GLOSSARY
- HP1
heterochromatin protein 1
- MAP
microtubule-associated protein
- MT
microtubule
References
- Aagaard L, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R.C, Javerzat J.P, Redhead N.J, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Allshire R.C, Nimmo E.R, Ekwall K, Javerzat J.P, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Amor D.J, Choo K.H. Neocentromeres: role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Goldshmidt I, Lyakhovetsky R, Cohen A. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics. 2000;156:983–994. doi: 10.1093/genetics/156.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J, Zegerman P, Partridge J.F, Miska E.A, Thomas J.O, Allshire R.C, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Baum M, Ngan V.K, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol. Biol. Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Allshire R. Centromeres become unstuck without heterochromatin. Trends Cell. Biol. 2002;12:419–424. doi: 10.1016/s0962-8924(02)02344-9. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure J.F, Partridge J.F, Genier S, Javerzat J.P, Allshire R.C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bjerling P, Silverstein R.A, Thon G, Caudy A, Grewal S, Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D, Sullivan B.A, Karpen G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.S, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon E.D. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell. Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Baum M, Marschall L.G, Ngan V.K, Steiner N.C. Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harb. Symp. Quant. Biol. 1993;58:687–695. doi: 10.1101/sqb.1993.058.01.076. [DOI] [PubMed] [Google Scholar]
- Cleveland D.W, Mao Y, Sullivan K.F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Cottarel G, Shero J.H, Hieter P, Hegemann J.H. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell Biol. 1989;9:3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowieson N.P, Partridge J.F, Allshire R.C, McLaughlin P.J. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- Dillon N, Festenstein R. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 2002;18:252–258. doi: 10.1016/s0168-9525(02)02648-3. [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald K.L, McIntosh J.R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast Schizosaccharomyces pombe. J. Cell. Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. The roles of histone modifications and small RNA in centromere function. Chromosome Res. 2004;12:535–542. doi: 10.1023/B:CHRO.0000036584.40567.e5. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Javerzat J.P, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo E.R, Javerzat J.P, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell. Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner B.M, Cranston G, Allshire R.C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire R.C. Novel fission yeast mutants which alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics. 1999;153:1153–1169. doi: 10.1093/genetics/153.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Garcia M.A, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Hahnenberger K.M, Baum M.P, Polizzi C.M, Carbon J, Clarke L. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA. 1989;86:577–581. doi: 10.1073/pnas.86.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger K.M, Carbon J, Clarke L. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol. Cell. Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I.M, Shankaranarayana G.D, Noma K, Ayoub N, Cohen A, Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hall I.M, Noma K, Grewal S.I. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl Acad. Sci. USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson D, Baum M, Stryker J, Carbon J, Clarke L. A centromere DNA-binding protein from fission yeast affects chromosome segregation and has homology to human CENP-B. J. Cell. Biol. 1997;136:487–500. doi: 10.1083/jcb.136.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- James T.C, Eissenberg J.C, Craig C, Dietrich V, Hobson A, Elgin S.C. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell. Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Jin Q.W, Pidoux A.L, Decker C, Allshire R.C, Fleig U. The Mal2p protein is an essential component of the fission yeast centromere. Mol. Cell. Biol. 2002;22:7168–7183. doi: 10.1128/MCB.22.20.7168-7183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Hamilton A.J, Voinnet O, Thomas C.L, Maule A.J, Baulcombe D.C. RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H, Allshire R.C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- Kniola B, O'Toole E, McIntosh J.R, et al. The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell. 2001;12:2767–2775. doi: 10.1091/mbc.12.9.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R.M, Clarke L, Carbon J. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc. Natl Acad. Sci. USA. 1991;88:1306–1310. doi: 10.1073/pnas.88.4.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck A.A, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Marschall L.G, Clarke L. A novel cis-acting centromeric DNA element affects S. pombe centromeric chromatin structure at a distance. J. Cell. Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Murakami S, Niwa O, Yanagida M. Construction and characterization of centric circular and acentric linear chromosomes in fission yeast. Curr. Genet. 1990;18:331–335. [Google Scholar]
- Measday V, Hailey D.W, Pot I, et al. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 2002;16:101–113. doi: 10.1101/gad.949302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone B.G, Allshire R.C. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Mellone B.G, Ball L, Suka N, Grunstein M.R, Partridge J.F, Allshire R.C. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr. Biol. 2003;13:1748–1757. doi: 10.1016/j.cub.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Mette M.F, Aufsatz W, van der Winden J, Matzke M.A, Matzke A.J. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E, Allory Y, Worman H.J, Courvalin J.C, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Lee J.K, Hurwitz J, et al. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Klar A.J, Grewal S.I. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice J.C, Strahl B.D, Allis C.D, Grewal S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F, Poleksic A. PSI-BLAST searches using hidden markov models of structural repeats: prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucl. Acids Res. 2000;28:3570–3580. doi: 10.1093/nar/28.18.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan V.K, Clarke L. The centromere enhancer mediates centromere activation in Schizosaccharomyces pombe. Mol. Cell. Biol. 1997;17:3305–3314. doi: 10.1128/mcb.17.6.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A, Haraguchi T, Hiraoka Y, et al. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2002;2:463–476. doi: 10.1016/s1534-5807(02)00144-2. [DOI] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Yanagida M. Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol. Gen. Genet. 1986;203:397–405. [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal S.I, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis C.D, Grewal S.I. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Partridge J.F, Borgstrom B, Allshire R.C. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Partridge J.F, Scott K.S, Bannister A.J, Kouzarides T, Allshire R.C. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- Perrod S, Gasser S.M. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell. Mol. Life Sci. 2003;60:2303–2318. doi: 10.1007/s00018-003-3246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A, Allshire R. Chromosome segregation: clamping down on deviant orientations. Curr. Biol. 2003;13:R385–R387. doi: 10.1016/s0960-9822(03)00316-6. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L, Uzawa S, Perry P.E, Cande W.Z, Allshire R.C. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell. Sci. 2000;113:4177–4191. doi: 10.1242/jcs.113.23.4177. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L, Richardson W, Allshire R.C. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell. Biol. 2003;161:295–307. doi: 10.1083/jcb.200212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L, Allshire R.A. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- Polizzi C, Clarke L. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J. Cell. Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K.P, Petronczki M, Javerzat J.P, Genier S, Chwalla B, Schleiffer A, Tanaka T.U, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J, Bartel D.P. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Reuter G, Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Richards E.J, Elgin S.C. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell. Dev. Biol. 2003;14:67–75. doi: 10.1016/s1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- Schramke V, Allshire R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science. 2003;301:1069–1074. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- Schramke V, Allshire R. Those interfering little RNAs! Silencing and eliminating chromatin. Curr. Opin. Genet. Dev. 2004;14:174–180. doi: 10.1016/j.gde.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Shankaranarayana G.D, Motamedi M.R, Moazed D, Grewal S.I. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 2003;13:1240–1246. doi: 10.1016/s0960-9822(03)00489-5. [DOI] [PubMed] [Google Scholar]
- Steiner N.C, Clarke L. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 1994;79:865–874. doi: 10.1016/0092-8674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Steiner N.C, Hahnenberger K.M, Clarke L. Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol. Cell. Biol. 1993;13:4578–4587. doi: 10.1128/mcb.13.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A, Blower M.D, Karpen G.H. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Sun X, Le H.D, Wahlstrom J.M, Karpen G.H. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Niwa O, Yanagida M. A large number of tRNA genes are symmetrically located in fission yeast centromeres. J. Mol. Biol. 1991;218:13–17. doi: 10.1016/0022-2836(91)90867-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen E.S, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal S.I, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A, Kidner C, Hall I.M, Teng G, Grewal S.I, Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton G.L, White S.A, Teng G, Martienssen R.A, Allshire R.C. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream M.A, Lyne M, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- Warburton P.E, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–914. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Wreggett K.A, Hill F, James P.S, Hutchings A, Butcher G.W, Singh P.B. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell. Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
- Zinkowski R.P, Meyne J, Brinkley B.R. The centromere-kinetochore complex: a repeat subunit model. J. Cell. Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]




